Abstract
The cellular protease caspase-8 activates extrinsic apoptosis and also functions to promote monocyte-to-macrophage differentiation. Differentiation-induced alterations to antiviral caspase-8-dependent cell death pathways are unclear. Here, we show THP-1 monocyte-to-macrophage differentiation alters the specific cell death pathways activated in response to human cytomegalovirus (HCMV) infection. Employing viruses with mutations in UL36, the gene that encodes the viral inhibitor of caspase-8 activation (vICA), our data indicate that both caspase-dependent and -independent death pathways are activated in response to infection. Activation of caspase-dependent and -independent cell death responses restricted growth of vICA-deficient viruses, and vICA/pUL36 inhibited either response. Thus, these studies also reveal that the UL36 gene controls a caspase-independent cell death pathway. The impact of caspases on control of antiviral responses differed at early and late stages of macrophage differentiation. Early in differentiation, vICA-deficient virus-induced cell death was dependent on caspases and inhibited by the pan-caspase inhibitor z-VAD(OMe)-fluoromethyl ketone. In contrast, virus-induced death at late times of differentiation was caspase independent. Additional unlabeled and fluorescent inhibitors indicated that caspase-8 promoted death from within infected cells at early but not late stages of differentiation. These data highlight the multifunctional role of vICA/pUL36 as HCMV encounters various antiviral responses during macrophage differentiation.
Based on the frequency of infected cells in patients with disseminated disease, monocytes and monocyte-derived macrophages are important to the pathogenesis of human cytomegalovirus (HCMV) (8, 24, 32). Further, peripheral blood monocytes and granulocyte-macrophage progenitors are suggested to be latent reservoirs as healthy seropositive individuals carry HCMV DNA in these cells, a result consistent with the maintenance of latent genomes within experimentally infected progenitors (6, 37, 38, 58, 72, 76, 79, 80). Infected monocytes do not produce new virus; however, replication follows monocyte-macrophage differentiation whether induced by growth or proinflammatory cytokines or allogeneic stimulation. Evidence suggests that monocyte infections promote differentiation to a permissive, proinflammatory macrophage (14, 74). Thus, the production of new progeny is highly dependent on cellular differentiation. Monocytes, cells undergoing macrophage differentiation, and mature macrophages differ in the expression patterns of specific genes, including those important to cell death control (44, 49). Whether any change in expression of cell death control genes influences cell death pathways activated by HCMV infection is unclear. The current study addresses this question by evaluating the requirement for one viral cell death suppressor during productive infection of monocytes at different stages of macrophage differentiation.
HCMV and the related murine cytomegalovirus (MCMV) encode several cell death suppressor proteins (50, 52). Studies with MCMV indicate that at least four genes, M36, M45, m38.5, and m41.1, are critical to prevent infection-induced cell death in macrophages (10, 11, 47, 57). The roles of these genes during infection of differentiating cells are unclear. CMVs have coevolved with the host and are species restricted. These viruses are colinear, share a number of sequence-similar genes, and encode many unique genes (9, 67). Current nomenclature employs “M” to indicate MCMV open reading frames (ORFs) with sequence similarity to an HCMV ORF or “m” for unique genes to reflect these properties. As a functional class, the cell death suppressor proteins provide examples of conserved functions encoded by CMV-unique genes as well as divergent functions of CMV-common genes. Thus, m38.5 provides the function of the unrelated HCMV UL37 exon 1 (UL37x1) (9, 51) while, on the other hand, UL45 lacks the M45-encoded protein-protein interaction domain required to inhibit cell death signaling of receptor-interacting proteins (RIP) (89, 90). Evidence thus far indicates that the CMV-common gene products M36 and UL36 prevent cell death by a conserved mechanism (51, 54, 57, 73), prompting the current evaluation of UL36 in differentiated macrophages.
M36-deficient viruses are severely debilitated in vivo and induce apoptosis of tissue macrophages (17, 18). In vitro studies indicated that M36-deficient virus replicates in fibroblasts or endothelial cells but not macrophage cell lines (57), results that may contribute to replication in some organs of the infected animal (18). Previous studies have established that the HCMV UL36 gene is dispensable for replication in fibroblasts (20, 64). None of these studies has evaluated the role of UL36 in macrophages or of UL36/M36 in monocytes differentiating to macrophages. Overall, these reports suggest that UL36 may be important to HCMV replication in differentiated macrophages. The specific mechanism of cell death suppression prompted our evaluation of UL36-deficient viruses in monocytes differentiating to macrophages.
Apoptosis is a well-characterized cell death pathway activated in response to cell-intrinsic and immune-regulated antiviral defenses and, consequently, the target of specific viral proteins and RNAs (4, 25, 50, 69). The caspase family of cellular cysteine proteases is activated in response to prodeath signals and is critical to the control of apoptosis (15, 21, 29). Caspase-8, also known as Fas-associating protein with death domain (FADD)-like interleukin-1β-converting enzyme, or FLICE, is required for initiation of apoptosis in response to extrinsic, immune-regulated death factors including Fas or tumor necrosis factor (TNF) that induce formation of a multiprotein complex, the death-inducing signaling complex (DISC). Protein-protein interaction domains known as death effector domains (DED) in the prodomain of procaspase-8 and in the adaptor protein FADD are required for FADD recruitment of the proenzyme to the DISC. Recruitment promotes dimerization of caspase-8, required for proteolytic activity, and autoproteolytic processing that increases the stability of the dimer (66). Caspase-8-dependent activation of the effector caspases follows these events either directly or as the result of mitochondrial alterations. The UL36 gene function viral inhibitor of caspase-8 activation (vICA), binds the prodomain of procaspase-8 to control proenzyme function and cell death (73). Unlike cellular and viral FLICE-inhibitory proteins, c- or v-FLIPs, respectively, that also function to prevent procaspase-8 activation, vICA/pUL36 does not include an apparent DED to mediate procaspase-8 binding. M36-deficient virus infection of differentiated macrophages activates caspase-8, and either dominant negative FADD or the pan-caspase inhibitor z-VAD(OMe)-fluoromethyl ketone (zVAD-fmk) restores growth, indicating that apoptosis limits replication (17, 18, 57).
In addition to extrinsic apoptosis, caspase-8 also controls monocyte-to-macrophage differentiation and other cellular processes (19, 23, 33, 40, 41, 63, 68, 71, 75). Mechanisms that regulate these different roles are incompletely understood but may include both modifications of the enzyme and functions of antiapoptotic proteins. Thus, mutations that prevent autoproteolysis of procaspase-8 inhibit extrinsic apoptosis but do not prevent macrophage differentiation (34). Further, macrophages have increased levels of c-FLIP proteins relative to monocytes that may function to increase resistance to extrinsic apoptosis (65). Whether monocytes, monocytes differentiating to macrophages, and monocyte-derived macrophages have similar or different caspase-8-dependent, prodeath responses to HCMV is unclear. Here, we report that HCMV requires vICA/pUL36 function for replication in THP-1 monocytes differentiating to macrophages and in macrophages. Our data suggest that cell death pathways that become activated by HCMV infection are altered as monocytes differentiate to macrophages. Further, the roles of vICA/pUL36 and caspases differ depending on the stage of THP-1 monocyte-to-macrophage differentiation. Cells infected early but not late in differentiation activated caspases in response to infection while vICA/pUL36 was required for replication with or without caspase activation. Thus, HCMV infection of THP-1 macrophages yielded unexpected results based on previous studies of MCMV-induced macrophage death, and, importantly, the data indicate that vICA/pUL36 controls both caspase-dependent and -independent cell deaths.
MATERIALS AND METHODS
Cells, viruses, and BACmid clones.
