Abstract
APOBEC3G (A3G) is a host cytidine deaminase that serves as a potent intrinsic inhibitor of retroviral replication. A3G is packaged into human immunodeficiency virus type 1 virions and deaminates deoxycytidine to deoxyuridine on nascent minus-strand retroviral cDNA, leading to hyper-deoxyguanine-to-deoxyadenine mutations on positive-strand cDNA and inhibition of viral replication. The antiviral activity of A3G is suppressed by Vif, a lentiviral accessory protein that prevents encapsidation of A3G. In this study, we identified dominant negative mutants of Vif that interfered with the ability of wild-type Vif to inhibit the encapsidation and antiviral activity of A3G. These mutants were nonfunctional due to mutations in the highly conserved HCCH and/or SOCS box motifs, which are required for assembly of a functional Cul5-E3 ubiquitin ligase complex. Similarly, mutation or deletion of a PPLP motif, which was previously reported to be important for Vif dimerization, induced a dominant negative phenotype. Expression of dominant negative Vif counteracted the Vif-induced reduction of intracellular A3G levels, presumably by preventing Vif-induced A3G degradation. Consequently, dominant negative Vif interfered with wild-type Vif's ability to exclude A3G from viral particles and reduced viral infectivity despite the presence of wild-type Vif. The identification of dominant negative mutants of Vif presents exciting possibilities for the design of novel antiviral strategies.
Human immunodeficiency virus type 1 (HIV-1) is a complex retrovirus that, in addition to the structural and enzymatic proteins encoded by all retroviruses (Gag, Pol, and Env), encodes six accessory proteins (Vif, Vpr, Tat, Rev, Vpu, and Nef). Replication of HIV-1 in most primary cells and some immortalized T-cell lines is dependent on the expression of a functional Vif protein. In the absence of Vif, virus replication is restricted by host deaminases, most notably APOBEC3G (A3G) (56). In the absence of Vif, A3G is incorporated into virus particles, where it causes editing of the viral cDNA during reverse transcription (29, 35, 36, 77). The conversion of deoxycytidine to deoxyuridine on the minus-strand cDNA results in deoxyguanine-to-deoxyadenine changes on the viral plus-strand cDNA to yield highly mutated viral genomes. Virus replication may be inhibited through accumulation of mutations in the viral genome or through degradation of the deaminated viral cDNA via a cellular DNA repair mechanism (reviewed in reference 31). Alternatively, A3G may inhibit virus replication through deamination-independent mechanisms (4, 7, 16, 20, 30, 33, 38, 45, 74).
Vif is a 23-kDa basic protein that is expressed late during infection. Previous immunocytochemical analysis revealed that Vif is largely localized in the cytoplasm (15, 24, 59). Vif is also incorporated into HIV virions, most prominently during productive infection, through an interaction with viral genomic RNA and associates with the viral nucleoprotein complex (21, 24, 25, 32). Vif has the remarkable ability to inhibit encapsidation of A3G. The precise mechanism through which Vif accomplishes this task is still under investigation. However, there is strong evidence that there is a physical interaction of Vif and A3G, which can lead to A3G degradation by the host proteasome machinery (for a review, see reference 14). Other studies, however, suggest the intracellular degradation of A3G may not be the sole mechanism by which Vif neutralizes its antiviral activity (22, 23, 36, 39, 47).
Mutational analysis of Vif has led to the characterization of several distinct binding domains in Vif for assembly of an E3 ubiquitin ligase complex, as well as for interaction with A3G. One of the domains involved is a highly conserved motif near the C terminus of Vif, referred to as the SLQ motif. The SLQ(Y/F)LA sequence resembles a conserved motif in the BC box of the suppressors of cytokine signaling (SOCS) proteins and was found to mediate binding of Vif to elongin C (39, 40, 75, 76), a homolog of the yeast Skp1 protein and a known component of E3 ubiquitin ligase complexes. In addition, a highly conserved H-X5-C-X17-18C-X3-5-H motif (also referred to as the HCCH motif) located upstream of the BC box was found to mediate interaction with cullin 5 (Cul5) (34, 39, 76). Furthermore, two cysteine residues that are part of the HCCH motif are critical components of a zinc finger domain (41, 49, 69, 70). Zinc binding appears to be important for Vif function, since chelation of zinc inhibited HIV Vif activity, presumably by affecting the proper folding of the protein (49, 68). Thus, together with the SLQ motif, the HCCH domain enables Vif to recruit a ubiquitin ligase (E3) complex containing elongin C, elongin B, cullin 5, and Rbx1 (39, 41, 75, 76). Finally, a conserved PPLP motif (residues 161 to 164) was shown to be important for Vif dimerization (3, 43, 73). This region was also shown to affect A3G binding (11, 43), association with Cul5 (61), and interaction with the Pr55gag precursor (5) and to be important for interaction with the cellular tyrosine kinase HCK (17).
The exact A3G binding domain in Vif is still incompletely defined and most likely consists of several discontinuous subdomains (13, 37, 42, 53-55, 60, 64). Site-directed mutagenesis identified residues 40 to 44 (YRHHY) in HIV-1 Vif as important for binding of A3G (42, 53, 54, 71, 78). Moreover, K26 in Vif was found to be critical for A3G interaction (6, 10). In addition, a stretch of hydrophobic amino acids comprising residues 69 to 72 in HIV-1 Vif is important for interaction with A3G (18, 50). Finally, analysis of patient-derived HIV-1 Vif sequences demonstrated the importance of residues K22, Y40, and E45 for A3G recognition (60).
The ability of Vif to bind A3G via its N-terminal domain and to assemble an E3 ubiquitin ligase complex through its C-terminal HCCH and BC box motifs was shown to accelerate polyubiquitination of A3G and resulted in the degradation of A3G by the 26S proteasome (8, 37, 39, 57, 62, 75, 76). One recent report suggests that Vif-induced degradation of A3G can also occur without A3G polyubiquitination (9). Irrespective of what mechanism applies, the role of Vif in A3G degradation appears to be that of an adapter molecule. Based on this model, we hypothesized that Vif mutants lacking either (but not both) of the A3G and E3 ubiquitin ligase binding domains may exhibit dominant negative properties and prevent A3G degradation by competing with wild-type (wt) Vif for binding to A3G or the E3 ubiquitin ligase through their remaining binding domains. For example, Vif mutants capable of binding A3G but functionally inactive due to mutations in other critical regions of the protein should compete for binding of wild-type Vif to A3G and thus interfere with A3G degradation by wild-type Vif. Such mutants would be expected to inhibit the infectivity of wild-type HIV-1 due to increased levels of virus-associated A3G.
