Abstract
The cytoplasmic domain of glycoprotein B (gB) from herpes simplex virus type 1 (HSV-1) is an important regulator of membrane fusion. C-terminal truncations of the cytoplasmic domain lead to either hyperfusion or fusion-null phenotypes. Currently, neither the structure of the cytoplasmic domain nor its mechanism of fusion regulation is known. Here we show, for the first time, that the full-length cytoplasmic domain of HSV-1 gB associates stably with lipid membranes, preferentially binding to membranes containing anionic head groups. This interaction involves a large increase in helical content. However, the truncated cytoplasmic domains associated with the hyperfusion phenotype show a small increase in helical structure and a diminished association with lipid membranes, whereas the one associated with the fusion-null phenotype shows no increase in helical structure and only a minimal association with lipid membranes. We hypothesize that stable binding to lipid membranes is an important part of the mechanism by which the cytoplasmic domain negatively regulates membrane fusion. Moreover, our experiments with truncated cytoplasmic domains point to two specific regions that are critical for membrane interactions. Taken together, our work provides several important new insights into the architecture of the cytoplasmic domain of HSV-1 gB and its interaction with lipid membranes.
Herpes simplex virus type 1 (HSV-1) is an enveloped, double-stranded DNA virus that causes diseases ranging from mild oral sores to fatal encephalitis. Viral entry, which is the first step in the infection process, is accomplished by the fusion of the viral envelope with the target cell membrane. Four viral surface glycoproteins, glycoprotein D (gD), gB, gH, and gL, are necessary for viral entry. gD specifically binds one of its cellular receptors (49). This binding event triggers membrane fusion, in which gB and the gH/gL complex are key participants. These four glycoproteins can also induce the fusion of cell membranes when coexpressed from plasmid vectors in the absence of any other viral components, as long as the gD receptor is expressed on the surface of target cells (5, 38, 53).
gB is highly conserved among different herpesviruses (22). HSV-1 gB is a 904-amino-acid membrane-anchored protein that consists of a secretory signal (residues 1 to 30), a large extraviral domain or ectodomain (residues 31 to 773), a transmembrane anchor (residues 774 to 795), and an intraviral or cytoplasmic domain (residues 796 to 904) (Fig. 1). Previously, we have determined the crystal structure of the HSV-1 gB ectodomain (22). gB is a class III viral fusion protein (2), presumably directly participating in fusion by bringing the viral and the host cell membranes together. Unlike other members of this class, i.e., glycoprotein G of vesicular stomatitis virus (48) and baculovirus gp64 (28), gB cannot function on its own and requires viral glycoproteins gD and gH/gL (21).
FIG. 1.
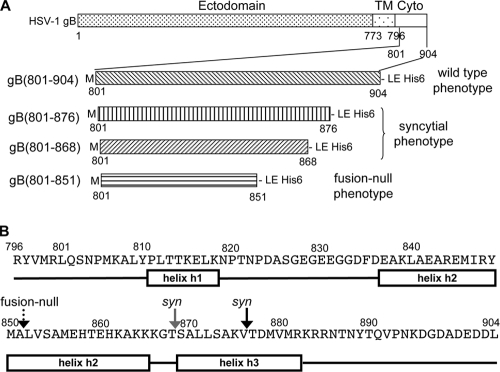
Different truncations of the HSV-1 gB cytoplasmic domain and their associated fusion phenotypes. (A) Schematic representation of HSV-1 gB, with marked domain boundaries (top). TM is the transmembrane region, and cyto is the cytoplasmic domain. Below, different gB cytoplasmic domain constructs used in the study. All recombinant proteins start with Met followed by L801 of the gB cytoplasmic domain. A His6 tag is fused to the C terminus of each protein. Associated fusion phenotypes of HSV-1 gB proteins are shown on the left. (B) Primary sequence and secondary structure prediction for the cytoplasmic domain of HSV-1 gB, residues 796 to 904. Secondary structure prediction was done using Jpred3 (9). Rectangular boxes represent helical regions, and black lines represent random coil regions. Arrows mark truncation sites in the constructs used in this work: black arrow, residue 876; gray arrow, residue 868; dotted arrow, residue 851.
Although HSV-infected cells normally do not fuse with uninfected neighboring cells (13), HSV mutants that induce rampant formation of syncytia (large multinucleated cells) occur with considerable frequency (47). These mutant viruses are referred to as syncytial or syn mutants. Interestingly, the first indication that gB participates in membrane fusion came from the observation that virus strains with point or deletion mutations within the cytoplasmic domain of gB display a syncytial phenotype (6). In addition to gB, viruses with syn phenotypes can contain mutations in gK (25), UL20 (36), or UL24 (27). Yet, the majority of the syn mutations occur within the 109-amino-acid cytoplasmic domain of gB (3, 6, 7, 11, 14, 18). These include point mutations, insertions, and C-terminal truncations. One of the most striking syn mutant viruses contains an amber mutation in codon 877 of gB, which causes gB to be truncated after residue 876 (3).
The effect of syn mutations in gB on its fusion activity can also be studied using cell-cell fusion assays. Transfection of effector cells with plasmids expressing HSV gD, gH, gL, and gB is sufficient to induce fusion with target cells expressing one of the HSV entry receptors (38, 45, 53). In such cell-cell fusion assays, transfection of the effector cells with a plasmid bearing a syn mutation in gB results in a hyperfusion phenotype (17). Thus, the same mutations that cause a syn phenotype in infected cells can cause a hyperfusion phenotype in transfected cells.
Perhaps the best-studied syn mutations of gB are the C-terminal truncations. In HSV-1 gB, truncations of 28 (gB876) or 36 (gB868) residues result in increased cell fusion (3, 17). In contrast, truncation of the gB cytoplasmic domain by 16 amino acids (gB888) has no effect on cell fusion (17). These observations point at a regulatory sequence within residues 877 to 888 of the C-terminal domain that negatively regulates, or restricts, membrane fusion. C-terminal truncations of the cytoplasmic domain of gB from HSV-2 by 25 to 49 residues result in a similar hyperfusion activity and syncytial phenotype (16).
Importantly, larger C-terminal truncations result in a protein that fails to function in either viral entry or cell fusion. For example, gB truncated after amino acid 851 fails to complement a gB-null virus (7). A truncation form lacking the last 49 amino acids (gB855) also fails to complement a gB-null virus (3). Thus, residues 856 to 888 appear to be critical for membrane fusion. Similar observations were made with HSV-2 gB. Truncation of 53 residues or more from the C terminus of HSV-2 gB resulted in a loss of fusion activity (16). These findings suggest that the cytoplasmic domains of gBs from closely related herpesviruses may share a conserved fusion regulation mechanism. Taken together, these data provide strong evidence that the cytoplasmic domain of gB plays an essential role both in regulating membrane fusion and in membrane fusion itself.
Although these mutants have been characterized in functional assays, the molecular mechanism by which the cytoplasmic domain of gB regulates membrane fusion is unknown. In a recent study, Park and colleagues (43) expressed a recombinant form of the full-length cytoplasmic domain of gB from Epstein-Barr virus (EBV), a herpesvirus. They showed that the cytoplasmic domain of EBV gB has a propensity to interact with lipid membranes. However, the cytoplasmic domains of the EBV and the HSV gB proteins have relatively low sequence identity, 17 to 18%; thus, whether the cytoplasmic domain of HSV gB has similar properties is yet unknown.