Human fibroblasts (HFs) and 293T cells were cultured as previously described (55). THP-1 cells, obtained from Patricia Kowalczyk, Emory University, were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Norcross, GA). To induce differentiation, cells were treated with 20 nM 12-O-tetradecanoylphorbol 13-acetate (TPA) and 50 μM hydrocortisone (HC) (both from Sigma, St. Louis, MO) as previously reported for HCMV infections (42, 88, 91). Isolation of ΔUL36, Towne-BAC (where BAC is bacterial artificial chromosome), and RC2940 strains and plaque purification of TownevarRIT3 from TownevarRIT were previously reported (20, 28, 51, 73).
Plasmids.
pUL36/vICA encoded by the laboratory strain AD169varATCC is inactive while an early-passage variant, AD169varDE, encodes intact pUL36/vICA (73). The plasmids pON2790 and pON2791 (gUL36-1) were derived by subcloning from AD169varATCC viral DNA or plasmid pZP8 derived from an AD169varDE cosmid (62) (kindly provided by David Anders, Wadsworth Center, New York State Department of Health) by digestion with EcoRI and HindIII and ligation to EcoRI-HindIII-digested pGEM3Zf+ DNA modified by collapse of sequence between SacI and SphI. Sequencing (Stanford University, Protein and Nucleic Acid [PAN] Facility) indicated that the UL36 gene in pON2790 and pON2791 differed by a single nucleotide, consistent with nonfunctional and functional vICA, respectively (73). The selection plasmid pON2792 was obtained from pON2791 (gUL36-1) by SnaBI digestion and ligation with HpaI-SmaI-digested DNA from plasmid EGFP-puro encoding enhanced green fluorescent protein (EGFP) fused to puromycin N-acetyltransferase (1) (kindly provided by Michael McVoy, Virginia Commonwealth University). The subclones gUL36-2 (pON2793) and gUL36-3 (pON2794) were each derived from gUL36-1 (pON2791) by PCR amplification with primers 5′-GCGAATTCGATATCAATAAAAATACCGTCGACGTGG-3′ and 5′-GCGAAGCTTTACGTACCACGGTACGGACGTTAATCGC-3′ or 5′-GCGAAGCTTTACGTACGTTGGGAAGAAACAAACGTCAC-3′, digestion with EcoRI and HindIII, and ligation to EcoRI-HindIII-restricted pGEM3Zf+. Expression plasmid UL36 (pON2795) was generated from gUL36-1 (pON2791) following PCR amplification with primers 5′-GGGAGATCTTAATTAAATGGACGACCTACGGGACACG-3′ and 5′-GTTCGTTAACTCGAGCGTTGTTCATGTAAACGTGTGG-3′, digestion with PacI and XhoI restriction enzymes, and ligation to LNCX-3myc (51) digested with the same restriction enzymes. Expression plasmid UL36mut (pON2796) was derived by PCR with the primers used to generate pON2795 and template pON2790, followed by PacI and XhoI digestion and ligation of gel-purified product to LNCX-3myc digested with the same enzymes. Expression plasmid UL36Stop (pON2797) was generated by PCR amplification of pON2795 with primers 5′-GCCCACGAAGGGCCCTTCAAGGGCGCTATGCGCGTTAACGGAGCTG-3′ and 5′-CTTATGGGACTTTCCTACTTGGCAGTACATCTACGTATTAGTCATCGC-3′, digestion with ApaI and SnaBI, and ligation to gel-purified ApaI- and SnaBI-digested plasmid pON2795. To derive the lentiviral vectors pON2798, pON2799, pON2800, and pON2801, DNA sequences encoding the UL36, UL36mut, UL36Stop, or DsRED (Discosoma sp. red fluorescent protein) protein were amplified by PCR from templates pON2795, pON2796, and pON2797 with primers 5′-CTACCCCTCTAGAATGGACGACCTACGGGACACG-3′ and 5′-GACGCGTGGATCCGATCAGTTGTTCATGTAAACG-3′ or from template pDsRed-Monomer-N1 (Clontech) with primers 5′-CTACCCCTCTAGAATGGACAACACCGAGGACGTCATC and 5′-GACGCGTGGATCCCTACTGGGAGCCGGAGTGGCGGGC. Restriction endonuclease products generated from the amplified sequences by XbaI and BamHI were ligated to the vector pLV-EF1α-MCS-IRES-puro (where MCS is multiple cloning site and IRES is internal ribosome entry site) (Biosettia, San Diego, California).
Recombinant viruses and analyses of viral genomes.
Plasmid pON2792 was used to generate the viruses RC2792 and RQ2792. For RQ2792, pON2792 was digested with ScaI to linearize the plasmid within the vector. For RC2792, pON2792 was digested with SacI to remove sequence required to repair UL36. Following transfection of digested plasmid DNA by calcium phosphate, HF cultures were infected with the plaque-purified isolate TownevarRIT3 (73) for 5 days. Fresh HF were infected with virus recovered from lysed cells and supernatant, and recombinants were selected by the addition of 4 μg/ml puromycin (Sigma, St. Louis, MO) to the culture medium. Puromycin-resistant recombinants were recovered from supernatant and plaque purified three times by identification of GFP-expressing virus.
DNA obtained from supernatant virions and purified as described elsewhere (55) was used to evaluate genomic structure of all viruses. To confirm replacement of UL36x2 with the kanamycin resistance (Kanr) cassette, DNA was amplified by PCR with primers 5′-CCGTGCTCGTCATCCTCTCC-3′ and 5′-GCTTCGTCGAGGGTCCGGGC-3′. Probe DNA for Southern analysis was obtained by gel purification of EcoRV-SnaBI-digested pON2791 followed by random-hexamer-primed Klenow (Roche Applied Science, Indianapolis, IN) fill-in reactions in the presence of [32P]dCTP (GE Healthcare, Piscataway, NJ). Conditions for Southern analysis of viral DNA have been reported previously (51). Restriction fragment length polymorphism analyses followed digestion with EcoRI, HindIII, BamHI, EcoRV, KpnI, XbaI, or PstI and agarose gel electrophoresis.
Viral yields, infected-cell numbers, and impact of protease inhibitors.
For growth curves, HFs or THP-1 cells were infected in triplicate cultures by ΔUL36, Towne-BAC, RC2792, or RQ2792. Input multiplicity was determined from GFP-positive (GFP+) plaques formed on HFs. Total virus included cell supernatant and sonicated infected cells processed and analyzed as previously described (51). Infected-cell numbers were assessed from GFP fluorescence by live-cell microscopy or by indirect immunofluorescence assays employing antibodies specific to viral antigens. To evaluate the impact of specific proteases on infected-cell viability and viral yield, inhibitors including the pan-caspase inhibitor zVAD-fmk, the caspase-8 inhibitor z-IE(OMe)TD(OMe)-fmk (zIETD-fmk), or cathepsin inhibitor z-FA-fmk (all from Calbiochem, La Jolla, CA) resuspended in dimethyl sulfoxide (DMSO; Sigma, St. Louis, MO) were added from 1 h (h) postinfection.
Antibodies, fluorescence reagents, immunodetection, flow cytometry, and imaging.
Monoclonal mouse antibodies (MAbs) used were to CMV immediate-early proteins IE1 and IE2 (MAb810; Chemicon, Temecula, CA), pUL36 (64) (kindly provided by Thomas Shenk, Princeton University), and β-actin (AC-74; Sigma, St. Louis, MO). Secondary antibodies were Texas Red-conjugated horse anti-mouse immunoglobulin G (IgG) or peroxidase-conjugated horse anti-mouse IgG (both from Vector, Burlingame, CA). For analyses of macrophage differentiation, adherent THP-1 cells, recovered by mechanical detachment, were stained with antibodies PacBlue-CD11b (clone ICRF44; BD Biosciences, San Jose, CA), allophycocyanin (APC)-CD14 (clone TuK4; Invitrogen, Carlsbad, CA), or the appropriate isotype controls and 7-aminoactinomycin D (7-AAD; BD Biosciences) prior to fixation with 2% formaldehyde. Data were collected by LSRII flow cytometer (BD Biosciences) and processed by FlowJo (TreeStar, Ashland, OR) software. Immunofluorescence assays (IFA), immunoblotting, and imaging were as previously described (53).