The goal of the current study was to test this hypothesis by identifying dominant negative mutants of Vif capable of interfering with the ability of wild-type Vif to neutralize A3G activity. Previous studies identified a Vif variant (F12 Vif) with dominant negative properties (51, 65); however, the dominant negative effect of F12 Vif was A3G independent (51, 65). This resembles our own observation of the A3G-independent inhibition of viral infectivity by high-level expression of Vif (2). Another previous attempt to identify dominant negative variants of Vif using a multiround replication assay was unsuccessful (12). Here, we employed single-cycle infectivity assays using Vif mutants expressed in trans to compete for the function of wild-type Vif encoded by full-length NL4-3. We found that Vif variants carrying mutations or deletions in the HCCH and SOCS box domains of Vif were nonfunctional but retained their ability to interact with A3G. These A3G binding-competent mutants of Vif potently interfered with the ability of wild-type Vif to produce infectious virions in A3G-expressing cells. Furthermore, mutation or deletion of the conserved PPLP motif induced a dominant negative phenotype similar to that of the HCCH and SOCS box mutants. Interestingly, however, the PPLP mutants had lost their ability to interact with A3G, suggesting that competitive binding to A3G of nonfunctional Vif variants in virus-producing cells may not be the sole mechanism of interference with the function of wild-type Vif. Our data provide proof of principle that it is possible to interfere with the ability of wild-type Vif to target A3G in virus-producing cells, thus opening a new avenue for the design of Vif-based novel antiviral strategies.
MATERIALS AND METHODS
Cell culture and transfections.
293T cells were propagated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. LuSIV cells are derived from CEMx174 cells and contain a luciferase indicator gene under the control of the SIVmac239 long terminal repeat (LTR). These cells were obtained through the NIH AIDS Research and Reference Reagent Program and were maintained in complete RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and hygromycin B (300 μg/ml). For transfection, 293T cells were grown in 25-cm2 flasks to about 80% confluence. The cells were transfected using Lipofectamine Plus (Invitrogen Corp., Carlsbad, CA), following the manufacturer's recommendations. A total of 5 to 6 μg of plasmid DNA per 25-cm2 flask was used. Where appropriate, empty-vector DNA [pcDNA3.1(−)MycHis (Invitrogen)] was used to adjust total DNA amounts. Samples were harvested 24 to 48 h posttransfection.
Antibodies.
A peptide antibody to human A3G was prepared as described previously (27) and is available through the NIH AIDS Research and Reference Reagent Program (catalog no. 10082). A monoclonal antibody to Vif (MAb 319) was used for all immunoblot analyses and was obtained from Michael Malim through the NIH AIDS Research and Reference Reagent Program (catalog no. 6459). A monoclonal antibody to alpha-tubulin (Sigma-Aldrich, Inc., St. Louis, MO; catalog no. T-9026) was used as a loading control. An HIV-positive patient serum was used for the identification of HIV-1 capsid (CA) protein.
Plasmids.
The infectious molecular clone pNL4-3 has been previously described (1) and is available through the NIH AIDS Research and Reference Reagent Program (catalog no. 114). A vif-defective variant, pNL4-3vif(−), carries a 178-bp out-of-frame deletion in the vif gene, as previously reported (24). For the expression of Vif in trans, we employed pcDNA-hVif, encoding codon-optimized NL4-3 Vif (46). This plasmid and its nucleotide sequence are available through the NIH AIDS Research and Reference Reagent Program (catalog no. 10077). In-frame deletions and point mutations were introduced by PCR-based site-directed mutagenesis, as indicated in Fig. 1A. A3G carrying a C-terminal MycHis tag was expressed from pcDNA-APO3GMycHis (23). A vector for the expression of epitope tag-free human A3G was described previously (48). Construction of pCul5-Rbx-HA encoding a dominant negative form of cullin 5 was reported elsewhere (13).
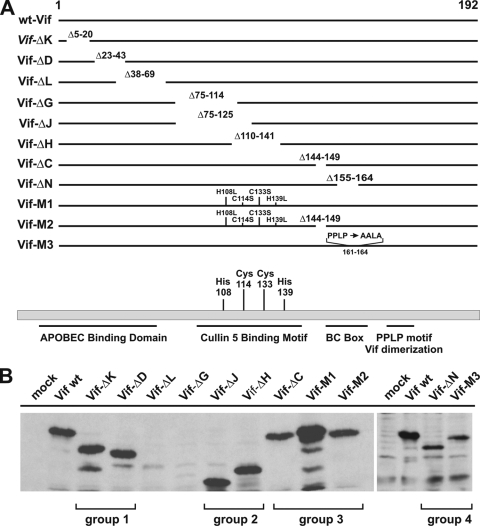
FIG. 1.
Construction and expression of Vif mutants. (A) A series of in-frame deletions mapping to different regions in Vif was created by PCR-based site-directed mutagenesis in the backbone of pcDNA-hVif (46). The positions of the deleted amino acid residues are shown for each mutant. The Vif-M1 mutant carries four point mutations affecting the conserved HCCH motif (H108L, C114S, C133S, and H139L) and was constructed by PCR-based mutagenesis. Mutant Vif-M2 carries the same four amino acid changes as Vif-M1, as well as an in-frame deletion of residues 144 to 149 affecting the conserved BC (SOCS) box of Vif. Mutant M3 carries three amino acid changes, resulting in the change of P161PLP164 to AALA. (B) Cells (5 × 106) were transfected with 5 μg each of the individual Vif mutants. Cells were harvested 24 h posttransfection, and whole-cell lysates were analyzed by immunoblotting them with a monoclonal antibody to Vif. A mock-transfected sample was included as a background control.
Immunoblotting.
For immunoblot analysis of cell-associated proteins, whole-cell lysates were prepared as follows. Cells were washed once with phosphate-buffered saline (PBS), suspended in PBS (200 μl/5 × 106 cells), and mixed with an equal volume of sample buffer (4% sodium dodecyl sulfate, 125 mM Tris-HCl, pH 6.8, 10% 2-mercaptoethanol, 10% glycerol, and 0.002% bromophenol blue). To analyze virus-associated proteins, cell-free filtered supernatants from transfected 293T cells (5 to 6 ml) were pelleted (75 min; 35,000 rpm) through a 20% sucrose cushion (4 ml) in an SW41 rotor. The concentrated virus pellet was suspended in PBS (150 μl) and mixed with an equal volume of sample buffer. Proteins were solubilized by heating them for 10 to 15 min at 95°C with occasional vortexing. Cell and virus lysates were subjected to SDS-PAGE; proteins were transferred to polyvinylidene difluoride (PVDF) membranes and reacted with appropriate antibodies as described above. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences, Piscataway, NJ), and the proteins were visualized by enhanced chemiluminescence (ECL) (Amersham Biosciences).
Virus preparation.
Virus stocks were prepared by transfection of 293T cells with appropriate plasmid DNAs. Virus-containing supernatants were harvested 24 to 48 h after transfection. Cellular debris was removed by centrifugation (5 min; 1,500 rpm), and the clarified supernatants were filtered (0.45 μm) to remove residual cellular contamination. The filtered virus stocks were further purified by pelleting them through a 20% sucrose cushion (75 min; 4°C at 35,000 rpm in an SW41 rotor).
Viral-infectivity assay.