In this study, we sought to characterize the biochemical properties of the cytoplasmic domain of HSV-1 gB and its interactions with membranes. Moreover, we wanted to determine if the biochemical and biophysical properties of the C-terminally truncated mutants correlate with their functional phenotypes. To this end, we expressed and purified recombinant proteins corresponding to the full-length cytoplasmic domain of HSV-1 gB as well as the C-terminally truncated mutants associated with either the syncytial or the fusion-null phenotype. We used circular dichroism and gel filtration to characterize the secondary structure and the oligomeric state of the recombinant proteins and to examine their interaction with lipid membranes. We found that the cytoplasmic domain of gB is a trimer in solution and that residues important for the trimerization reside within the region from residue 801 to 851. Further, we found that the full-length cytoplasmic domain stably interacts with anionic lipid membranes with a concomitant increase in helical structure. In contrast, interactions of all truncated cytoplasmic domains with anionic lipids were diminished. Moreover, the truncated mutants corresponding to a syncytial phenotype, gB(801-876) and gB(801-868), displayed only a modest increase in helical structure in the presence of anionic lipids, whereas the truncated mutant corresponding to a fusion-null phenotype, gB(801-851), showed no such increase. Based on these observations, we propose a model whereby residues 869 to 882 form a helix in the presence of anionic lipid membranes, which accompanies a stable association between the cytoplasmic domain and the lipid membranes. We hypothesize that this event plays an important role in negative regulation of membrane fusion by the cytoplasmic domain.
MATERIALS AND METHODS
Reagents.
Cholesterol (Chol), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate monosodium salt (PA), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE) were purchased from Avanti Polar Lipids Inc. (AL). 1,6-Diphenyl-1,3,5-hexatriene (DPH) was purchased from Sigma Aldrich Corp. (St. Louis, MO). All other reagents used in the study are of analytical grade.
Cloning of the full-length and the truncated HSV-1 gB cytoplasmic domains.
Nucleotide sequences corresponding to HSV-1 (KOS) gB wild-type cytoplasmic domain (residues 801 to 904) or its C-terminal truncations, gB(801-876), gB(801-868), and gB(801-851), were cloned into an Escherichia coli expression vector, pET24b. Nucleotide sequences corresponding to the above-mentioned regions were amplified by PCR from the full-length HSV-1 (KOS) gB gene. The same forward primer 5′-CGCGCGCATATGCTGCAGAGCAACCCCATG-3′ (an NdeI site is underlined) was used for all constructs. The reverse primers were 5′-CGCGCGCTCGAGCAGGTCGTCCTCGTC-3′ for gB(801-904), 5′-GATCCTCGAGGACCTTGGCGCTGAGCAG-3′ for gB(801-876), 5′-CGCGCTCGAGCGTGCCCTTCTTCTT-3′ for gB(801-868), and 5′-CGCGCTCGAGGGCCATGTACCGTAT-3′ for gB(801-851), with an XhoI site underlined in each. Individual PCR products were cloned into the NdeI and XhoI restriction sites of pET24b vector (Novagen). The resulting constructs encode respective proteins with C-terminal His6 tags.
Expression and purification of the recombinant gB cytoplasmic domains.
The full-length cytoplasmic domain gB(801-904) and the truncated cytoplasmic domains gB(801-876), gB(801-868), and gB(801-851) were expressed in the Rosetta strain of E. coli (Novagen). Protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an optical density at 600 nm (OD600) of 0.6. Cells were harvested after a 3-hour induction and lysed by ultrasonication in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). Lysates were clarified by centrifugation at 18,000 × g. Cell lysis and all further purifications steps were done at 4°C. Clarified lysates were passed over an Ni-Sepharose 6B fast-flow column (GE Healthcare), and the proteins were eluted with 300 mM imidazole. The eluates were concentrated using Ultra-4 concentrators (Millipore) and further purified by size exclusion chromatography using a Superdex 200 column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM EDTA (TNE buffer). Purified proteins were concentrated and their purity assessed by SDS-PAGE and Coomassie blue G250 staining. Protein concentration was estimated from absorbance at 280 nm (42).
Preparation of lipid SUVs.
Small unilamellar lipid vesicles (SUVs) were prepared by ultra-sonication of hydrated lipid mixtures of different compositions (4). We used 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate monosodium salt (PA), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE), and cholesterol. The SUVs were composed of either pure PC, PC mixed with PA in 1:1 molar ratio (PC/PA), PC mixed with PE in 1:1 molar ratio (PC/PE), or PC mixed with cholesterol in 1:2 molar ratio (PC/Chol). To prepare SUVs, chloroform stocks of lipids were mixed in appropriate ratios in glass vials. Chloroform was then removed by drying under an N2 gas stream, and the lipid film was hydrated in TNE buffer at room temperature overnight with agitation. Hydrated lipid layers were then downsized by ultrasonication until suspension became clear. After sonication, large multilamellar vesicles and titanium debris from the sonication tip were removed from suspension by centrifugation at 18,000 × g for 20 min in a tabletop centrifuge. Lipid SUVs prepared by this method are generally 20 to 50 nm in diameter (30). Lipid SUVs were stored at 4°C and used within 1 week of preparation.
Interaction of gB cytoplasmic domains with membrane mimetics, studied by far-UV CD spectroscopy.
We used two types of membrane mimetics: detergent micelles of either anionic (sodium dodecyl sulfate [SDS]) or nonionic (Tween 20) nature or water-alcohol mixtures (water-ethanol [EtOH] or water-trifluoroethanol [TFE]). The proteins were incubated with detergents or with water-alcohol mixtures for 1 h at room temperature before far-UV circular dichroism (CD) spectra were recorded. To study the interactions of the gB cytoplasmic domain with lipid membranes, proteins were incubated with SUVs of different lipid composition at different protein-to-lipid molar ratios for 3 h at room temperature before recording far-UV CD spectra.
Far-UV CD spectra of gB cytoplasmic domain proteins in buffer alone or in the presence of different membrane mimetic solvents or lipid SUVs were recorded in 20 mM sodium phosphate buffer (pH 7.6) and 100 mM NaF at a final protein concentration of 0.2 mg/ml, at room temperature, using a Jasco 810 circular dichroism spectropolarimeter. The scan speed was set to 50 nm/min. Band pass and response time were set to 1 nm and 1 s, respectively. All spectra were averaged over three accumulations, blank corrected, and processed for noise elimination. Ellipticity values are reported as mean residue weight ellipticity, or θ. Initially, we estimated secondary structure from the CD spectra of the protein in buffer alone and in the presence of SDS, TFE, or ethanol using the program CONTINLL in the CDPro suite (http://lamar.colostate.edu/∼sreeram/CDPro/). However, CD spectra in the presence of lipid SUVs had a very low signal-to-noise ratio, below 200 nm, which resulted in unrealistic secondary structure estimates in CDPro. Hence, the helical content of the protein in the presence of liposomes was estimated from θ222 values using the formula % helix = [(θ222 − θ2220)/(θ222H − θ2220)] × 100%, as described previously (32), and using θ222H = −38,000° cm2 dmol−1 and θ2220 = 4,000° cm2 dmol−1 as θ222 values for 100% helix and 100% random coil, respectively (34, 37). Both methods gave similar estimates of helical content for gB(801-904) in buffer alone as well as in the presence of detergent micelles or alcohol-water mixtures, validating the use of either method for estimation of secondary structure in our experiments. Therefore, for consistency, secondary structure content under all tested conditions was reported from θ222 values.
Size exclusion chromatography.
The oligomeric state of the gB cytoplasmic domain proteins was analyzed by size exclusion chromatography on a Superose 12 10/30 HR column (GE Healthcare). The column was equilibrated with TNE buffer. Approximately 0.5 mg of each protein was loaded onto the column. Elution of proteins was monitored by absorbance at 280 nm. The column was calibrated with molecular mass standards catalase (232 kDa), bovine serum albumin (BSA) (66 kDa), ovalbumin (44 kDa), β-lactoglobulin (36 kDa), chymotrypsinogen (25 kDa), and lysozyme (14 kDa). The void volume of the column was determined using blue dextran.
Assay to study stable association of gB cytoplasmic domains with PC-PA SUVs.