RNA analyses.
Total RNA was isolated from mock- or CMV-infected HF or THP-1 cells at 8 or 24 h postinfection (Qiagen RNAeasy). Five micrograms of purified RNA, separated by denaturing agarose gel electrophoresis and transferred to Bright Star nylon membrane (Ambion), was hybridized to strand-specific RNA probes prepared as previously described (54). Ethidium bromide staining was used to monitor transfer, and transcript sizes were predicted by migration relative to Millenium RNA markers (Ambion). Probes were generated by in vitro transcription of PCR products derived from plasmid pON2791 and primers TAATACGACTCACTATAGGGATGTCTCCAGTCTACGTGAATC and TTAACCCTCACTAAAGGGCTGGTGAGACTGCTGGGGGCCG or TAATACGACTCACTATAGGGTCGGTGCGGCCCACGCGTCAGC and TTAACCCTCACTAAAGGGGACCACGACCACCATCTGTACC that included promoter sequences for T7 or T3 polymerases (underlined) and viral sequences specific to UL37x1 and UL38, respectively, or from plasmid pON303G (59) with primers TAATACGACTCACTATAGGGGTGACATCCTCGCCCAGGCTGTC and TTAACCCTCACTAAAGGGCTGAGACTTGTTCCTCAGGTCCTG including the same promoter sequences (underlined) and viral sequences specific to UL122.
Cell viability assays.
For extrinsic cell death assays in HF, cultures were infected at a multiplicity of infection (MOI) of 3 for 24 h prior to addition of α-Fas antibody 7C11 (Beckman Coulter, Fullerton, CA) at 0.2 μg/ml or TNF-α at 10 ng/ml and cycloheximide (CH) at 10 μg/ml (both from Sigma, St. Louis, MO) to the culture medium for 20 h. Lipopolysaccharides (LPS) (Sigma, St. Louis, MO) resuspended in THP-1 culture medium were added to cells at 1 μg/ml for 96 h. Survival for either cell type was evaluated following 3.7% formaldehyde (Sigma, St. Louis, MO) fixation, reaction with Hoechst 33342 (Anaspec, San Jose, CA), and immunofluorescence analyses completed as previously described (55). Surviving cells remained attached to the coverslip and appeared nonapoptotic based on cell size, membrane morphology, and DNA staining. Dead HF detached from the monolayer. For transient complementation assays, HF cells were cotransfected with pDsRED-Monomer-N1 (DsRED; Clontech, Mountain View, CA) and plasmids UL36 and UL36mut or vector control (pcDNA3myc) by calcium phosphate as previously reported (55). At 24 h posttransfection, cells were infected, and extrinsic apoptosis was induced as described above. Infected THP-1 cells were evaluated by immunofluorescent detection of viral nuclear antigen with MAb810 or by GFP fluorescence. Caspase-8 labeling of live-cell cultures employed the Red-IETD-fmk reagent (Calbiochem, La Jolla, CA) according to the manufacturer's protocols. Percentages of Red-IETD-fmk-labeled cells that were GFP+ or IE2-GFP+ were determined from evaluation of >100 infected cells within multiple microscopic fields. Transient complementation in THP-1 cells was mediated by plasmids prepared as endotoxin free (Qiagen) and transfected using the Amaxa Nucleofector I Device, using Nucleofector Kit V reagents according to the manufacturer's protocols (Amaxa, Gaithersburg, MD). Following transfection, cultures were differentiated and rested for 24 h prior to infection. Lentiviral vector particles prepared as recommended (Biosettia) were used to transduce THP-1 cells at 7 days postdifferentiation. Transduced cells were rested 1 to 2 days prior to infection. Evaluations of GFP+, IE-positive (IE+), or Hoechst-positive (Hoechst+) cells were evaluated by live-cell microscopy or following fixation, as indicated in the text, and included assessment of multiple fields from multiple infected wells, and evaluations were repeated in multiple experiments. In some experiments, cell viability was determined by trypan blue exclusion from cells counted using a hemacytometer following recovery of adherent cells from the culture dish by trypsin and of nonadherent cells from the supernatant by centrifugation.
RESULTS
vICA-deficient virus replication and cell death suppressor expression in HF.
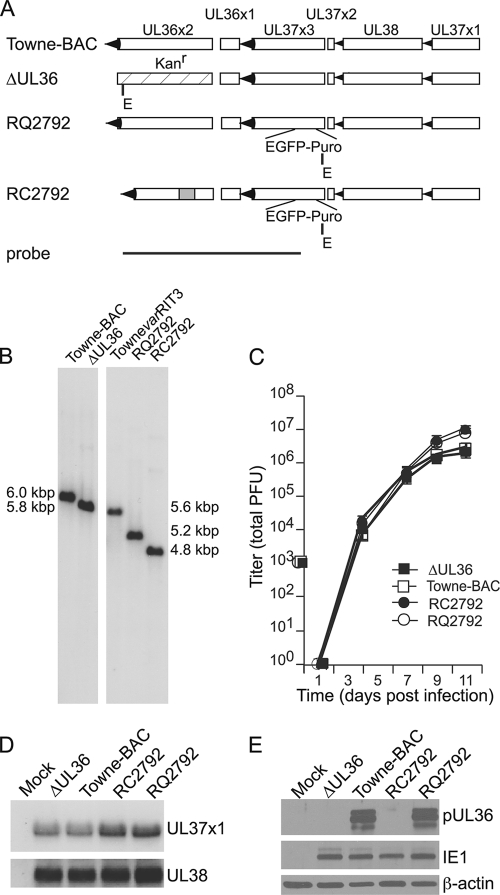
Previously, the antiapoptotic role of HCMV vICA was evaluated following infection with distantly related viruses carrying adventitious mutations in UL36 (73). To determine whether vICA-deficient viruses were replication defective in monocytes and macrophages, we began by confirming expectations from these early studies with directly related viruses. Specifically, we compared the properties of two vICA-deficient viruses, ΔUL36 and RC2792, to the viruses Towne-BAC and RQ2792 that encode native and rescued UL36 genes, respectively (Fig. 1 and Table 1). ΔUL36, derived from Towne-BAC, includes a selection cassette in place of UL36x2 and grows like the parent in fibroblasts (HF) (20) (Fig. 1A). RC2792 and RQ2792 were each derived from TownevarRIT3, a plaque-purified viral strain variant with an adventitious deletion impacting amino acids Leu156 to His295 of vICA/pUL36 (73). In previous analyses, TownevarATCC was more resistant to extrinsic apoptosis than TownevarRIT3 in vICA/pUL36-specific assays in HF (73), indicating the likelihood that Towne-BAC, derived from TownevarATCC (48), would also encode functional vICA. The deletion within TownevarRIT3 UL36 was repaired to generate the rescued virus RQ2792 while RC2792 was generated as the control vICA-deficient virus (Fig. 1 and Table 1). Viral DNAs were analyzed by hybridization to confirm genome structures (Fig. 1B). Digestion of Towne-BAC viral DNA with the restriction enzyme EcoRI produced a 6.0-kbp fragment that hybridized to UL36 sequence while the deletion in the TownevarRIT3 gene reduced the corresponding region to 5.6 kbp. Each selection cassette, Kanr and EGFP-puro, introduced a novel EcoRI site to the region. Thus, the UL36 region was 5.8 kbp for ΔUL36 and 4.8 kbp for RC2792. Repair of the UL36 gene in RQ2792 increased the size of the hybridizing fragment to 5.2 kbp. Additional restriction length polymorphism and DNA hybridization analyses also confirmed the expected structure of these viruses (data not shown).