To determine viral infectivity, virus stocks were normalized for equal reverse transcriptase activities (67) and used to infect 5 × 105 LuSIV cells (52) in a 24-well plate in a total volume of 1.2 to 1.4 ml. Infection was allowed for 24 h at 37°C. Cells were then harvested and lysed in 150 μl of Promega 1× reporter lysis buffer (Promega Corp., Madison, WI). To determine the luciferase activity in the lysates, 50 μl of each lysate was combined with luciferase substrate (Promega Corp., Madison, WI) by automatic injection, and light emission was measured for 10 s at room temperature in a luminometer (Optocomp II; MGM Instruments, Hamden, CT).
Coimmunoprecipitation analyses.
293T cells were transfected with expression vectors for A3G and Vif as indicated in the text. Cells were harvested 24 h posttransfection, washed twice with cold PBS, lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% [vol/vol] Triton X-100, and Complete protease inhibitor cocktail) at 4°C for 30 min, and then clarified by centrifugation at 10,000 × g for 2 min. Ten percent of the lysate was used as an input control, and the remaining lysate was used for immunoprecipitation of Myc-tagged antigens. Precleared cell lysates were mixed with anti-Myc antibody-conjugated agarose beads (Sigma-Aldrich, Inc., St. Louis, MO) and incubated at 4°C for 1 h. Samples were then washed three times with wash buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, 0.05% [vol/vol] Tween 20). Proteins were eluted by boiling beads in sample buffer and subjected to immunoblot analysis with antibodies to A3G and Vif.
RESULTS
Construction and expression of codon-optimized Vif mutants.
A series of Vif mutants was constructed based on pcDNA-hVif, a codon-optimized expression vector carrying the vif gene from NL4-3, allowing Rev- and Tat-independent expression of Vif (46). We introduced in-frame deletions in vif designed to remove potential functionally important regions of the protein as depicted in Fig. 1A. A total of eight in-frame deletion mutants were analyzed, with deletions ranging from 6 (Vif-ΔC) to 50 (Vif-ΔJ) amino acids and spanning the N-terminal 164 residues of the 192-residue protein as outlined in Fig. 1A. In addition, two mutants were created carrying point mutations to replace a conserved HCCH motif (H108L, C114S, C133S, and H139L) either alone (Vif-M1) or in combination with the deletion of the SOCS box domain encompassing residues 144 to 149 (Vif-M2). The HCCH motif was previously found to be critical for Vif-Cul5 interactions (41, 75, 76), while the BC (SOCS) box has been implicated in the association of Vif with elongin B/C (39, 76). Finally, Vif-M3 was constructed by replacing the PPLP sequence (residues 161 to 164) with AALA.
To investigate the expression of our Vif mutants, cells were transfected with each of the constructs and subjected to immunoblot analysis. Mock-transfected cells served as a negative control. Whole-cell lysates were prepared and subjected to immunoblot analysis using a Vif-specific monoclonal antibody as described in Materials and Methods (Fig. 1B). All of the mutants, with the exception of Vif-ΔL and Vif-ΔG, were expressed well and identified by a monoclonal antibody to Vif. Since Vif-ΔL and Vif-ΔG were only poorly expressed, they were excluded from further analysis. Shorter protein products were observed for some of the mutants. It is unclear if they represent degradation intermediates or are caused by internal initiation, as previously reported (66); however, they are frequently observed in transient-expression studies (23, 26).
Functional characterization of Vif mutants.
To investigate the functionality of the Vif deletion mutants, we investigated their effects on both metabolic stability and packaging of A3G into virions. We also assessed their abilities to rescue the infectivity of Vif-defective NL4-3 produced in the presence of A3G. For simplicity, the mutants depicted in Fig. 1 were analyzed in four separate groups. Every group included a negative control [(−)] and a positive control (wt). Group 1 included Vif-ΔK and Vif-ΔD and represented N-terminal deletions that were likely to affect Vif's ability to interact with A3G. Group 2 represented the central region of Vif and included Vif-ΔJ and Vif-ΔH. Group 3 included Vif-ΔC, Vif-M1, and Vif-M2. These mutants were expected to be incapable of assembling a functional Cul5-E3 ubiquitin ligase complex but should have retained the ability to interact with A3G. Finally, group 4 included Vif-ΔN and Vif-M3 with deletion or mutation of the conserved PPLP motif, respectively.
293T cells were transfected with constant amounts of pNL4-3vif(−) and pcDNA-A3G, along with codon-optimized vectors encoding wt or mutant Vif proteins. An equivalent amount of empty-vector DNA was added to the Vif(−) controls. Cells and virus-containing culture supernatants were collected 24 h posttransfection. Whole-cell lysates were prepared, and virus-containing supernatants were purified, concentrated, and subjected to immunoblot analysis using antibodies to A3G, Vif, CA, and tubulin (tub), as described in Materials and Methods.
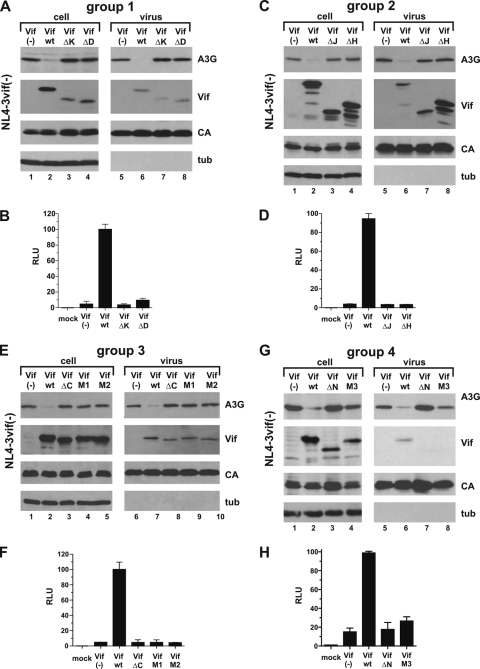
Analysis of group 1 mutants is shown in Fig. 2A and B. As expected, expression of Vif wt caused a severe reduction in cellular A3G levels (Fig. 2A, compare lanes 1 and 2). In contrast, neither Vif-ΔK nor Vif-ΔD affected cellular expression of A3G (Fig. 2A, lanes 3 and 4). A3G was packaged into Vif-deficient viruses (Fig. 2A, lane 5) but was excluded from virions in the presence of Vif wt (Fig. 2A, lane 6); neither Vif-ΔK nor Vif-ΔD was capable of inhibiting encapsidation of A3G (Fig. 2A, lanes 7 and 8). Thus, deletion of residues 5 to 20 or 23 to 43 in Vif abolishes its ability to induce degradation and inhibit encapsidation of A3G. Consistent with the protein data, analysis of viral infectivity revealed that only virus produced in the presence of Vif wt was infectious, while neither Vif-ΔK nor Vif-ΔD was able to overcome the antiviral effect of A3G (Fig. 2B). Thus, deletion of residues 5 to 20 or 23 to 43 leads to the functional inactivation of Vif.
FIG. 2.