To test binding of the gB cytoplasmic domain proteins to lipid SUVs, 0.3 mg of each protein were mixed with a 30 M excess of PC/PA SUVs, incubated at room temperature for 3 h, and loaded onto a Superose 12 10/30 HR column, at 4°C. Protein elution was monitored by absorbance at 280 nm, and fractions of 0.5 ml were collected throughout the run. PC/PA SUVs alone and protein alone served as controls. The elution volume of PC/PA SUVs was determined by adding 10 μM 1,6-diphenyl-l,3,5-hexatriene (DPH) to each fraction and recording fluorescence intensity in a fluorimeter with excitation and emission wavelengths set to 358/430 nm. DPH is a lipophilic dye that fluoresces intensely only when bound to lipids. Aliquots of equal volume from fractions in the void peak and the protein peak from each gel filtration run were analyzed by using SDS-PAGE and Coomassie blue G250 staining. The amount of protein that coeluted with PC/PA SUVs was determined by densitometry analysis of scanned gel images. For each protein, band intensities of protein that coeluted with SUVs were integrated and expressed as a percentage of the total integrated intensity of all bands, i.e., protein coeluted with SUVs plus unbound protein. Each experiment was done in triplicate, and the mean result was reported. All differences, except that between gB(801-876) and gB(801-868), are statistically significant, with P < 0.05.
RESULTS
Expression and purification of the full-length and the truncated cytoplasmic domains.
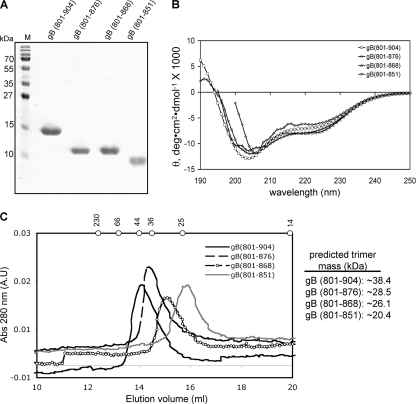
We expressed constructs gB(801-904), gB(801-876), gB(801-868), and gB(801-851) (Fig. 1A) with C-terminal His6 tags in E. coli. All proteins were purified to homogeneity by metal affinity and size exclusion chromatographies (Fig. 2A). We initially expressed the entire cytoplasmic domain of HSV-1 gB, residues 796 to 904. However, this protein was rapidly cleaved by a contaminating protease between residues R800 and L801, as determined by N-terminal sequencing and mass spectrometry of the proteolytic product. To avoid heterogeneity in protein samples due to proteolysis, we excluded the first five residues of the cytoplasmic domain from all constructs. N-terminal methionine is not cleaved off, and as a result, all proteins used in our studies start with methionine followed by residue L801 (Fig. 1A). In addition to gB(801-904), we expressed gB(801-876) and gB(801-868), which correspond to a syncytial, or hyperfusion, phenotype, and gB(801-851), which corresponds to a fusion-null phenotype (Fig. 1A).
FIG. 2.
Biochemical characterization of the full-length and the truncated cytoplasmic domains of HSV-1 gB. (A) SDS-PAGE of purified recombinant gB(801-904), gB(801-876), gB(801-868), and gB(801-851). M, molecular weight markers (labeled on the left). (B) Overlays of the far-UV CD spectra. Averages of three replicates are reported. θ is mean residue ellipticity. (C) Overlays of size exclusion chromatograms on a Superose 12 HR 10/30 column. Elution positions of molecular mass standards catalase (230 kDa), bovine serum albumin (66 kDa), ovalbumin (44 kDa), β-lactoglobulin (36 kDa), chymotrypsinogen (25 kDa), and lysozyme (14 kDa) are marked by dots on top. Predicted molecular masses of trimeric proteins are shown on the right.
Secondary structures of the full-length and the truncated cytoplasmic domains in aqueous buffer.
Secondary structure prediction algorithms (9) strongly predict three helices within the gB cytoplasmic domain (Fig. 1B). We refer to these putative helices as helix h1, helix h2, and helix h3, respectively. Previously, helices at similar locations within the cytoplasmic domain of gB were predicted (41, 44). We wished to determine whether the differences in the fusion phenotypes of gB proteins containing the full-length or the truncated cytoplasmic domains could be due to differences in the secondary structures of their respective cytoplasmic domains.
Circular dichroism (CD) is routinely used to estimate secondary structure content in proteins. Different types of regular secondary structure found in proteins give rise to characteristic CD spectra in the far-UV region, below 250 nm (29). For example, α-helices yield two prominent negative peaks, at 208 nm and 222 nm, and a positive peak at 192 nm; the intensity of α-helical CD signal increases with increasing helical content. In contrast, CD spectra of β-sheets typically show a single negative peak at 215 nm and a positive peak at 198 nm, whereas random coils give rise to a negative peak at 200 nm. The CD spectrum of a protein that contains different types of secondary structure, i.e., a typical protein, is essentially a linear combination of CD spectra of different types of secondary structure in the protein determined by their relative contents (50). A number of algorithms exist which use data from the far-UV CD spectra to estimate the secondary structure composition of a given protein. Most procedures employ basis data sets that contain CD spectra of several proteins with various folds, for which crystal structures are available (50). In addition, the helical content of a protein or a peptide can be estimated more straightforwardly, even if less accurately, by using the value of the CD signal at 222 nm (8, 32). CD provides only an estimate of the secondary structure content, however, and does not assign secondary structure to specific protein regions (29).
We first recorded far-UV CD spectra of the full-length cytoplasmic domain, gB(801-904) (Fig. 2B). The CD spectrum of gB(801-904) showed minima at ∼222 nm and ∼205 nm, consistent with the presence of α-helical structure in the cytoplasmic domain. A secondary structure estimate from the CD signal at 222 nm yielded 26.0% α-helix. One of the minima was observed at 205 nm rather than at 208 nm, which is characteristic of helical structure. This is because the helical content is relatively low but the random-coil content is high, so the minimum is shifted away from 208 nm toward the random coil minimum at 200 nm. Similar CD spectra were observed for the cytoplasmic domain of EBV gB (43). Although the cytoplasmic domains of EBV and HSV gB proteins have relatively low sequence identity, 17 to 18%, their secondary structures appear to be similar.
Next, we recorded the CD spectra for gB(801-876) and gB(801-868) and found that they were not significantly different from the CD spectrum of gB(801-904) (Fig. 2B). Spectra for gB(801-851) could be collected only at wavelengths above 200 nm due to a noisy CD signal below 200 nm. The CD signal is very sensitive to protein aggregation, and even very small amounts of aggregates can result in noise at below 200 nm. Over time, a small fraction of gB(801-851) protein aggregates; thus, the noisy CD signal could have resulted from the presence of very small amounts of aggregates in freshly purified protein. Nevertheless, above 205 nm, the gB(801-851) spectrum was similar to that of gB(801-904). Therefore, we conclude that the C-terminal truncations in gB associated with the syncytial and fusion-null phenotypes do not result in dramatic changes in the secondary structure, at least in aqueous solutions.
The full-length and the truncated cytoplasmic domains are trimeric.
We determined the oligomeric state of the full-length cytoplasmic domain of HSV-1 gB by size exclusion chromatography. The calculated molecular mass of the gB(801-904) polypeptide is 12.8 kDa, and so the calculated molecular mass of a trimer is 38.4 kDa. On the Superose 12 column, gB(801-904) elutes in the same elution volume as ovalbumin (44 kDa) (Fig. 2C). Thus, gB(801-904) has an apparent molecular mass of approximately 44 kDa, which is close to the expected molecular mass of a trimer. For comparison, the cytoplasmic domain of EBV gB is also a trimer in solution (43). We next tested the truncated cytoplasmic domains. The predicted molecular masses of trimeric gB(801-876), gB(801-868), and gB(801-851) are 28.5 kDa, 26.1 kDa, and 20.4 kDa, respectively. On the size exclusion column, the elution volumes of gB(801-876), gB(801-868), and gB(801-851) corresponded to progressively smaller apparent molecular masses (Fig. 2C). This trend agreed with the calculated molecular masses. Thus, the full-length and the truncated cytoplasmic domains are trimers in solution. We conclude that truncations do not disrupt the cytoplasmic domain trimer and that the syncytial and the fusion-null phenotypes must be due to some other changes in the physical properties of the cytoplasmic domain. Importantly, none of the gB(801-851) protein eluted in the void volume of the size exclusion column, which suggests that it does not aggregate to any appreciable extent.