FIG. 1.
Cell death suppressor expression and viral yields of vICA-deficient, Towne-BAC, and rescue viruses grown on HFs. (A) The UL36-UL37 regions of Towne-BAC and rescue virus RQ2792 and the vICA-deficient viruses ΔUL36 and RC2792. The map indicates exon sequences (open rectangles), transcript orientation (arrowheads), relative locations of the KanMX4 (stippled rectangle) (20) and EGFP-puro cassettes (1), the deletion within UL36x2 (73) (gray rectangle), and probe sequences used for DNA hybridization studies. Novel EcoRI (E) restriction sites introduced with the selection cassettes are indicated. (B) Hybridization analyses of viral DNAs digested with EcoRI. Novel restriction sites within the KanMX4 and EGFP-puro cassettes and the deletion within UL36x2 each increase the electrophoretic mobility of the EcoRI fragment. Images shown are from samples separated on the same gel. (C) Mean viral titers (± standard deviation) recovered from HF cultures infected at an MOI of 0.01. Error bars fall within the symbols at all points. (D) Hybridization results with strand-specific probes for viral transcripts encoding the cell death suppressors vMIA/pUL37x1 and pUL38. (E) Immunoblot analyses of pUL36/vICA expression in uninfected HFs (mock) or HFs infected as indicated for 24 h. Loading controls IE1 and β-actin indicate equivalent viral infection and cell numbers, respectively.
TABLE 1.
Experimental viruses
| Virus | Parent virus | vICA | Distal gene alteration(s) | Reference or source |
|---|---|---|---|---|
| Towne-BAC | TownevarATCC | + | US1-12 replaced by BAC, GFP | 48 |
| ΔUL36 | Towne-BAC | − | US1-12 replaced by BAC, GFP | 20 |
| TownevarRIT3 | TownevarRIT | − | Reduced UL37x3 expression | 73 |
| RQ2792 | TownevarRIT3 | + | EGFP-puro disrupting UL37x3 | This study |
| RC2792 | TownevarRIT3 | − | EGFP-puro disrupting UL37x3 | This study |
The UL36-UL37 locus includes multiple cell death suppressor genes (26, 51, 61, 73, 84). UL36 is not required for replication in HFs (20, 64), and we used HF infections to evaluate growth and cell death suppressor expression levels for all viruses. Multistep growth analyses of ΔUL36 and RC2792 indicated indistinguishable yields compared to those of Towne-BAC and the rescue RQ2792 viruses, respectively (Fig. 1C). These data are consistent with earlier reports of vICA-deficient viruses (20, 64, 73) and the conclusion that vICA is not required for replication in HFs. In contrast, Towne-BAC and ΔUL36 yields were reduced slightly (<1 log) compared to those of RQ2792, RC2792, and parental TownevarRIT3 by 11 days postinfection (Fig. 1C and data not shown). Previous studies of UL37x3-deficient virus or deletions of the distal genes US1-US12, replaced by BAC and GFP sequences in Towne-BAC, have failed to reveal any impact on replication (20, 36). In combination, these data indicate that other differences in the viral genomes of the Towne-BAC and TownevarRIT3 parental viruses distal to the UL36-UL37x3 locus may contribute to the differences in the yields of Towne-BAC and ΔUL36 in comparison to RQ2792 and RC2792. Overall, these results confirmed that the UL36 gene is not essential for replication in HFs, suggesting that caspase-8 is not activated or that other viral cell death suppressors prevent apoptosis.
Cell death suppressors near the UL36 locus were evaluated for expression from the vICA-deficient, Towne-BAC, and rescue viruses. The UL37x1 and UL38 transcript levels were evaluated by RNA hybridization (Fig. 1D). Unspliced and spliced transcripts including UL37x1, UL37x2, and UL37x3 are expressed with immediate-early kinetics (81-83). Evidence suggests that the proximity of RNA sequences required for splicing and polyadenylation and the abundance of cellular factors required for these events promote overproduction of the unspliced UL37x1 transcript relative to splice variants (78). Moreover, the cell death suppressor activity of the UL37 gene, viral mitochondrial inhibitor of apoptosis (vMIA), is encoded entirely within UL37x1 (26). The UL37x1 probe indicated apparently similar quantities of the unspliced transcript in ΔUL36- and RC2792-infected cell RNA compared to quantities in Towne-BAC and RQ2792 infections, respectively (Fig. 1D, top panel). The UL38 ORF is expressed from an early transcript initiated from a unique promoter (81-83). The UL38 probe indicated apparently similar quantities of this transcript in ΔUL36- and RC2792-infected cell RNAs compared with quantities in Towne-BAC- and RQ2792-infected cells, respectively (Fig. 1D, bottom panel). Expression of vICA/pUL36 was evaluated by immunoblotting (Fig. 1E). As expected, the ΔUL36 and RC2792 viruses failed to express and the rescue virus RQ2792 did express vICA/pUL36, a 52-kDA protein (64) (Fig. 1E). Thus, the vICA-deficient viruses grew as well as control viruses in permissive HFs and expressed nearby cell death suppressor genes.
vICA/pUL36 prevents extrinsic apoptosis in infected HFs.
To directly evaluate whether vICA/pUL36 controls caspase-8-dependent apoptosis in HFs, α-Fas antibody or TNF was added following synchronous infection with mutant and control viruses (Fig. 2). Compared to uninfected cells (Fig. 2M to O), HFs infected with either Towne-BAC (Fig. 2D to F) or rescue (Fig. 2J to L) viruses were more resistant to these proapoptotic stimuli. In contrast, ΔUL36- and RC2792-infected cells (Fig. 2A to C and G and H) were less resistant than control virus infections (Fig. 2D to F and J to L). To confirm that these differences were related to vICA/pUL36 specifically, apoptosis was introduced following transient expression of vICA/pUL36 and infection with vICA-deficient viruses (Fig. 2P and data not shown). For the vICA-deficient viruses, RC2792 (Fig. 2P) and ΔUL36 (data not shown), transient expression of vICA/pUL36 restored resistance to Fas (Fig. 2P) or TNF (data not shown). In contrast, UL36mut including an inactivating, site-specific mutation of Cys131 to Arg131 that prevents procaspase-8 binding (73) failed to increase infected-cell survival. In combination, these data confirm earlier predictions from functional studies in the absence of infection and results obtained with distantly related viruses (73). Further, efficient growth of vICA-deficient viruses in the absence of caspase activation and increased sensitivity to induced caspase-dependent apoptosis indicated that these viruses could highlight any change in caspase-8-dependent antiviral responses during monocyte-to-macrophage differentiation.
FIG. 2.
vICA/pUL36 prevents extrinsic apoptosis of infected HFs. (A to O) Examples of Hoechst fluorescence from infected cultures exposed to anti-Fas (α-Fas) or TNF and cycloheximide (MOI of 3). (P) Resistance to Fas-mediated apoptosis of RC2792-infected HFs mediated by transfection of a plasmid encoding UL36, UL36mut, or a control (empty vector) along with DsRED. Graphed are numbers of surviving transfected/infected cells (indicated by DsRED) along with standard deviations.
The UL36 gene is required for replication in THP-1 monocyte-macrophages.