Vif mutants are nonfunctional. (A) 293T cells were transfected with 3 μg of pNL4-3vif(−) virus and 0.5 μg of pcDNA-A3G, together with 2.5 μg each of empty vector (lanes 1 and 5), pcDNA-hVif wt (lanes 2 and 6), Vif-ΔK (lanes 3 and 7), or Vif-ΔD (lanes 4 and 8). Cells and virus-containing supernatants were harvested 24 h posttransfection. Whole-cell lysates and virus-containing supernatants were prepared as described in Materials and Methods. Samples were analyzed by immunoblotting them using antibodies to A3G and Vif. The Vif blot was subsequently reprobed with an HIV-positive patient serum to detect viral capsid protein (CA), and the A3G blot was reprobed with a tubulin-specific monoclonal antibody (tub). (B) The infectivities of the viruses shown in panel A were determined in a single-cycle infectivity assay by infection of LuSIV indicator cells (52). Virus-induced activation of luciferase, as measured in relative light units (RLU), was quantified 24 h later in a standard luciferase assay as described in Materials and Methods. The relative infectivity of viruses produced in the presence of Vif wt was defined as 100%. Uninfected LuSIV cells were included as a negative control (mock). The error bars reflect standard deviations calculated from three independent infections. (C and D) 293T cells were transfected as for panel A, except that Vif-ΔJ and Vif-ΔH mutants were analyzed. Analysis of viral infectivity was done as for panel B. (E and F) 293T cells were transfected as for panel A, except that Vif-ΔC, Vif-M1, and Vif-M2 mutants were analyzed. The infectivities of the resulting viruses were determined as for panel B. (G and H) 293T cells were transfected as for panel A, except that Vif-ΔN and Vif-M3 mutants were analyzed. Analysis of viral infectivity was done as for panel B.
Group 2 to 4 Vif mutants were analyzed in a similar fashion (Fig. 2C to H). In all cases, Vif wt reduced intracellular expression of A3G and prohibited the encapsidation of A3G into viral particles. None of the Vif deletion mutants or the M1, M2, or M3 point mutants affected the expression or packaging of A3G compared to the Vif-negative control. Similarly, none of the Vif mutants analyzed here were able to overcome the inhibitory effects of A3G on viral infectivity (Fig. 2D, F, and H). Thus, all regions in Vif analyzed in this study are critical for interference with A3G, and deletion or mutation of any of these regions invariably leads to the loss of Vif function.
Group 1, 2, and 4 mutants, but not group 3 mutants, of Vif are defective for A3G binding.
The N-terminal region of Vif affected by group 1 mutants has previously been implicated in the interaction with A3G (37, 42, 53-55, 60, 64, 71, 78). In contrast, group 3 mutants affect the HCCH and BC box domains in Vif and were expected to be defective for interaction with the Cul5-E3 ubiquitin ligase complex but to retain the ability to bind A3G. Similarly, group 2 mutants were expected to retain their A3G binding capacities; however, because of their overlap with the HCCH and BC box motifs, group 2 mutants were expected to be defective for Cul5 binding. Finally, group 4 mutants were expected to be defective for A3G binding, since deletion of the PPLP motif in Vif was previously shown to affect A3G binding (11).
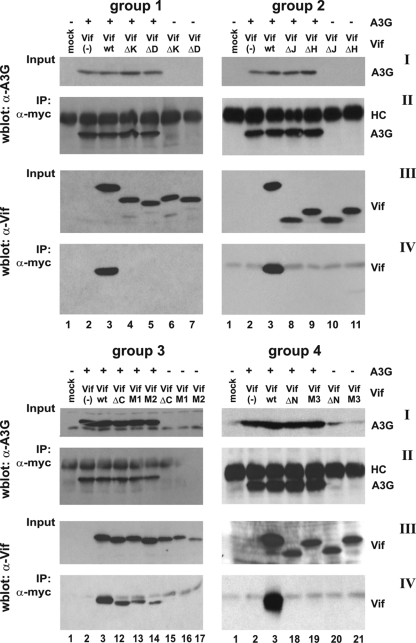
To assess the abilities of the Vif mutants to interact with A3G, we performed coimmunoprecipitation studies, in which we coexpressed epitope-tagged A3G and individual Vif variants. Since Vif wt can induce degradation of A3G, all experiments were done in the presence of dominant negative Cul5-Rbx (13, 75) to minimize signal fluctuations due to A3G degradation. A C-terminally Myc epitope-tagged A3G was used here (23) to minimize the risk of antibody interference with Vif-A3G interaction. 293T cells were cotransfected with pcDNA-A3G-Myc, pCul5-Rbx-HA, and expression vectors carrying codon-optimized vif genes. Cell lysates were prepared 24 h posttransfection, and protein complexes were immunoprecipitated with anti-Myc antibody-conjugated agarose beads. Input A3G and immunoprecipitated A3G were identified by immunoblotting with an A3G-specific antibody (Fig. 3I and II, respectively). Input samples and immunoprecipitates were also probed with a Vif-specific monoclonal antibody (Fig. 3III and IV, respectively). As expected, Vif wt exhibited a strong binding affinity for A3G (Fig.3IV, lanes 3), and group 1 mutants Vif-ΔK and Vif-ΔD did not interact with A3G (Fig.3IV, lanes 4 and 5). Surprisingly, Vif-ΔJ and Vif-ΔH mutants also did not interact with A3G (Fig.3IV, lanes 8 and 9). Consistent with a previous report (11), neither Vif-ΔN nor Vif-M3 interacted with A3G in the pull-down analysis (Fig.3IV, lanes 18 and 19). Only group 3 mutants retained the ability to interact with A3G, although the efficiency of interaction seemed to be reduced compared to Vif wt (Fig. 3, lanes 12 to 14). Interaction of group 3 Vif mutants with A3G was specific, as evidenced by the lack of Vif precipitation in the absence of A3G (Fig.3IV, lanes 15 to 17). Our results confirm that the N-terminal region in Vif contains sequences critical for interaction with A3G. However, our results also indicate that sequences in Vif important for interaction with A3G may extend beyond the region identified in previous studies (42, 50, 53, 54).
FIG. 3.
Only group 3 Vif mutants interact with A3G. Cells were either mock transfected (lanes 1) or transfected with 2.5 μg of pcDNA-A3G-Myc (lanes 2 to 5, 8 to 9, 12 to 14, and 18 to 19), together with 2.5 μg of empty-vector DNA [Vif(−)] (lanes 2), pcDNA-hVif wt (lanes 3), or Vif mutants from groups 1 to 4 (lanes 4 and 5, 8 and 9, 12 and 14, and 18 and 19). Mutant Vif proteins were also expressed in the absence of A3G to control for nonspecific binding to the antibody matrix (lanes 6 and 7, 10 and 11, 15 and 17, and 20 and 21). To prevent possible degradation of A3G by Vif, 1 μg of a dominant negative mutant of Cul5, Cul5ΔRbx-HA (75), was cotransfected in all samples. Cells were harvested 24 h after transfection. Cell lysates were immunoprecipitated with anti-Myc antibody conjugated to agarose beads as described in Materials and Methods. Aliquots of total cell lysates (Input) and immunoprecipitated proteins (IP: α-myc) were separated by SDS-PAGE and subjected to immunoblot analysis using antibodies to A3G (I and II) or Vif (III and IV). Proteins are identified on the right. HC, immunoglobulin heavy chain.