The cytoplasmic domain changes its conformation in the presence of membrane mimetics.
Earlier, Park et al. reported that the cytoplasmic domain of EBV gB has a propensity to interact with membranes (43). We wanted to determine if the same is true for the cytoplasmic domain of HSV-1 gB, given the low sequence identity between the two cytoplasmic domains. Detergent micelles and water-alcohol mixtures are commonly used membrane mimetics for studying protein-membrane interactions (23, 26). We chose the anionic detergent sodium dodecyl sulfate (SDS), the nonionic detergent Tween 20, and two alcohols, ethanol and trifluoroethanol (TFE).
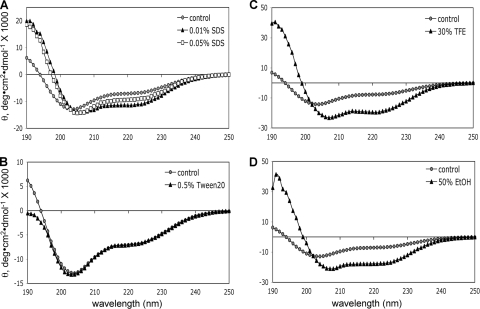
We recorded far-UV CD spectra of gB(801-904) in the presence of detergents or alcohols and compared them with the spectrum of gB(801-904) in aqueous buffer alone (Fig. 3). In buffer alone, gB(801-904) had 26.0% of helix as estimated from its CD spectrum (Fig. 3A; Table 1). In the presence of 0.01% SDS (below the critical micelle concentration [CMC]), however, helical content increased to 36.4% (Fig. 3A; Table 1). In the presence of a higher SDS concentration, 0.05% SDS (around the CMC), gB(801-904) showed a smaller increase in helical content, to 31.7%. Currently, it is unclear why monomeric SDS (at 0.01%) resulted in a higher increase in helicity than SDS micelles (0.05%). Perhaps this is due to the denaturing effect of SDS at high concentrations.
FIG. 3.
Far-UV CD spectra of gB(801-904) in the presence of detergent micelles and water-alcohol mixtures. (A) SDS; (B) Tween 20, (C) 30% TFE; (D) 50% EtOH. Control spectra (gray filled circles) in each panel are of protein in buffer alone.
TABLE 1.
Secondary structure estimates for gB(801-904) obtained from θ222 valuesa
| Additive | θ222, ° cm2 dmol−1 | % Helicityb | Absolute increase in helicity (%) |
|---|---|---|---|
| Protein alone (control) | −6,916.3 | 26.0 | |
| 0.01% SDS | −11,281.5 | 36.4 | 10.4 |
| 0.05% SDS | −9,312.5 | 31.7 | 5.7 |
| 0.5% Tween 20 | −6,921.8 | 26.0 | None |
| 30% TFE | −19,482.3 | 55.9 | 29.9 |
| 50% EtOH | −17,540.1 | 51.3 | 25.3 |
The CD spectra of gB(801-904) recorded in the presence of 0.5% nonionic detergent Tween 20 overlapped those recorded in buffer alone (Fig. 3B). Thus, the helical content of gB(801-904) was not affected by the presence of neutrally charged Tween 20 micelles. This suggests that the negative charge on SDS micelles may be important for inducing higher helicity in the cytoplasmic domain.
Alcohols stabilize helices in proteins by promoting intramolecular hydrogen bonds and lowering the dielectric constant of the solvent (26). In the presence of 30% TFE, just as in the presence of SDS, the molar ellipticity at 222 nm increased with a concomitant shift of the negative minimum from 205 nm to 208 nm (Fig. 3C). Both trends indicate an increase in helical content in TFE, which was estimated as 55.9% (Table 1). The same effect was observed in 50% EtOH (Fig. 3D), where the helical content increased to 51.3% (Table 1). In summary, gB(801-904) showed a significant increase in helical structure in a more hydrophobic environment, i.e., anionic detergent micelles or water-alcohol mixtures. These data suggest that the cytoplasmic domain of HSV-1 gB may interact with membranes, an interaction that increases its helical content.
The cytoplasmic domain changes its conformation in the presence of anionic lipid SUVs.
We further studied the interaction of the cytoplasmic domain with membranes using small unilamellar lipid vesicles (SUVs), a more realistic membrane model. We chose to use small instead of large unilamellar vesicles (LUVs) because SUVs are more compatible with spectroscopic studies (35). Lipid SUVs containing various amounts of PC (a neutrally charged lipid), PE (a neutrally charged lipid), cholesterol (a sterol), and PA (an anionic lipid) were used to evaluate the effect of lipid composition on membrane interaction. PC, cholesterol, and PE are relatively abundant in eukaryotic membranes (55). In addition, membrane sites with a high abundance of cholesterol, such as lipid rafts, are preferred sites of interaction for many viral fusion proteins (52). PA was chosen to test the effect of the negative charge of the head group.
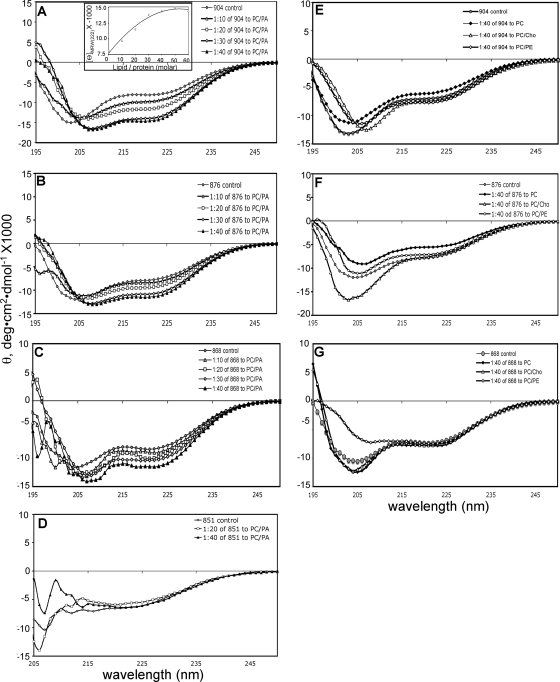
We recorded the CD spectra of gB(801-904) in the presence of increasing amounts of lipid SUVs of four different compositions (PC alone, PC/Chol, PC/PE, and PC/PA) (Fig. 4A and E). Large changes in the CD spectra of gB(801-904) were seen with increasing concentrations of PC/PA SUVs (Fig. 4A). Specifically, the molar ellipticity at 222 nm increased with the increasing lipid-to-protein ratio and reached a maximum at a molar ratio of 50:1 (Fig. 4A, inset). The helical content increased from 28.6% in buffer alone to 43.9% in the presence of a 40-fold molar excess of PC/PA (Table 2). This large change indicates that in the presence of PC/PA-containing SUVs, some disordered parts of the polypeptide chain fold into helices due to interaction between the protein and the lipid. In contrast, the CD spectra of gB(801-904) in the presence of PC, PC/Chol, or PC/PE were similar to the protein spectrum in buffer alone, even at a molar lipid-to-protein ratio of 40:1 (Fig. 4E). These lipids have a net neutral charge, and so we conclude that the cytoplasmic domain interacts only with anionic lipids. This conclusion is supported by our observation that gB(801-904) showed an increase in helical content in the presence of anionic but not nonionic detergent micelles (Fig. 3A and B).
FIG. 4.
Far-UV CD spectra of the full-length and the truncated cytoplasmic domains of HSV-1 gB in the presence of lipid SUVs. (A to D) Spectra recorded in the presence of increasing molar ratios of PC/PA SUVs to protein. (A) gB(801-904); (B) gB(801-876); (C) gB(801-868); (D) gB(801-851). (E to G) Spectra were recorded in the presence of a 40:1 molar ratio of PC, PC/PE, or PC/Chol SUVs to protein. (E) gB(801-904); (F) gB(801-876); (G) gB(801-868). Control spectra in each panel are of protein in buffer alone. The inset in panel A shows θ at 222 nm plotted against increasing molar ratios of PC/PA SUVs to protein.