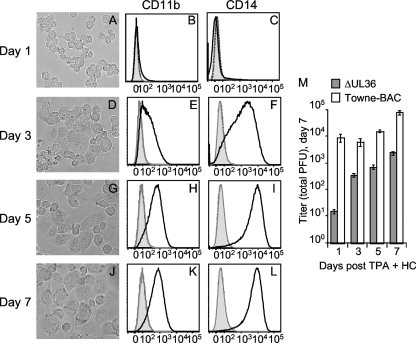
Differentiated THP-1 cells were employed to determine whether HCMV requires the UL36 gene for growth in monocyte-derived macrophages (Fig. 3). THP-1 monocytes differentiate into macrophage-like cells several days postaddition of phorbol ester through a process related to primary monocyte-to-macrophage differentiation (35, 44, 49, 85). Morphological and functional characteristics, expression of membrane antigens and receptors, and transcriptional analyses have all highlighted similarities to the differentiation pathway of primary cells (2, 35, 44, 49, 85, 86). When treated with the phorbol ester TPA and HC to optimize HCMV replication (42, 88, 91, 92), THP-1 cells became loosely adherent within the first 24 h and remained spherical (Fig. 3A). Morphological alterations of adherent cells were evident by 3 days and became most apparent by 5 to 7 days, when the majority of cells were flat and strongly adherent (Fig. 3D, G, and J). Increased cell size and granularity observed by microscopy were confirmed by flow cytometry (data not shown). These features were most evident in cultures differentiated for 7 days. Cell surface expression of the adhesion molecule CD11b and the LPS receptor CD14 increased by day 3, with peak levels reached by days 5 to 7 (Fig. 3B, C, E, F, H, I, K, and L). Similar results were obtained from cultures treated with TPA only but were not apparent in cultures treated by HC only (data not shown). Cultures treated with the pan-caspase inhibitor zVAD-fmk or the caspase-8-specific inhibitor, zIETD-fmk, adhered within 24 h but failed to progress to the late stages of differentiation, consistent with a caspase-dependent process (data not shown). Earlier studies have demonstrated THP-1 cells are permissive for HCMV replication within 24 h of treatment with TPA and HC (42, 88, 91, 92). Combined, these observations suggested that THP-1 cells could be used to evaluate responses to HCMV infection within monocytes at different stages of caspase-dependent differentiation to macrophages.
FIG. 3.
vICA-deficient virus replication is defective throughout monocyte-to-macrophage differentiation. (A to L) Phase-contrast images of THP-1 cells cultured in medium with TPA and HC for the indicated times (A, D, G, and J) are shown at a magnification of ×200. The relative cell surface expression levels of CD11b (B, E, H, and K) and CD14 (C, F, I, and L) of 7-AAD-negative (live) cells are shown as follows: fluorescence from unstained controls, dashed line; isotype controls, filled gray line; and CD11b or CD14, solid dark line (M) Mean viral titers (± standard deviation) recovered from THP-1 cultures cultured in medium with TPA and HC for the indicated times prior to infection for 7 days.
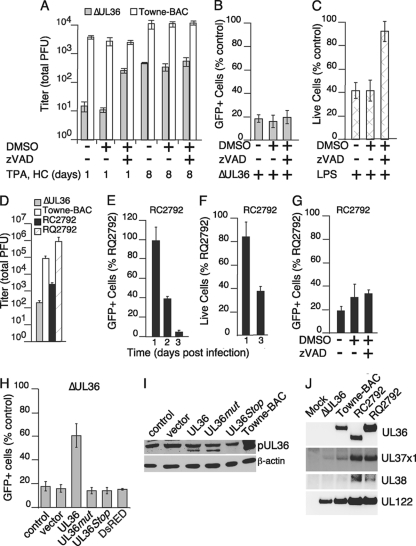
To evaluate the impact of vICA/pUL36 on HCMV replication in THP-1 cells, cultures were differentiated for 1, 3, 5, or 7 days prior to infection with ΔUL36 and Towne-BAC viruses for 7 days. Viral yields were subsequently determined on permissive HFs (Fig. 3E). Undifferentiated cultures did not produce virus (data not shown), as expected from previous studies with THP-1 cells (42, 88, 91, 92). In contrast, Towne-BAC was recovered from THP-1 cells differentiated for 1, 3, 5, or 7 days prior to infection (Fig. 3 M). vICA-deficient virus was also recovered; however, quantities were reduced relative to the control virus for each of these times. More specifically, quantities of ΔUL36 were reduced by 3 logs compared to the quantity of control virus when infection was initiated at 24 h postaddition of TPA and HC and by <2 logs at later times. Overall, these data are consistent with an important role for the UL36 gene in infections of monocytes differentiating to macrophages.
vICA/pUL36 sustains replication in THP-1 cells infected early in macrophage differentiation.
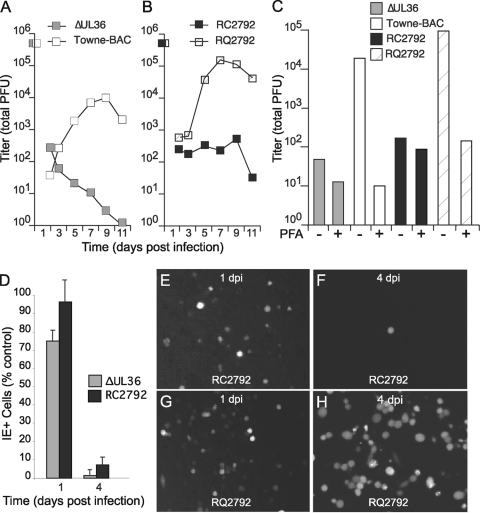
Initially, the role of vICA was evaluated in THP-1 cells infected within 24 h of differentiation since virus replication was apparently most dependent on the UL36 gene in cells at this early stage (Fig. 3M). In addition, previous work with MCMV characterized defects in fully differentiated macrophages rather than in cells undergoing differentiation (18, 57), making the outcome of these experiments of broader interest. To confirm the role of the UL36 gene in cells at early times postdifferentiation, THP-1 cells were treated with TPA and HC for 24 h and then infected with vICA-deficient, Towne-BAC, and rescue viruses (Fig. 4). For Towne-BAC, new virus was produced at 3 to 5 days postinfection, and peak levels were obtained at 7 to 9 days postinfection (Fig. 4A). In contrast, levels of virus recovered from cultures infected with either of the vICA-deficient viruses, ΔUL36 or RC2792, remained low throughout 11 days (Fig. 4A and B). The specific role of the UL36 gene was confirmed by infection with the rescue virus RQ2792. Similar to Towne-BAC, RQ2792 produced peak levels of new virus at 7 to 9 days postinfection. Differences in the peak levels of Towne-BAC and RQ2792 may reflect differences observed following infection in HFs that result from genes mapping outside the UL36 locus (Fig. 1). The viral DNA synthesis inhibitor phosphonoformate (PFA) was used to evaluate replication of all viruses. PFA reduced the levels of Towne-BAC and rescue viruses to those obtained from vICA-deficient viruses (Fig. 4C). In contrast, virus quantities recovered from cultures infected with ΔUL36 or RC2792 were similar in the presence or absence of PFA. These results support the conclusion that HCMV is highly dependent on UL36 for replication in THP-1 cells at early times postdifferentiation.
FIG. 4.
vICA-deficient viruses do not replicate within monocytes infected at early times of macrophage differentiation. (A and B) Mean viral titers recovered from THP-1 cells during a time course analysis. The results shown were obtained from an experiment done in parallel. (C) Viral titers recovered from cultures infected in duplicate for 7 days in the presence or absence of the viral DNA synthesis inhibitor PFA. Shown are the average values from an experiment that has been repeated. (D) IE+ cells in cultures infected with vICA-deficient viruses ΔUL36 and RC2792 expressed as the percentage of IE+ cells in cultures infected with the control viruses Towne-BAC and RQ2792, respectively. (E to H) Images of GFP+ fluorescence from infected THP-1 cells. Original magnification, ×100. For all experiments, an MOI of 3 was used following treatment with TPA and HC for 24 h.