Group 3 and 4 Vif mutants dominantly interfere with the function of wild-type Vif.
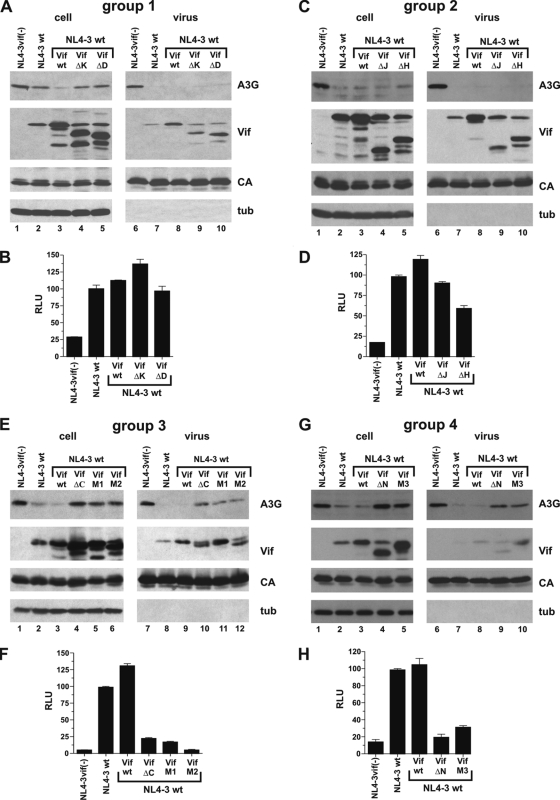
Having identified a series of nonfunctional Vif mutants and having characterized their A3G binding properties, we next wanted to assess their potentials to act as dominant negative inhibitors of wild-type Vif function. To test this, cells were transfected with the full-length HIV molecular clone NL4-3 encoding wild-type Vif, together with A3G either alone or in combination with codon-optimized vectors for expression of potential dominant negative Vif mutants in trans. As a control, the effect of A3G on Vif-defective NL4-3vif(−) was analyzed in parallel. Cells and virus-containing supernatants were harvested 48 h posttransfection and processed for immunoblot analysis as described for Fig. 2. As expected, A3G was packaged into Vif-deficient virions (Fig. 4A, C, and G, lanes 6, and E, lane 7). However, in all cases, NL4-3 Vif efficiently inhibited the packaging of A3G into viral particles (Fig. 4A, C, and G, lanes 7, and E, lane 8) even though virus-encoded Vif affected intracellular A3G levels to various degrees (Fig. 4A, C, E, and G, A3G, compare lanes 1 and 2 of groups 1 to 4). Therefore, under the conditions of this experiment, Vif expressed from NL4-3 was necessary and sufficient to overcome the antiviral effects of A3G (Fig. 4B, D, F, and H). Indeed, viral infectivity was only slightly improved relative to NL4-3 wt by coexpression of additional Vif from the codon-optimized vector (Fig. 4B, D, F, and H, compare NL4-3 wt and Vif wt).
FIG. 4.
Group 3 and group 4 Vif mutants exhibit dominant negative characteristics and interfere with the function of wild-type Vif. The experiment essentially mimicked the one shown in Fig. 2, except that Vif mutants were coexpressed with NL4-3 encoding wild-type Vif. (A, C, E, and G) 293T cells were cotransfected with 3 μg of pNL4-3 wt, 0.5 μg of pcDNA-A3G, and 2.5 μg each of empty vector (NL4-3 wt), Vif wt (wt), or Vif mutants as indicated. Cells transfected with vif-defective NL4-3 and A3G were included as a control [NL4-3vif(−)]. Cell lysates and virus-containing supernatants were processed for immunoblotting as described for Fig. 2. (B, D, F, and H) The infectivities of the viruses shown in panels A, C, E, and G were determined in a single-cycle infectivity assay as described for Fig. 2. The error bars reflect the standard deviations calculated from three independent infections.
Expression of Vif mutants from the codon-optimized vectors was apparent from the immunoblot analysis of the increased Vif signal in the cases of Vif wt, Vif-M1, and Vif-M3 or from the appearance of additional Vif-specific bands in the cases of the deletion mutants (Fig. 4A, C, E, and G). Invariably, Vif mutants expressed in trans were copackaged with virus-encoded wild-type Vif (Fig. 4A, C, E, and G, virus). Importantly, we did not observe evidence for competitive inhibition of wild-type Vif packaging by any of the Vif mutants. Analysis of A3G encapsidation revealed that neither group 1 nor group 2 Vif mutants had the ability to interfere with the function of virus-encoded Vif to inhibit A3G packaging (Fig. 4A and C). Consequently, none of the group 1 and group 2 Vif mutants inhibited the rescue of viral infectivity by NL4-3-encoded wild-type Vif (Fig. 4B and D). The only exception was Vif-ΔH, which caused moderate interference with viral infectivity (Fig. 4D). Group 3 and group 4 Vif mutants, on the other hand, interfered with the activity of wild-type Vif, leading to increased cellular A3G levels and resulting in the encapsidation of A3G at levels approaching those of vif-deficient virions (Fig. 4E and G). As a result, group 3 and group 4 Vif mutants strongly inhibited the infectivity of wild-type NL4-3 (Fig. 4F and H). Thus, group 3 and group 4 Vif mutants carrying mutations or deletions in the conserved HCCH, SLQ, and PPLP motifs have the ability to dominantly interfere with the function of virus-encoded wild-type Vif.
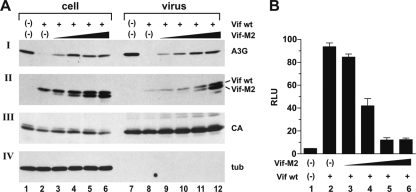
The dose dependence of dominant negative interference with wild-type Vif function was studied using the Vif-M2 mutant as a representative example (Fig. 5). The amount of NL4-3 DNA was kept constant, and the ratios of NL4-3 to Vif-M2 DNA varied over a 10-fold range, as indicated in the legend to Fig. 5. Total amounts of transfected DNA were kept constant by adding empty-vector DNA as appropriate. NL4-3vif(−) and NL-4-3 wt produced in the absence of Vif-M2 served as negative and positive controls, respectively (Fig. 5, lanes 1 and 2 and lanes 7 and 8). Cell lysates and viral pellets were analyzed by immunoblotting using antibodies to A3G, Vif, CA, and tubulin. All samples produced comparable amounts of viral Gag proteins, and viral pellets were devoid of cellular debris, as evidenced by the absence of tubulin in the viral fractions. As expected, NL4-3-encoded Vif prevented encapsidation of A3G into HIV-1 virions (Fig. 5, lane 8). In contrast, coexpression of Vif-M2 resulted in a dose-dependent increase in virus-associated A3G (Fig. 5, lanes 9 to 12) and was accompanied by increased cellular A3G levels (Fig. 5A, lanes 3 to 6). Consistent with these results, the infectivity of cell-free viruses was reduced in a dose-dependent manner (Fig. 5B, lanes 3 to 6). Maximum inhibition of viral infectivity was achieved when NL4-3 Vif and Vif-M2 were expressed at roughly equimolar levels (Fig. 5A and B, lane 5). Interestingly, increasing expression of Vif-M2 appeared to also facilitate the packaging of Vif wt. The reason for this phenomenon is unclear. However, we previously reported that virus-associated Vif is subject to proteolytic processing by the viral protease (26). It is therefore conceivable that packaging of increasing amounts of Vif-M2 leads to stabilization of Vif wt through substrate competition.