TABLE 2.
Secondary structure estimates obtained from θ222 valuesa
| gB cytoplasmic domain | Ratio of protein to PC/PA | θ222, ° cm2 dmol−1 | % Helicityb | Absolute increase in helicity (%) |
|---|---|---|---|---|
| gB(801-904) | Protein alone (control) | −8,053.5 | 28.6 | |
| 1:10 | −9,691.7 | 32.6 | 4.0 | |
| 1:20 | −11,508.6 | 36.9 | 8.3 | |
| 1:30 | −13,849.7 | 42.5 | 13.9 | |
| 1:40 | −14,466.7 | 43.9 | 15.3 | |
| gB(801-876) | Protein alone (control) | −7,743.0 | 27.9 | |
| 1:10 | −8,336.2 | 29.4 | 2.5 | |
| 1:20 | −9,319.6 | 31.7 | 3.8 | |
| 1:30 | −10,751.7 | 35.1 | 7.2 | |
| 1:40 | −11,346.5 | 36.5 | 8.6 | |
| gB(801-868) | Protein alone (control) | −8,418.9 | 29.6 | |
| 1:10 | −9,077.9 | 31.1 | 1.5 | |
| 1:20 | −10,000.8 | 33.3 | 3.7 | |
| 1:30 | −10,363.4 | 34.1 | 4.5 | |
| 1:40 | −11,535.7 | 36.9 | 7.3 | |
| gB(801-851) | Protein alone (control) | −6,706.0 | 25.5 | |
| 1:20 | −5,713.7 | 23.1 | None | |
| 1:40 | −6,417.4 | 24.8 | None |
To summarize, the CD data demonstrate that the cytoplasmic domain of gB preferentially interacts with anionic lipid SUVs, which is accompanied by the formation of new helices. Our findings are consistent with the observations made with the cytoplasmic domain of EBV gB (43), where changes in CD spectra were observed in the presence of PC/PG anionic LUVs.
Truncated cytoplasmic domains show diminished conformational changes in the presence of lipid membranes.
We next investigated whether truncated cytoplasmic domains show a similar interaction with lipid SUVs. The CD spectra of gB(801-876) and gB(801-868) showed increased ellipticity in the presence of increasing concentrations of PC/PA SUVs (Fig. 4B and C), but the extent of this increase in both cases was lower than that observed for gB(801-904). At a molar lipid-to-protein ratio of 40:1, the helical content of gB(801-876) increased from 27.9% to 36.5% whereas the helical content of gB(801-868) increased from 29.6% to 36.9% (Table 2). The CD spectra of these two proteins in the presence of PC, PC/Chol, and PC/PE did not show any significant changes (Fig. 4F and G). We conclude that the removal of residues 877 to 904 in the cytoplasmic domain of gB diminishes its ability to interact with anionic lipid SUVs, probably because residues 877 to 904 are important for membrane interaction. The removal of additional residues 869 to 876 did not result in a further decrease in interactions.
Far-UV CD spectra of the gB(801-851) protein in the presence of lipid SUVs were very noisy in the 195- to 210-nm region, which could be due to aggregation. Over time, a small fraction of gB(801-851) aggregates in solution, as mentioned earlier. Although we do not know the aggregation mechanism, it is possible that truncation results in an exposure of a hydrophobic patch which is normally buried in the full-length cytoplasmic domain. This aggregation behavior of the protein is probably enhanced in the presence of lipid SUVs compared to protein alone in solution. Nevertheless, in the 215- to 260-nm range, gB(801-851) showed no appreciable change in its CD spectra in the presence of PC/PA SUVs (Fig. 4D; Table 2). Had the protein aggregated completely, the spectrum would have shown noise throughout the scanned wavelength range, and the signal would also have been significantly reduced. Therefore, we conclude that gB(801-851) does not change its secondary structure in the presence of anionic lipid SUVs. This further suggests that residues that undergo disorder-to-helical structure transition in the presence of anionic lipids may be located within the C-terminal region encompassing residues 852 to 904. Alternatively, residues within the region from position 801 to 851 can undergo a conformational change in the presence of lipids but only when downstream residues are present as well.
The full-length cytoplasmic domain stably associates with anionic lipid membranes.
Our next question was whether the cytoplasmic domain stably associates with anionic lipid SUVs, which we tested by using size exclusion chromatography. Due to their relatively large size, 20 to 50 nm, free SUVs should elute in the void volume of a Superose 12 column. In contrast, the cytoplasmic domain falls within the inclusion limit of the column and elutes much later than the void (Fig. 2C). Coelution of protein with SUVs would thus indicate a stable association.
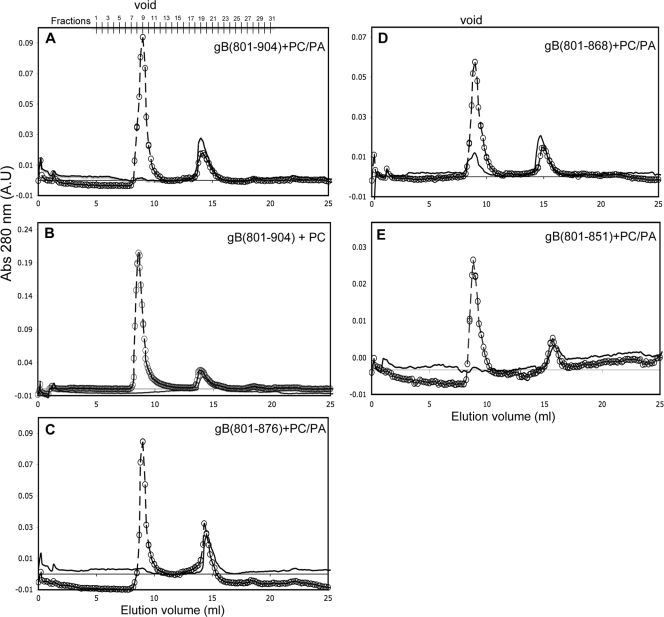
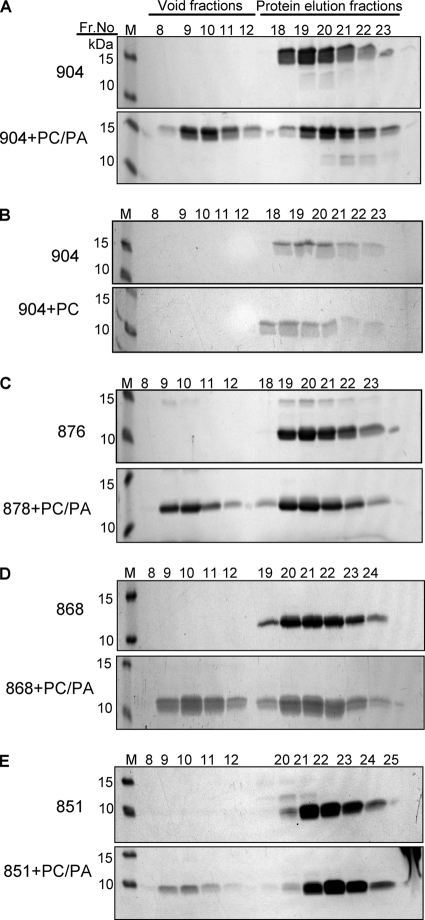
The full-length cytoplasmic domain, gB(801-904), was incubated with a 30-fold molar excess of PC/PA SUVs, and the mixture was analyzed on a Superose 12 column. In the absence of lipid SUVs, no protein eluted in the void volume, consistent with earlier experiments (Fig. 2C), and only the peak corresponding to free protein was seen (Fig. 5A). However, in the presence of a 30-fold molar excess of PC/PA SUVs, we observed a large void peak (Fig. 5A). This void peak, however, represents both lipid-bound protein and free SUVs, since the latter cause strong light scattering at 280 nm. We confirmed that SUVs elute in the void peak by probing the fractions corresponding to the peak with the lipophilic dye 1,6-diphenyl-1,3,5-hexatriene (DPH) (data not shown). To confirm the presence of protein in the void peak, fractions were analyzed by SDS-PAGE (Fig. 6A), and the amount of protein that coeluted with SUVs was determined by densitometry of scanned gel images. On average, 41.6% of gB(801-904) coeluted with PC/PA SUVs, which indicates that nearly half of gB(801-904) stably associates with PC/PA SUVs. The remaining free protein is probably in equilibrium with lipid-bound protein under the conditions of the experiment.