To evaluate the replication defect of vICA-deficient viruses, numbers of infected cells that expressed immediate-early proteins IE1 and IE2 and the GFP indicator were compared in time course analyses (Fig. 4D and data not shown). Following infection with Towne-BAC or RQ2792, numbers of IE-positive cells increased between days 1 and 4 postinfection. However, numbers on day 1 postinfection (24 h) were only slightly lower than on day 4 (75 to 85% positive by day 1), indicating that the majority of infections were initiated by day 1. By day 4, ≥80% of IE+ cells were also GFP+, suggesting that the indicator was expressed in the majority of infected cells. In comparison, vICA-deficient viruses produced only slightly lower numbers of IE+ cells than vICA-expressing viruses on day 1 postinfection. In contrast, numbers of IE+ cells were greatly reduced compared to numbers in control vICA-positive (vICA+) infections by 4 days, consistent with the GFP+ cell numbers. These results suggest that vICA-deficient viruses initiated but failed to sustain infection through late times, consistent with decreased yields (Fig. 4A to C). These GFP expression results, obtained from fixed cultures, were compared to results in live-cell cultures (Fig. 4E to H and data not shown). RC2792- and RQ2792-infected cultures produced similar numbers of GFP+ cells by 24 h postinfection, as expected since an immediate-early promoter regulates the gene (Fig. 4E and G). By day 4, vICA-mutant virus-infected cultures had greatly reduced numbers of adherent GFP+ cells, and nonadherent GFP+ cells did not account for these differences (Fig. 4F and H and data not shown), consistent with viral yields (Fig. 4A to C). Towne-BAC did not reliably express GFP until 3 days postinfection, a result consistent with the simian virus early promoter and the impact of DNA replication on expression levels (48, 53). At late times of infection, ΔUL36-infected cell cultures included fewer GFP+ cells than Towne-BAC-infected cultures, demonstrating that the indicator could reveal common, vICA-dependent defects (data not shown). Thus, fixed or live cultures each gave results indicating severe replication defects in the vICA-deficient viruses. Altogether, these data the suggest that replication in monocytes differentiating to macrophages requires vICA/pUL36 to sustain infection.
vICA-deficient viruses induce premature death in THP-1 cells infected early in macrophage differentiation.
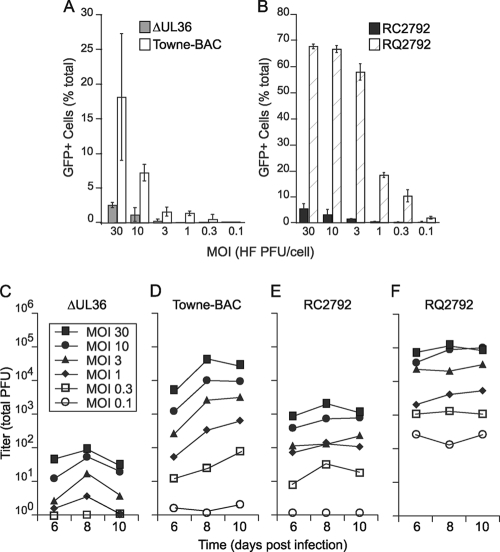
Declining infected-cell numbers suggested that premature cell death prevented completion of vICA-deficient virus replication. To evaluate cell death with population-based methods, it was first necessary to determine an appropriate multiplicity of infection since THP-1 cells were less permissive to infection than HFs. Input multiplicity was determined from HF infections because plaques did not emerge in THP-1 cultures. Both infected-cell numbers and viral yield increased with multiplicity for both Towne-BAC and RQ2792 (Fig. 5A, B, D, and F). These assays indicated that RQ2792 infections at an MOI of ≥10 were appropriate for population studies of cell death promoted by vICA-deficient virus. Specifically, 60 to 70% of the THP-1 culture was GFP+ by day 3 at MOIs of ≥10 following infection with RQ2792 (Fig. 5B). In contrast, Towne-BAC infections at an MOI of 30 were insufficient to infect the majority of cells (Fig. 5A), consistent with additional analyses of IE1 and IE2 expression (data not shown). Viral yields also indicated that RQ2792, but not Towne-BAC, reached a plateau between MOIs of 3.0 and 10.0 (Fig. 5D and F), confirming that THP-1 cells were more permissive to RQ2792 than to Towne-BAC infection. Thus, additional viral genes mapping outside the UL36 locus may contribute to infection of THP-1 cells if Towne-BAC and RQ2792 are compared directly, but THP-1 cells were permissive for either virus. For the vICA-deficient viruses, infection of THP-1 cells also increased with increased multiplicity (Fig. 5A and B). However, the quantity of recovered virus was 2 to 3 logs lower for vICA-deficient virus than for controls at all multiplicities (Fig. 5C and E). Thus, neither increasing nor decreasing multiplicity overcame the requirement for vICA, indicating that THP-1 cells could be infected at a high MOI to determine whether premature cell death contributed to the growth defect of vICA-deficient viruses.
FIG. 5.
The vICA-deficient virus replication defect in monocytes infected early in differentiation is multiplicity independent. (A and B) Infected cell number (GFP+) at 3 days postinfection graphed as the percentage of total cell number before infection (data are means ± standard deviations). (C to F) Viral titers were recovered on day 7 following infection at the indicated MOIs. For all experiments, THP-1 cells were treated with TPA and HC for 24 h prior to infection.
Infected cell viability was determined at days 1 to 4 postinfection (Fig. 6A). For cultures infected with vICA-expressing RQ2792, total viable cell numbers, including adherent and nonadherent populations, remained constant at days 1 to 4 (data not shown). In comparison, viability of cells infected with the vICA-deficient virus was greatly decreased by day 2 of infection, consistent with observations of viral and marker gene expression (Fig. 4 and 5). Nonadherent viable cells in cultures infected with the rescue virus were consistently <2% of the total on days 1 through 4 of infection. Over these same days, the vast majority of viable, vICA-deficient virus-infected cells also remained adherent. The nonadherent population ranged from 0.4% (day 1) to 6% (day 4), indicating that a minor population of viable cells detached. Overall, these data suggest that vICA-deficient virus replication is inhibited because infected cells die.
FIG. 6.
vICA-deficient viruses induce caspase-dependent death of monocytes infected at early times of macrophage differentiation. (A) Total live cell numbers determined by trypan blue exclusion in RC2792-infected cultures (MOI of 10) and expressed as the percentage of live-cell numbers in RQ2792 cultures. (B and C) Numbers of GFP+ cells and viral titers recovered (7 days postinfection) following infection of THP-1 cells and culture in the presence or absence of 25 μM zVAD-fmk or the solvent DMSO added from 1 h postinfection. (D) Titration of zVAD-fmk and the impact on numbers of GFP+ infected cells. (E) Numbers of GFP+ cells in the presence of cathepsin-specific inhibitor (z-FA-fmk), zVAD-fmk, or the caspase-8-specific inhibitor zIETD-fmk, all at 10 μM, or the solvent DMSO. For panels B, D, and E, GFP+ cell numbers were determined at 4 days postinfection from five microscopic fields. Data are means ± the standard deviations. For all experiments, THP-1 cells were treated with TPA and HC for 24 h prior to infection.
Premature cell death induced by vICA-deficient viruses early in macrophage differentiation is caspase dependent.