FIG. 5.
Interference with wild-type Vif function by dominant negative Vif is dose dependent. (A) 293T cells were transfected with 3 μg of pNL4-3 Vif(−) DNA (lane 1) or 3 μg of pNL4-3 wt (lane 2) plus 0.5 μg of pcDNA-A3G and 2.5 μg of empty-vector DNA each. In lanes 3 to 6, cells were transfected with 3 μg of pNL4-3 wt, 0.5 μg of pcDNA-A3G, and increasing amounts of Vif-M2 (lane 3, 0.1 μg; lane 4, 0.25 μg; lane 5, 0.5 μg; lane 6, 1.0 μg). The total amounts of DNA in each sample were adjusted to 6 μg using empty-vector DNA as appropriate. Cells and virus-containing supernatants were harvested 48 h posttransfection. Concentrated virus preparations, along with the whole-cell lysates, were analyzed by immunoblotting them using antibodies to A3G (I) or Vif (II). The A3G blot was subsequently reprobed with an HIV-positive patient serum (III) to detect viral capsid protein (CA); the Vif blot was reprobed with a tubulin-specific monoclonal antibody (IV, tub) to control for loading errors and to verify the absence of cellular contaminants in the viral samples. (B) The infectivities of virions in panel A were determined in a single-cycle infectivity assay by infection of LuSIV indicator cells. Relative virus-induced activation of luciferase (RLU) was quantified 24 h later in a standard luciferase assay. The error bars reflect the standard deviations calculated from three independent infections.
DISCUSSION
The recent identification of potent antiviral host restriction factors and the ability of viruses to outsmart these defense mechanisms highlight the ongoing struggle between viruses and host cells. Thus, continued intensified efforts are needed to understand the mechanisms of viral resistance. The realization that viral accessory proteins, such as Vif, Vpu, and Nef, are prominently involved in counteracting host defense mechanisms (for a review, see references 14 and 63) has put new emphasis on the functional analysis of these accessory proteins. In particular, the ability of Vif to counteract the antiviral effects of A3G has been studied in great detail and has been attributed to several mechanisms, including proteasomal degradation of A3G and degradation-independent mechanisms (reviewed in references 14 and 63). Most functional models of Vif require a physical interaction with A3G. Thus, inhibition of Vif-A3G interaction by pharmacological means, by small molecule inhibitors, or by dominant negative forms of Vif has great potential as an effective tool to prevent the spread of HIV-1 in infected patients. Several studies have provided proof of principle for this notion. For instance, peptides targeting the PPLP domain in Vif were found to inhibit Vif dimerization, increase packaging of A3G into Vif-expressing virus, and inhibit replication of wild-type HIV-1 (43, 72). Furthermore, the zinc chelator TPEN was shown to inhibit recruitment of Cul5 by Vif and to prevent Vif-induced A3G degradation (68). Next, Nathans et al. identified a small-molecule inhibitor, RN-18, that enhances degradation of Vif in the presence of A3G through an unknown mechanism (44). Finally, protease-derived peptides were found to inhibit Vif function in nonpermissive H9 cells but had no effect on virus replication in permissive SupT1 cells (19).
Our idea to test functionally inactive Vif proteins for their potential as dominant negative inhibitors of HIV-1 was based on the recent identification of distinct domains in Vif responsible for interaction with A3G on one side and the Cul5-E3 ubiquitin ligase complex on the other. Formation of the A3G-Vif-Cul5-E3 ternary complex results in A3G ubiquitination and subsequent proteasomal degradation of the protein (8, 28, 37, 40, 57, 58, 75, 76). We hypothesized that nonfunctional Vif mutants capable of interacting with either A3G or E3 ligase, but not both, should be able to inhibit the function of wild-type Vif through competitive binding to a common ligand. Indeed, the group 3 mutants analyzed in this study had lost the ability to assemble a functional E3 ligase complex but retained A3G binding activity and inhibited wild-type Vif function. As a result, wild-type NL4-3 produced from A3G-expressing cells in the presence of group 3 Vif mutants was noninfectious. Surprisingly, group 1 Vif mutants, which had lost the ability to bind A3G but had intact binding domains for Cul5-E3 ubiquitin ligase complexes, did not exhibit dominant negative characteristics and were unable to inhibit the function of wild-type Vif. The reason for this is unclear, but it could be due to subsaturating levels of Vif or to conformational constraints lowering the relative affinity of mutant Vif proteins for E3 ligase components compared to wild-type Vif. In contrast, group 4 mutants, which also did not interact with A3G in our in vitro binding assay (Fig. 3), exhibited dominant negative properties and inhibited the function of wild-type Vif. The mechanism of interference by group 4 mutants is currently unclear. Our own attempts to test the dimerization properties of these mutants were inconclusive. Nevertheless, deletion of residues 151 to 164 in Vif, which is similar to our Vif-ΔN mutant and includes the PPLP motif, was previously found to reduce dimerization of Vif (73), making interference through formation of nonfunctional Vif heterodimers unlikely. In contrast, Donahue et al. demonstrated that mutation of the PPLP motif did not interfere with Vif's ability to bind Cul5 and elongin C (11). It is therefore possible that our group 4 Vif mutants exert their dominant negative phenotype through binding to Cul5 and/or elongin C, thereby contributing to the depletion of these components of the Cul5-E3 ubiquitin ligase complex.
Overall, our results strongly suggest that the dominant negative phenotype of group 3 and group 4 Vif mutants is specific and due to interference with wild-type Vif rather than affecting viral infectivity through binding to Gag, as previously described (2). This is further supported by the observation that group 3 mutants did not inhibit the infectivity of Vif-deficient HIV-1 virions in the absence of A3G (data not shown). Also, the relatively low levels of mutant Vif required to reveal the dominant negative effect suggest that the interference is specific. Finally, group 2 Vif mutants were predicted to have dominant negative properties because the A3G binding domain had been mapped to regions upstream of the mutations. Surprisingly, neither Vif-ΔJ (lacking residues 75 to 125), Vif-ΔH (lacking residues 110 to 141), nor Vif-ΔN (lacking residues 155 to 164) interacted with A3G in our pull-down experiment (Fig. 3). Thus, it appears that regions in Vif important for A3G interaction cover a much larger area of the Vif protein than previously reported. It is possible that deletions in the central and C-terminal parts of Vif are not directly involved in A3G binding but instead impose conformational constraints, preventing proper folding of the protein and thereby indirectly precluding A3G interaction. This possibility will have to be addressed in future studies by analyzing smaller deletions or point mutations. Also surprisingly, Vif-ΔD, lacking residues 23 to 43, failed to interact with A3G in our coimmunoprecipitation analysis (Fig. 3). This result is consistent with the reported importance of residues 40 to 44 in Vif for interaction with A3G (42, 53, 54, 71, 78) but conflicts with our own previous observation that a similar deletion mutant was able to interact with A3G in vitro (13). It should be noted that the Vif mutants employed in the current study, including Vif-ΔD, are based on the NL4-3 vif sequence while the previous in vitro study used HXB2-based vif vectors. The NL4-3 and HXB2 Vif proteins differ by 15 amino acids (9%), including 4 nonconservative changes in the vicinity of residues 23 to 44. It is therefore possible that the ability or inability of Vif-ΔD to interact with A3G is governed by amino acid residues in the area surrounding the deletion, again suggesting that conformational changes imposed by these amino acid differences affect the Vif-A3G interaction.