FIG. 5.
Association of the full-length and the truncated cytoplasmic domains of HSV-1 gB with SUVs. (A) gB(801-904) and PC/PA SUVs; (B) gB(801-904) and PC SUVs; (C) gB(801-876) and PC-PA SUVs; (D) gB(801-868) and PC-PA SUVs; (E) gB(801-851) and PC-PA SUVs. Protein in buffer alone (solid line) or a mixture of SUVs and protein at a 30:1 molar ratio (dashed line with open circles) was applied to the Superose 12 HR 10/30 column. For each protein, the resulting chromatograms of the protein alone and protein-lipid mixtures were overlaid to match the baselines.
FIG. 6.
Association of the full-length and the truncated cytoplasmic domains of HSV-1 gB with SUVs, analyzed by SDS-PAGE. (A) gB(801-904) and PC/PA SUVs; (B) gB(801-904) and PC SUVs; (C) gB(801-876) and PC/PA SUVs; (D) gB(801-868) and PC/PA SUVs; (E) gB(801-851) and PC/PA SUVs. Equal-volume aliquots from the respective fractions from the Superose 12 HR 10/30 column (Fig. 5) were analyzed by SDS-PAGE and stained with Coomassie blue G250. M stands for molecular weight markers, which are labeled on the left. Each gel is representative of three experiments.
Stable association was specific to PC/PA SUVs because in the presence of PC SUVs, no gB(801-904) was found in the void peak (Fig. 5B and 6B). Thus, the cytoplasmic domain stably associates only with anionic lipid membranes, a result that is consistent with our observation that gB(801-904) shows an increase in helical content in the presence of anionic but not neutrally charged lipid SUVs (Fig. 3A and B).
Truncated cytoplasmic domains have diminished binding to anionic lipid membranes.
We next tested the three truncated cytoplasmic domains in the liposome association experiment (Fig. 5C to E and Fig. 6C to E). Significantly less of gB(801-876) (34.2%) and gB(801-868) (30.5%) protein coeluted with PC/PA liposomes compared to gB(801-904) (Fig. 6C and D). Interestingly, only 12% of gB(801-851) bound to the PC/PA liposomes (Fig. 6E). Even this small amount may represent only the nonspecifically bound protein, because mixing of gB(801-851) with SUVs caused a small fraction of the lipids to precipitate. Thus, each of the truncations diminished association with anionic lipid membranes, and the largest truncation had the biggest effect. Our overall conclusion from the size exclusion experiments is that the cytoplasmic domain can stably associate with anionic liposomes, and C-terminal truncations diminish this association. This association was nearly eliminated for gB(801-851), and thus we conclude that residues 877 to 904 may be required for stable interaction with anionic liposomes while residues 852 to 868 may additionally contribute to this interaction. Nonetheless, we cannot exclude the possibility that residues within the region from position 801 to 851 can undergo a conformational change in the presence of lipids but require the presence of residues in the region from position 852 to 904.
DISCUSSION
The cytoplasmic domain of HSV-1 gB functions as an important regulator of cell membrane fusion in herpesviruses, but its structure and mechanism of action are still unknown. Previous work (43) showed that the full-length cytoplasmic domain of gB from a related EBV is trimeric in solution and can interact with anionic membranes and detergent micelles, with a concomitant increase in helical content. However, due to low sequence identity between the cytoplasmic domains of EBV and HSV gBs, we could not predict whether the latter would display the same properties as the former. Thus, our first goal was to determine the oligomeric state of the cytoplasmic domain of gB and its ability to interact with membranes. Moreover, we wanted to characterize the C-terminal truncation mutants of the gB cytoplasmic domain, so as to determine the biochemical basis of their hyperfusion or fusion-null phenotypes.
The data presented here provide several important new insights into the architecture of the cytoplasmic domain of HSV gB and its interaction with lipid membranes. For the first time, we show that the full-length cytoplasmic domain of HSV-1 gB interacts stably with lipid membranes, preferentially binding to membranes containing anionic head groups. This membrane interaction involves a dramatic increase in helical content. Moreover, our experiments with truncated cytoplasmic domains point to two specific regions, residues 852 to 868 and 877 to 904, which are critical for these changes to occur. Finally, we show that residues within the region from position 801 to 851 are important for trimerization.
Secondary structure and the oligomeric state of the cytoplasmic domain in solution.
Secondary structure algorithms (9) strongly predict three helices within the cytoplasmic domain of HSV-1 gB: helix h1, residues 811 to 818; helix h2, residues 837 to 865; and helix h3, residues 869 to 882 (Fig. 1). Previously, helices at similar locations within the cytoplasmic domain of gB were predicted (41, 44). The full-length cytoplasmic domain of gB, gB(801-904), has a characteristic helical signature in its CD spectrum when in aqueous buffer. However, the secondary structure content, between 26 and 28.6% as estimated from CD spectra, is below the expected helical content of 45% if all predicted helices actually form in aqueous solution. Further, the truncated cytoplasmic domain proteins gB(801-876), gB(801-868), and gB(801-851) have CD spectra similar to that of gB(801-904). Despite differences in polypeptide length, the helical content of the truncated gB cytoplasmic domain proteins is similar to that of the full-length domain. This suggests that some of the predicted helices do not fully form in aqueous solution.
We also determined that the full-length and the truncated gB cytoplasmic domains gB(801-876), gB(801-868), and gB(801-851) are trimers in solution. Thus, residues important for trimerization reside within the region from position 801 to 851.
The cytoplasmic domain of HSV-1 gB interacts with lipid membranes.
gB(801-904) stably bound anionic liposomes, coeluting with the liposomes in a size exclusion experiment. Moreover, the helical content of the full-length cytoplasmic domain, gB(801-904), increased from 28.6% to 43.9% in the presence of anionic lipid SUVs, which is close to the predicted helical content value for the cytoplasmic domain. It has been previously observed that disordered segments in proteins adopt a regular secondary structure upon binding to their biological targets (12, 54). Accordingly, we found a disorder-to-order change when the cytoplasmic domain was incubated with anionic lipid membranes. Our data suggest that this conformational change is brought about by a stable interaction between the protein and the lipid membranes.
The biochemical properties of gB(801-876) and gB(801-868) in our assays were similar. Both retained some ability to associate with anionic liposomes (less than the full length), and showed a small increase in helical content. In contrast, the shortest form, gB(801-851), did not associate with liposomes to an appreciable degree and did not show any increase in helical content in their presence. These results suggest that residues 877 to 904 are required for stable interaction with lipid membranes, while residues 852 to 868 additionally contribute to this interaction.
Based on these results, we propose that in the presence of lipid membranes, residues 869 to 882 undergo a disorder-to-helix transition, which is necessary for stable association of the cytoplasmic domain with membranes. When the predicted helix h3 is either shortened at its C terminus, as in gB(801-876), or deleted, as in gB(801-868), the ability of the cytoplasmic domain to associate with membranes is diminished. How can a shorter helix result in diminished lipid binding? In one possible explanation, in the absence of residues 877 to 882, the remainder of the predicted helix h3 is less stable. Alternatively, the C terminus of the predicted helix h3 is directly involved in interactions with liposomes. The last residue in this helix is R882, which is followed by the sequence K883-R884-R885. This stretch of four positively charged amino acids could be critical for association with anionic liposomes. Importantly, R882 and R884 are highly conserved among gB proteins of other herpesviruses, suggesting that they are functionally important.