To evaluate the role of caspases in limiting vICA-deficient virus replication in THP-1 cells differentiating to macrophages, we employed broad-spectrum and specific inhibitors (Fig. 6B to E). The pan-caspase inhibitor zVAD-fmk increased the numbers of vICA-deficient virus-infected cells that survived to late times of infection (Fig. 6B). In contrast, Towne-BAC and rescue viruses were neither inhibited by nor benefited from addition of zVAD-fmk. Yields of vICA-deficient viruses were also increased by addition of zVAD-fmk (Fig. 6C). In contrast, replication of Towne-BAC and rescue viruses remained the same in the presence or absence of zVAD-fmk. In addition to caspases, cathepsins, calpains, and serine proteases can also impact cell death (7, 16, 43, 45). zVAD-fmk inhibits both caspases and cathepsins at concentrations as low as 50 μM (70). To confirm the specific role of caspases in CMV-induced death of THP-1 cells, various doses of zVAD-fmk and other inhibitors were evaluated. Concentrations of zVAD-fmk as low as 6 μM were sufficient to inhibit CMV-induced death (Fig. 6D) and increase replication (data not shown), suggesting that the restriction was specifically related to caspases. The potential contribution of cathepsins to infection-induced cell death was more directly evaluated by addition of the specific inhibitor, z-FA-fmk. This cathepsin-specific inhibitor failed to increase infected cell viability (Fig. 6E). In contrast, the caspase-8 specific inhibitor zIETD-fmk increased infected cell viability as well as the pan-caspase inhibitor zVAD-fmk (Fig. 6E), suggesting an important role for this caspase in death induced by vICA-deficient virus.
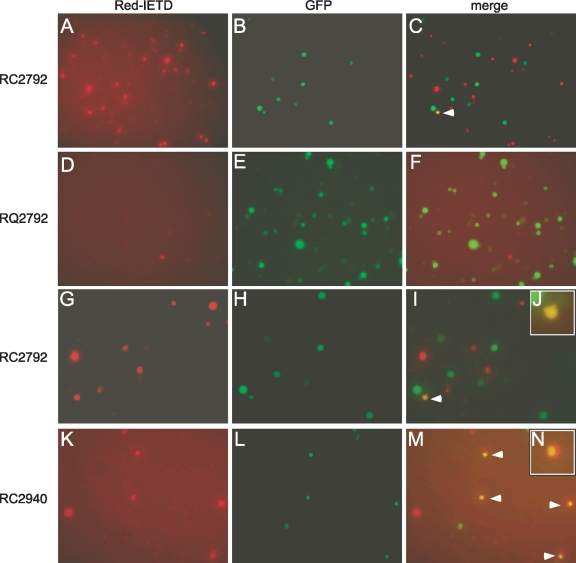
To determine whether caspase-8 was active within infected cells, cultures were labeled with the specific fluorescent inhibitor Red-IETD-fmk (Fig. 7). Cells that bound the inhibitor were more numerous within cultures infected with vICA-deficient viruses RC2792 (Fig. 7A) or ΔUL36 (data not shown) than within rescue (Fig. 7D) or Towne-BAC (data not shown) virus-infected cultures, suggesting increased levels of active caspase-8. However, GFP+ cells within the vICA-deficient virus-infected cultures were infrequently labeled (Fig. 7C) even when evaluated as early as 24 h postinfection when most cells remained viable (Fig. 4 and 6). The vICA-deficient virus RC2940 was employed to clarify these results. RC2940 was derived from TownevarRIT3 and expresses IE2-GFP, a fusion protein of GFP and the viral nuclear, immediate-early protein IE2 (28, 77). Similar to the other vICA-deficient viruses, RC2940-infected THP-1 cells died prior to virus production, and both infected-cell viability and replication were rescued by the addition of zVAD-fmk (data not shown). At 24 h postinfection, RC2940 expressed nuclear IE2-GFP, and when labeled with Red-IETD-fmk, the majority of cells (>60% in evaluations of >100 labeled cells) were IE2-GFP+ (Fig. 7K to N). These results indicate that the vICA-deficient virus RC2940 activates caspase-8, similar to RC2792 and ΔUL36. Localization of active caspase-8 and IE2-GFP+ within the same cell indicates that caspase-8 promotes death from within the infected cell. Consistent with this suggestion, numbers of cells labeled with Red-IETD-fmk directly correlated with the input multiplicity of ΔUL36, RC2792, or RC2940 at multiplicities of 0.3, 3.0, and 30, also suggesting that active caspase-8 resided within the infected cells with little or no contribution to the numbers from uninfected, bystander cells (Fig. 5 and data not shown). In combination, these data suggest that IE2-GFP was retained within the nucleus during cell death while diffusely localized GFP expressed by ΔUL36 and RC2792 was either insufficiently expressed prior to death or rapidly released from the cell following activation of caspase-8. Nonetheless, these data indicate that all vICA-deficient viruses activate caspase-8. Overall, these results suggest that vICA/pUL36 controls caspase-induced death to promote viral replication in THP-1 cells infected at early times of differentiation to macrophages.
FIG. 7.
vICA-deficient virus activates caspase-8 in monocytes early in differentiation to macrophages. Images from Red-IETD-fmk and GFP+ fluorescence from THP-1 cultures treated with TPA and HC for 24 h prior to infection for 24 h with RC2792 (A to C and G to J), RQ2792 (D to F), or the IE2-GFP-expressing, vICA-deficient virus RC2940 (K to N). Original magnification for all images, ×100. For panels G to N, images obtained at the original magnification were processed in Photoshop to select specific regions to highlight IE2-GFP.
The UL36 gene is sufficient to prevent caspase-dependent death in THP-1 cells early in macrophage differentiation.
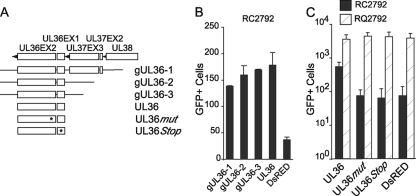
RNA analyses of ΔUL36 and RC2792 indicated that the genomic alterations within UL36 did not alter transcription of the nearby cell death suppressor genes UL37x1 and UL38 (Fig. 1). Nonetheless, the cell death suppressor region encodes multiple functions that could potentially contribute to replication defects. Therefore, transient complementation was used to confirm the specific contribution of the UL36 gene (Fig. 8). Initial evaluation included genomic clones of the intact UL36 gene plus upstream sequences (gUL36-1 to gUL36-3) in comparison to an expression clone that encoded the UL36 ORFs under the control of a heterologous promoter (Fig. 8A). In comparison to control transfections (DsRED), each of these clones increased cell viability (Fig. 8B) and replication (data not shown). None of these plasmids altered infected-cell viability of Towne-BAC virus (Fig. 8C and data not shown). Remaining differences in the numbers of infected cells comparing vICA-deficient and control virus after reintroduction of the UL36 gene may be due to transient gene expression, transfection efficiency, or contributions from nearby cell death suppressor genes. Nonetheless, these data indicate that the UL36 gene promoted infected-cell viability and replication. Next, mutations were introduced into the UL36 plasmid to evaluate the role of UL36-p52 generally and procaspase-8 binding specifically. The 478-amino-acid (aa) coding sequence of UL36 was altered to introduce translation termination codons after aa 49 (UL36Stop), and the role of procaspase-8 binding was evaluated by employing UL36mut (Cys131 to Arg131). Based on previous analyses of the inactive UL36 encoded by the laboratory strain AD169varATCC (54, 73) and analyses of transient complementation (Fig. 2P), this mutant protein cannot bind procaspase-8 and fails to prevent caspase-dependent death. UL36Stop and UL36mut were no different than DsRED when evaluated in infected-cell viability (Fig. 8C) or replication (data not shown) assays in THP-1 cells. Thus, replication in THP-1 cells early in differentiation to macrophages apparently requires an intact UL36 gene encoding a procaspase-8 binding protein, consistent with a role in controlling caspase-dependent death.
FIG. 8.
vICA/pUL36 inhibits HCMV-induced death of monocytes infected at early times of macrophage differentiation. (A) Representation of UL36-encoding plasmids used to complement growth of UL36-deficient virus in THP-1 cells. The asterisk indicates the relative location of introduced mutations. (B and C) Numbers of GFP+ cells on day 4 postinfection with RC2792 or RQ2792 of THP-1 cells transfected with UL36-encoding or control plasmids prior to treatment with TPA and HC for 24 h. Data are means ± standard deviations.
vICA/pUL36 prevents caspase-independent death of THP-1 cells infected late in macrophage differentiation.