Dominant negative interference by both group 3 and group 4 Vif mutants involved the stabilization of intracellular A3G and resulted in increased encapsidation of the deaminase into wild-type HIV-1 virions. Although mechanistic details have yet to be defined for both group 3 and group 4 Vif mutants, the most likely scenario seems to be that group 3 mutants act by competitive binding to A3G while group 4 mutants may inhibit wild-type Vif function through competitive binding to components of the Cul 5-E3 ubiquitin ligase complex. Another possible mode of dominant negative interference is the formation of Vif-Vif heterodimers. Vif dimerization was previously reported to be important for biological activity (43, 72, 73), and it is conceivable that wild-type Vif function is inhibited through the formation of heterodimers between wild-type Vif and mutant Vif. Such a mechanism could, in theory, apply to group 3 and group 4 mutants, since it would not require competition for Vif binding sites on A3G and would be consistent with our observation that Vif can prevent the encapsidation of degradation-resistant A3G (47). However, the fact that mutation or deletion of the PPLP motif inhibits Vif dimerization (73) makes such a model less likely, at least for the group 4 mutants. Our own attempts to assess hetero-oligomerization of Vif have remained inconclusive due to problems with the solubility of Vif mutants (data not shown), and more sensitive assays will have to be developed to satisfactorily address this issue in future experiments.
It is unclear why a previous study failed to identify dominant negative Vif mutants (12). However, Fujita et al. analyzed Vif mutants carrying single amino acid changes, while our dominant negative mutants contained multiresidue deletions and/or multiple amino acid changes. It is therefore possible that single amino acid changes, as analyzed by Fujita et al., are insufficient to confer a dominant negative phenotype. The identification of dominant negative mutants of Vif presents exciting possibilities for the design of novel antiviral strategies. Future studies will have to test the robustness of the dominant negative phenotype of our Vif mutants and determine if and how fast HIV-1 can develop resistance to dominant negative interference.
Acknowledgments
We are grateful to Amy Andrew for valuable suggestions and for critical comments on the manuscript. We thank Xiao-Fang Yu for plasmid pCul5-HA.
This work was supported in part by a grant from the NIH Intramural AIDS Targeted Antiviral Program to K.S. and by the Intramural Research Program of the NIH, NIAID.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari, H., M. Fujita, S. Kao, M. A. Khan, M. Shehu-Xhilaga, A. Adachi, and K. Strebel. 2004. High level expression of human immunodeficiency virus type-1 Vif inhibits viral infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J. Biol. Chem. 279:12355-12362. [DOI] [PubMed] [Google Scholar]
- 3.Auclair, J. R., K. M. Green, S. Shandilya, J. E. Evans, M. Somasundaran, and C. A. Schiffer. 2007. Mass spectrometry analysis of HIV-1 Vif reveals an increase in ordered structure upon oligomerization in regions necessary for viral infectivity. Proteins 69:270-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouyac, M., M. Courcoul, G. Bertoia, Y. Baudat, D. Gabuzda, D. Blanc, N. Chazal, P. Boulanger, J. Sire, R. Vigne, and B. Spire. 1997. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J. Virol. 71:9358-9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, G., Z. He, T. Wang, R. Xu, and X. F. Yu. 2009. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVX4YX9Y motif influences its interaction with APOBEC3G. J. Virol. 83:8674-8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 8.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 9.Dang, Y., L. M. Siew, and Y. H. Zheng. 2008. APOBEC3G is degraded by the proteasomal pathway in a Vif-dependent manner without being polyubiquitylated. J. Biol. Chem. 283:13124-13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang, Y., X. Wang, T. Zhou, I. A. York, and Y. H. Zheng. 2009. Identification of a novel WXSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J. Virol. 83:8544-8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donahue, J. P., M. L. Vetter, N. A. Mukhtar, and R. T. D'Aquila. 2008. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology 377:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita, M., S. Matsumoto, A. Sakurai, N. Doi, M. Miyaura, A. Yoshida, and A. Adachi. 2002. Apparent lack of trans-dominant negative effects of various vif mutants on the replication of HIV-1. Microbes Infect. 4:1203-1207. [DOI] [PubMed] [Google Scholar]
- 13.Goila-Gaur, R., M. Khan, E. Miyagi, S. Kao, S. Opi, H. Takeuchi, and K. Strebel. 2008. HIV-1 Vif promotes the formation of high molecular mass APOBEC3G complexes. Virology 372:136-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goila-Gaur, R., and K. Strebel. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goncalves, J., P. Jallepalli, and D. H. Gabuzda. 1994. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J. Virol. 68:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of tRNA3Lys-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassaine, G., M. Courcoul, G. Bessou, Y. Barthalay, C. Picard, D. Olive, Y. Collette, R. Vigne, and E. Decroly. 2001. The tyrosine kinase Hck is an inhibitor of HIV-1 replication counteracted by the viral Vif protein. J. Biol. Chem. 276:16885-16893. [DOI] [PubMed] [Google Scholar]
- 18.He, Z., W. Zhang, G. Chen, R. Xu, and X. F. Yu. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381:1000-1011. [DOI] [PubMed] [Google Scholar]
- 19.Hutoran, M., E. Britan, L. Baraz, I. Blumenzweig, M. Steinitz, and M. Kotler. 2004. Abrogation of Vif function by peptide derived from the N-terminal region of the human immunodeficiency virus type 1 (HIV-1) protease. Virology 330:261-270. [DOI] [PubMed] [Google Scholar]
- 20.Iwatani, Y., D. S. Chan, F. Wang, K. S. Maynard, W. Sugiura, A. M. Gronenborn, I. Rouzina, M. C. Williams, K. Musier-Forsyth, and J. G. Levin. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao, S., H. Akari, M. A. Khan, M. Dettenhofer, X. F. Yu, and K. Strebel. 2003. Human immunodeficiency virus type 1 Vif is efficiently packaged into virions during productive but not chronic infection. J. Virol. 77:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, S., R. Goila-Gaur, E. Miyagi, M. A. Khan, S. Opi, H. Takeuchi, and K. Strebel. 2007. Production of infectious virus and degradation of APOBEC3G are separable functional properties of human immunodeficiency virus type 1 Vif. Virology 369:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karczewski, M. K., and K. Strebel. 1996. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J. Virol. 70:494-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan, M. A., H. Akari, S. Kao, C. Aberham, D. Davis, A. Buckler-White, and K. Strebel. 2002. Intravirion processing of the human immunodeficiency virus type 1 Vif protein by the viral protease may be correlated with Vif function. J. Virol. 76:9112-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan, M. A., S. Kao, E. Miyagi, H. Takeuchi, R. Goila-Gaur, S. Opi, C. L. Gipson, T. G. Parslow, H. Ly, and K. Strebel. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 79:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi, M., A. Takaori-Kondo, Y. Miyauchi, K. Iwai, and T. Uchiyama. 2005. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem. 280:18573-18578. [DOI] [PubMed] [Google Scholar]