The predicted helix h3 is not the only region within the cytoplasmic domain that becomes more ordered upon interaction with lipid membranes. Residues 852 to 868 within the C-terminal half of the predicted helix h2 also may undergo a disorder-to-order transition in the presence of anionic lipid membranes. gB(801-868), which lacks the predicted helix h3, showed a small increase in helicity when incubated with anionic lipid membranes. However, gB(801-851), which lacks both the predicted helix h3 and the C-terminal half of the predicted helix h2, did not show any increase in helicity. Thus, both predicted helices h2 and h3 become ordered upon interaction with lipid membranes and contribute to stable association of the cytoplasmic domain with lipid membranes.
We hypothesize that the interactions with lipid membranes mediated by residues from the predicted helix h3 may play an important role in regulating the fusion activity of gB. It is known that gB mutants truncated after residue 876 or 868, gB876 and gB868, have a hyperfusion phenotype in transfected cells (17). Moreover, HSV-1 mutant virus encoding gB876 has a syncytial phenotype in infected cells (3). In these hyperfusion mutants, the predicted helix h3 is either missing (gB868) or shortened (gB876). Therefore, the intact predicted helix h3 may be necessary for regulating the fusion activity of gB such that its removal or shortening leads to a hyperfusion phenotype. Along this line of reasoning, a helix-destabilizing mutation in the predicted helix h3 would be predicted to result in the hyperfusion phenotype. Indeed, gB containing a “helix-breaking” mutation, A874P, has been reported to have a mild hyperfusion phenotype in a cell-cell fusion assay (17).
The fusion-null phenotype displayed by gB truncated after residue 851 (gB851) is more difficult to explain. gB(801-851) shows no increase in helical structure in the presence of anionic lipid membranes and does not stably associate with them. Thus, we conclude that some weak interaction with membranes, such as seen for gB(801-868), may be necessary for fusion. Also, gB851 may lack regions that are important for proper folding. gB(801-851) is aggregation prone and slowly precipitates out of solution over time; thus, it may simply be somewhat unstable. Although gB(801-851) has a helical structure and is trimeric in solution, its structural integrity may be compromised, thereby affecting its function.
How does the cytoplasmic domain interact with the lipid membranes?
The precise mode of interaction with membranes is yet unknown. However, the predicted helix h3 is unlikely to span the lipid bilayer because it has low hydrophobicity and a suboptimal length, approximately 14 amino acid residues, whereas a transmembrane helix has, on average, 21 amino acid residues (15). The predicted helix h3 could be oriented parallel to the membrane surface, which has been observed in other membrane-associated proteins (e.g., viral glycoproteins) (58). However, this type of membrane interaction typically involves an amphipathic helix (10), and the predicted helix h3 is not amphipathic. As another possibility, the cytoplasmic domain could interact with the membrane in a manner reminiscent of the HIV matrix protein. This trimeric protein is involved in HIV assembly and associates with the inner leaflet of the viral envelope by means of a myristoyl group and several basic residues. The crystal structure of the matrix protein highlights a flat membrane-binding surface in which several large basic patches formed by exposed basic residues could cooperate with the myristoyl groups to anchor the protein to the acidic inner membrane of the virus (24, 46). Although the cytoplasmic domain of gB is not myristoylated, it does contain two conserved arginines, R882 and R884, and several other basic residues. These conserved and nonconserved basic residues in the gB cytoplasmic domain could be important for membrane association.
Possible mechanism of fusion regulation.
How interaction with lipids with concomitant ordering of helix h3 restricts membrane fusion is unknown. Cytoplasmic domains of several viral fusion proteins have been shown to inhibit membrane fusion. For example, F proteins from different isolates of the paramyxovirus simian virus 5 (SV5) have either short (20-amino-acid) or long (42-amino-acid) cytoplasmic domains. F proteins with short cytoplasmic domains cause extensive cell-cell fusion whereas F proteins with long cytoplasmic domains cause very limited cell fusion, if any. Thus, the C-terminal 22 amino acid residues in F proteins with long cytoplasmic domains are thought to function as a negative regulatory sequence. The F proteins are trimeric, and the C-terminal 22 amino acids are thought to stabilize the structure of the cytoplasmic domain through self-association within F trimers (57). Importantly, the addition of a triple-stranded coiled coil from an unrelated protein to the short cytoplasmic domain of F suppressed cell-cell fusion (56, 57). Thus, although the precise mechanism involved in regulation is unknown, stabilization of the structure of the cytoplasmic domain through intramolecular protein-protein interactions leads to suppression of cell-cell fusion.
As another example, in several retroviruses, e.g., Moloney murine leukemia virus (MLV), cleavage of 16 amino acids from the C terminus of the cytoplasmic domain of the Env protein upon particle formation is required for Env to become fusogenic (19). In this way, the 16-residue region, called the R peptide, inhibits this activity of Env until the virus has left the cell. Although the mechanism by which the R peptide inhibits fusion is unknown, it has been proposed that the uncleaved MLV Env cytoplasmic domain forms a trimer in which the R peptide forms a triple-stranded coiled coil that suppresses fusion by stabilizing Env in its fusion-inactive form (51).
The presumed mechanisms of inhibition in both examples appear to be relevant to the one used by the HSV-1 gB cytoplasmic domain. In all three cases, stabilization of the inactive conformation of a fusion protein through stabilization of its cytoplasmic domain appears to play a key role. However, the mechanisms used to achieve this common goal are somewhat different. In the cases of SV5 F and MLV Env, this stabilization of an inactive form is achieved through protein-protein interactions within the cytoplasmic domain, whereas in case of HSV-1 gB, it is achieved through protein-lipid interactions. Even then, we cannot exclude the possibility that protein-protein interactions, perhaps through formation of a triple-stranded coiled coil, are involved in fusion inhibition by HSV-1 gB.
Ultimately, in all of the cases discussed here, the mechanism of regulation is thought to involve transmission of a signal from the cytoplasmic domain to the ectodomain in what can be described as inside-out signaling. In SV5, several mutations in the ectodomain of F protein increased the extent of syncytia; this effect was suppressed in the presence of the long cytoplasmic domain (57). Also, the antibody reactivity of the ectodomain differed between the F proteins with long and short cytoplasmic domains (56). In MLV, removal of the R peptide affected antibody reactivity of the ectodomain (1) as well as isomerization of the intersubunit disulfide of the Env SU/TM complex (33). In HSV-1 gB, truncations in the cytoplasmic domain affected the antibody reactivity of the ectodomain (31). All of these data point to communication between the ectodomain and the cytoplasmic domain, probably through the transmembrane region.
In summary, based on our results, we propose that putative helix-forming regions in the C-terminal part of gB cytoplasmic domain, helices h2 and h3, become folded in the presence of lipid membranes and are necessary for stable association of the cytoplasmic domain with membranes (Fig. 7). Because we observed this effect only in the presence of anionic liposomes, we propose that the net negative charge of the lipid surface is an important determinant of this interaction. We hypothesize that the formation of the helices and the concomitant association of the cytoplasmic domain with membranes play an important part in the mechanism of fusion regulation. In the absence of the predicted helix h3, the gB cytoplasmic domain does not interact strongly with membranes, and this leads to unregulated fusion, i.e., the hyperfusion phenotype (Fig. 7). When both the predicted helix h3 and the C terminus of the predicted helix h2 are absent, the interaction between the cytoplasmic domain and membranes is lost, leading to a fusion-null phenotype. Thus, a specific and stable association between the cytoplasmic domain of gB and membranes may be required for fusion.
FIG. 7.
Proposed model of gB fusion activity regulation by the cytoplasmic domain.
Although the cytoplasmic domains of gB proteins from less related herpesviruses share very few conserved residues, they may serve conserved functions. For example, truncations in the cytoplasmic domain of gB from a closely related HSV-2 have very similar phenotypes (16, 39). Further, truncations in the cytoplasmic domains of two other, less related, herpesviruses, Epstein-Barr virus and pseudorabies virus, result in hyperfusion and fusion-null phenotypes (20, 40). Sites of these truncations are located at positions equivalent to residues 868 and 851 of HSV-1. Cytoplasmic domains of these and other herpesvirus gB proteins are also predicted to contain helices. Thus, our proposed regulatory mechanism could also apply to other herpesviruses. To conclude, our observations, for the first time, provide a biochemical rationale for the syncytial and fusion null phenotypes observed with gB mutants containing cytoplasmic domain truncations.