- 29.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 30.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 282:32065-32074. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 286:1897-1905. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H., X. Wu, M. Newman, G. M. Shaw, B. H. Hahn, and J. C. Kappes. 1995. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 69:7630-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 81:7238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo, K., Z. Xiao, E. Ehrlich, Y. Yu, B. Liu, S. Zheng, and X. F. Yu. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U. S. A. 102:11444-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 36.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 37.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 38.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 41.Mehle, A., E. R. Thomas, K. S. Rajendran, and D. Gabuzda. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281:17259-17265. [DOI] [PubMed] [Google Scholar]
- 42.Mehle, A., H. Wilson, C. Zhang, A. J. Brazier, M. McPike, E. Pery, and D. Gabuzda. 2007. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J. Virol. 81:13235-13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, J. H., V. Presnyak, and H. C. Smith. 2007. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology 4:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nathans, R., H. Cao, N. Sharova, A. Ali, M. Sharkey, R. Stranska, M. Stevenson, and T. M. Rana. 2008. Small-molecule inhibition of HIV-1 Vif. Nat. Biotechnol. 26:1187-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen, K. L., M. Llano, H. Akari, E. Miyagi, E. M. Poeschla, K. Strebel, and S. Bour. 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319:163-175. [DOI] [PubMed] [Google Scholar]
- 47.Opi, S., S. Kao, R. Goila-Gaur, M. A. Khan, E. Miyagi, H. Takeuchi, and K. Strebel. 2007. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 81:8236-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Opi, S., H. Takeuchi, S. Kao, M. A. Khan, E. Miyagi, R. Goila-Gaur, Y. Iwatani, J. G. Levin, and K. Strebel. 2006. Monomeric APOBEC3G is catalytically active and has antiviral activity. J. Virol. 80:4673-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul, I., J. Cui, and E. L. Maynard. 2006. Zinc binding to the HCCH motif of HIV-1 virion infectivity factor induces a conformational change that mediates protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 103:18475-18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pery, E., K. S. Rajendran, A. J. Brazier, and D. Gabuzda. 2009. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J. Virol. 83:2374-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porcellini, S., L. Alberici, F. Gubinelli, R. Lupo, C. Olgiati, G. P. Rizzardi, and C. Bovolenta. 2009. The F12-Vif derivative Chim3 inhibits HIV-1 replication in CD4+ T lymphocytes and CD34+-derived macrophages by blocking HIV-1 DNA integration. Blood 113:3443-3452. [DOI] [PubMed] [Google Scholar]
- 52.Roos, J. W., M. F. Maughan, Z. Liao, J. E. Hildreth, and J. E. Clements. 2000. LuSIV cells: a reporter cell line for the detection and quantitation of a single cycle of HIV and SIV replication. Virology 273:307-315. [DOI] [PubMed] [Google Scholar]
- 53.Russell, R. A., and V. K. Pathak. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell, R. A., J. Smith, R. Barr, D. Bhattacharyya, and V. K. Pathak. 2009. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J. Virol. 83:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schrofelbauer, B., T. Senger, G. Manning, and N. R. Landau. 2006. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 80:5984-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 57.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 58.Shirakawa, K., A. Takaori-Kondo, M. Kobayashi, M. Tomonaga, T. Izumi, K. Fukunaga, A. Sasada, A. Abudu, Y. Miyauchi, H. Akari, K. Iwai, and T. Uchiyama. 2006. Ubiquitination of APOBEC3 proteins by the Vif-Cullin5-ElonginB-ElonginC complex. Virology 344:263-266. [DOI] [PubMed] [Google Scholar]
- 59.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanley, B. J., E. S. Ehrlich, L. Short, Y. Yu, Z. Xiao, X. F. Yu, and Y. Xiong. 2008. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J. Virol. 82:8656-8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 63.Strebel, K., J. Luban, and K. T. Jeang. 2009. Human cellular restriction factors that target HIV-1 replication. BMC Med. 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian, C., X. Yu, W. Zhang, T. Wang, R. Xu, and X. F. Yu. 2006. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 80:3112-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vallanti, G., R. Lupo, M. Federico, F. Mavilio, and C. Bovolenta. 2005. T lymphocytes transduced with a lentiviral vector expressing F12-Vif are protected from HIV-1 infection in an APOBEC3G-independent manner. Mol. Ther. 12:697-706. [DOI] [PubMed] [Google Scholar]
- 66.Wang, H., A. Sakurai, B. Khamsri, T. Uchiyama, H. Gu, A. Adachi, and M. Fujita. 2005. Unique characteristics of HIV-1 Vif expression. Microbes Infect. 7:385-390. [DOI] [PubMed] [Google Scholar]
- 67.Willey, R. L., D. H. Smith, L. A. Lasky, T. S. Theodore, P. L. Earl, B. Moss, D. J. Capon, and M. A. Martin. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao, Z., E. Ehrlich, K. Luo, Y. Xiong, and X. F. Yu. 2007. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J. 21:217-222. [DOI] [PubMed] [Google Scholar]
- 69.Xiao, Z., E. Ehrlich, Y. Yu, K. Luo, T. Wang, C. Tian, and X. F. Yu. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349:290-299. [DOI] [PubMed] [Google Scholar]
- 70.Xiao, Z., Y. Xiong, W. Zhang, L. Tan, E. Ehrlich, D. Guo, and X. F. Yu. 2007. Characterization of a novel Cullin5 binding domain in HIV-1 Vif. J. Mol. Biol. 373:541-550. [DOI] [PubMed] [Google Scholar]
- 71.Yamashita, T., K. Kamada, K. Hatcho, A. Adachi, and M. Nomaguchi. 2008. Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes Infect. 10:1142-1149. [DOI] [PubMed] [Google Scholar]
- 72.Yang, B., L. Gao, L. Li, Z. Lu, X. Fan, C. A. Patel, R. J. Pomerantz, G. C. DuBois, and H. Zhang. 2003. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J. Biol. Chem. 278:6596-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang, S., Y. Sun, and H. Zhang. 2001. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J. Biol. Chem. 276:4889-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, Y., F. Guo, S. Cen, and L. Kleiman. 2007. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology 365:92-100. [DOI] [PubMed] [Google Scholar]
- 75.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 76.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang, W., G. Chen, A. M. Niewiadomska, R. Xu, and X. F. Yu. 2008. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PLoS One 3:e3963. [DOI] [PMC free article] [PubMed] [Google Scholar]