Acknowledgments
We thank Mike Berne at the Tufts University Core Facility for N-terminal protein sequencing, David King at the University of California at Berkeley for mass spectrometry, and Elisa Saldaña Prieto for help with cloning and protein purification. We are also grateful to Roselyn Eisenberg and Gary Cohen at the University of Pennsylvania for critical readings of the manuscript.
This work was supported by Public Health Service grant 1DP20D001996 from the National Institute of General Medicine and by the Pew Scholars Program in Biomedical Sciences (E.E.H.).
Footnotes
Published ahead of print on 3 March 2010.
REFERENCES
- 1.Aguilar, H. C., W. F. Anderson, and P. M. Cannon. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R Peptide. J. Virol. 77:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backovic, M., and T. S. Jardetzky. 2009. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baghian, A., L. Huang, S. Newman, S. Jayachandra, and K. G. Kousoulas. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barenholz, Y., D. Gibbes, B. J. Litman, J. Goll, T. E. Thompson, and R. D. Carlson. 1977. A simple method for the preparation of homogeneous phospholipid vesicles. Biochemistry 16:2806-2810. [DOI] [PubMed] [Google Scholar]
- 5.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 6.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 7.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, C. T., C. S. Wu, and J. T. Yang. 1978. Circular dichroic analysis of protein conformation: inclusion of the beta-turns. Anal. Biochem. 91:13-31. [DOI] [PubMed] [Google Scholar]
- 9.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197-W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornell, R. B., and S. G. Taneva. 2006. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr. Protein Pept. Sci. 7:539-552. [DOI] [PubMed] [Google Scholar]
- 11.Diakidi-Kosta, A., G. Michailidou, G. Kontogounis, A. Sivropoulou, and M. Arsenakis. 2003. A single amino acid substitution in the cytoplasmic tail of the glycoprotein B of herpes simplex virus 1 affects both syncytium formation and binding to intracellular heparan sulfate. Virus Res. 93:99-108. [DOI] [PubMed] [Google Scholar]
- 12.Dyson, H. J., and P. E. Wright. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6:197-208. [DOI] [PubMed] [Google Scholar]
- 13.Ejercito, P. M., E. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 14.Engel, J. P., E. P. Boyer, and J. L. Goodman. 1993. Two novel single amino acid syncytial mutations in the carboxy terminus of glycoprotein B of herpes simplex virus type 1 confer a unique pathogenic phenotype. Virology 192:112-120. [DOI] [PubMed] [Google Scholar]
- 15.Engelman, D. M., T. A. Steitz, and A. Goldman. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15:321-353. [DOI] [PubMed] [Google Scholar]
- 16.Fan, Z., M. L. Grantham, M. S. Smith, E. S. Anderson, J. A. Cardelli, and M. I. Muggeridge. 2002. Truncation of herpes simplex virus type 2 glycoprotein B increases its cell surface expression and activity in cell-cell fusion, but these properties are unrelated. J. Virol. 76:9271-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, T. P., J. M. Melancon, and K. G. Kousoulas. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 18.Gage, P. J., M. Levine, and J. C. Glorioso. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol. 67:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, N., T. M. Shinnick, O. Witte, A. Ponticelli, J. G. Sutcliffe, and R. A. Lerner. 1981. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc. Natl. Acad. Sci. U. S. A. 78:6023-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 21.Heldwein, E. E., and C. Krummenacher. 2008. Entry of herpesviruses into mammalian cells. Cell Mol. Life Sci. 65:1653-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 23.Henry, G. D., and B. D. Sykes. 1994. Methods to study membrane protein structure in solution. Methods Enzymol. 239:515-535. [DOI] [PubMed] [Google Scholar]
- 24.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. U. S. A. 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ionescu, R. M., and C. R. Matthews. 1999. Folding under the influence. Nat. Struct. Biol. 6:304-307. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 28.Kadlec, J., S. Loureiro, N. G. Abrescia, D. I. Stuart, and I. M. Jones. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024-1030. [DOI] [PubMed] [Google Scholar]
- 29.Kelly, S. M., T. J. Jess, and N. C. Price. 2005. How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751:119-139. [DOI] [PubMed] [Google Scholar]
- 30.Lichtenberg, D., and Y. Barenholz. 1988. Liposomes: preparation, characterization, and preservation. Methods Biochem. Anal. 33:337-462. [DOI] [PubMed] [Google Scholar]
- 31.Lin, E., and P. G. Spear. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 104:13140-13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindberg, M., J. Jarvet, U. Langel, and A. Graslund. 2001. Secondary structure and position of the cell-penetrating peptide transportan in SDS micelles as determined by NMR. Biochemistry 40:3141-3149. [DOI] [PubMed] [Google Scholar]
- 33.Loving, R., K. Li, M. Wallin, M. Sjoberg, and H. Garoff. 2008. R-peptide cleavage potentiates fusion-controlling isomerization of the intersubunit disulfide in Moloney murine leukemia virus Env. J. Virol. 82:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning, M. C. 1989. Underlying assumptions in the estimation of secondary structure content in proteins by circular dichroism spectroscopy—a critical review. J. Pharm. Biomed. Anal. 7:1103-1119. [DOI] [PubMed] [Google Scholar]
- 35.Mao, D., and B. A. Wallace. 1984. Differential light scattering and absorption flattening optical effects are minimal in the circular dichroism spectra of small unilamellar vesicles. Biochemistry 23:2667-2673. [DOI] [PubMed] [Google Scholar]
- 36.Melancon, J. M., T. P. Foster, and K. G. Kousoulas. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 78:7329-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merutka, G., and E. Stellwagen. 1989. Analysis of peptides for helical prediction. Biochemistry 28:352-357. [DOI] [PubMed] [Google Scholar]
- 38.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. :2017-2027. [DOI] [PubMed]
- 39.Muggeridge, M. I., M. L. Grantham, and F. B. Johnson. 2004. Identification of syncytial mutations in a clinical isolate of herpes simplex virus 2. Virology 328:244-253. [DOI] [PubMed] [Google Scholar]
- 40.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norton, D. D., D. S. Dwyer, and M. I. Muggeridge. 1998. Use of a neural network secondary structure prediction to define targets for mutagenesis of herpes simplex virus glycoprotein B. Virus Res. 55:327-348. [DOI] [PubMed] [Google Scholar]
- 42.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park, S. J., M. D. Seo, S. K. Lee, and B. J. Lee. 2008. Membrane binding properties of EBV gp110 C-terminal domain; evidences for structural transition in the membrane environment. Virology 379:181-190. [DOI] [PubMed] [Google Scholar]
- 44.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 46.Rao, Z., A. S. Belyaev, E. Fry, P. Roy, I. M. Jones, and D. I. Stuart. 1995. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature 378:743-747. [DOI] [PubMed] [Google Scholar]
- 47.Read, G. S., S. Person, and P. M. Keller. 1980. Genetic studies of cell fusion induced by herpes simplex virus type 1. J. Virol. 35:105-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 49.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 50.Sreerama, N., and R. W. Woody. 2004. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 383:318-351. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, G. M., and D. A. Sanders. 2003. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology 312:295-305. [DOI] [PubMed] [Google Scholar]
- 52.Teissier, E., and E. I. Pecheur. 2007. Lipids as modulators of membrane fusion mediated by viral fusion proteins. Eur. Biophys. J. 36:887-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uversky, V. N. 2002. What does it mean to be natively unfolded? Eur. J. Biochem. 269:2-12. [DOI] [PubMed] [Google Scholar]
- 55.van Meer, G., D. R. Voelker, and G. W. Feigenson. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waning, D. L., C. J. Russell, T. S. Jardetzky, and R. A. Lamb. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl. Acad. Sci. U. S. A. 101:9217-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waning, D. L., A. P. Schmitt, G. P. Leser, and R. A. Lamb. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J. Virol. 76:9284-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]