Comparison of viral yields from THP-1 cells at multiple stages of differentiation suggested a slight reduction in cellular control of vICA-deficient virus with increased time of differentiation (Fig. 3). With results suggesting that caspase-dependent death controlled replication in cells infected at early times of differentiation (Fig. 4 to 7), inhibitors were reemployed to determine whether the apparent increase in vICA-deficient virus growth correlated with an alteration in the cellular control mechanism. In contrast to control cultures infected early during differentiation, the pan-caspase inhibitor zVAD-fmk failed to increase ΔUL36 replication in THP-1 cells late in differentiation (Fig. 9A). Despite this outcome, infected-cell numbers (GFP+) were reduced at late times of infection with ΔUL36 compared to infection with control viruses (Fig. 9B), consistent with a continued role for vICA/pUL36 in promoting cell survival. However, zVAD-fmk also failed to increase numbers of ΔUL36-infected cells at late stages of differentiation (Fig. 9B). In contrast, LPS-induced cell death (27) remained zVAD-fmk responsive at late stages of differentiation, indicating that caspase-dependent cell death pathways were intact (Fig. 9C). Combined, these data suggest that vICA-deficient virus induces a caspase-independent process that limits numbers of infected cells at late stages of differentiation.
FIG. 9.
vICA/pUL36 inhibits caspase-independent death of macrophages. (A) Viral titers recovered from THP-1 cells cultured in medium with TPA and HC for the indicated time prior to infection for 7 days in the presence or absence of zVAD-fmk or the solvent DMSO added from 1 h postinfection. Numbers of ΔUL36 virus-infected cells at day 4 postinfection are expressed as a percentage of control, Towne-BAC virus-infected cells (B); numbers of THP-1 cells that survived 4 days of treatment with LPS at 1 μg/ml are expressed as a percentage of the number of viable cells in control, untreated cultures (C). (D) Viral titers recovered from THP-1 cells infected for 7 days at an MOI of 3. (E to G) GFP+ cell numbers (E and G) or viable cells (F) at 3 days postinfection (G) or as indicated (E and F) at an MOI of 3 (E and G) or 30 (F). RC2792-infected cell numbers are graphed as the percentage of RQ2792-infected cells. Total live-cell numbers were determined as described in the legend of Fig. 6A. (H) Numbers of GFP+ cells at 5 days postinfection with ΔUL36, expressed as the percentage of Towne-BAC-infected cell numbers, following infection of differentiated THP-1 cells transduced by UL36-encoding or control lentiviral particles. (I) Immunoblot analyses of pUL36/vICA expression in transduced cells along with a loading control (β-actin). (J) Hybridization results with strand-specific probes for viral transcripts encoding the cell death suppressors pUL36, vMIA/pUL37x1, and pUL38, along with the loading and infection control UL122. Data in graphs are means ± standard deviations. For experiments in panels B to J, THP-1 cultures were grown in the presence of TPA and HC for 7 days prior to transduction, infection, or addition of LPS. For panels B, C, and G, zVAD-fmk or the solvent DMSO was added from 1 h of infection or LPS treatment.
RC2792 and RQ2792 were employed to address the specific contribution of vICA/pUL36 to replication in THP-1 cells late in macrophage differentiation (Fig. 9D to G). Similar to ΔUL36, the yield of the vICA-deficient virus RC2792 was reduced compared to control virus yield in cells at late stages of macrophage differentiation (Fig. 9D). In contrast, yields of the rescue virus RQ2792 were increased relative to all other viruses (Fig. 9D). Differences between RQ2792 and Towne-BAC virus in these cells reflect growth properties in HFs and THP-1 cells at early stages of macrophage differentiation and may result from genes outside the UL36 locus (Fig. 1, 4, and 5). Importantly, increased growth of the rescue RQ2792 virus compared to that of the vICA-deficient virus RC2792 confirmed an important role for the UL36 gene in late-stage cells.
To evaluate whether caspase-independent events also limited replication of RC2792 in THP-1 cells at late stages of differentiation to macrophages, gene expression patterns, cell viability, and inhibitor impact were each evaluated (Fig. 9E to G). A time course analysis indicated that GFP cell numbers were similar for RC2792 and the RQ2792 rescue virus on day 1 postinfection (Fig. 9E). In contrast, infected-cell numbers were reduced for the mutant virus compared to those for the rescue virus by day 2 and at later times of infection, consistent with reduced viral yields. Analyses of IE nuclear antigen following infection with either vICA-deficient, Towne-BAC, or rescue virus were consistent with these results (data not shown) and suggested that sustained replication required vICA expression. Infections with multiplicities over the range 0.3 to 30 did not overcome the vICA-deficient virus defect but did increase both infected-cell number and viral yield (data not shown). To evaluate cell death, multiplicity was therefore increased to ensure that the majority of cells (≥70%) in the culture were infected. Dye exclusion indicated that the viability of the vICA-deficient virus RC2792 was reduced by day 3 (Fig. 9F), a result consistent with reduced GFP+ (Fig. 9E) and IE+ cell numbers (data not shown) and viral yields (Fig. 9D). Importantly, zVAD-fmk failed to increase infected-cell numbers when added to cultures infected with RC2792 at late stages of differentiation (Fig. 9G) but restored viability of cells infected early in differentiation (Fig. 6 and data not shown). The inhibitor also failed to increase viral yield in cells at late stages of differentiation (data not shown). Consistent with these results, vICA-deficient virus-infected cells could be differentially labeled with the caspase-8-specific Red-IETD-fmk only within cultures infected early after differentiation (Fig. 7 and data not shown), suggesting that levels of caspase-8 activated by HCMV infection differ depending on the time of differentiation prior to infection. Each of these results was consistent with a role for the UL36 gene in controlling a caspase-independent process that promoted death of infected, differentiated THP-1 macrophages.
The vICA-deficient viruses ΔUL36 and RC2792 each indicated a role of vICA/pUL36 in the control of caspase-independent cell death. Therefore, transient complementation by transduction of differentiated cells was used to confirm the specific contribution of the UL36 gene (Fig. 9H and I and data not shown). Transduced vICA/pUL36 increased infected-cell survival in comparison to empty vector or DsRed controls (Fig. 9H). The quantities of vICA/pUL36 expressed following transduction were reduced in comparison to infection and may have contributed to incomplete complementation (Fig. 9I). Next, mutations that terminate UL36 translation (UL36Stop) or inhibit procaspase-8 binding (UL36mut) were employed. The UL36Stop vector failed to express UL36 (Fig. 9I) and failed to rescue infected-cell viability (Fig. 9H). UL36mut and UL36 proteins were expressed to similar levels in transduced cells (Fig. 9I); nonetheless, UL36, not UL36mut, increased infected-cell viability (Fig. 9H). These data indicate that the UL36 protein has a specific role in promoting survival following infection of differentiated THP-1 macrophages. To confirm expression of nearby cell death suppressor genes, UL37x1 and UL38 transcripts were evaluated at 8 h postinfection of differentiated THP-1 cells (Fig. 9J). Infected cells included UL37x1 and UL38 transcripts of similar lengths and abundances in comparisons of ΔUL36 with Towne-BAC virus or of RC2792 with RQ2792 virus, confirming expectations derived from similar analyses of infected HFs (Fig. 1). Combined, these data indicate that vICA/pUL36 prevents caspase-independent cell death and promotes infected-cell survival in monocytes at late stages of macrophage differentiation. Overall, these results complete analyses that highlight an apparent change in the host response to HCMV infection dependent on the stage of macrophage differentiation as well as a previously unrecognized function of pUL36/vICA. Thus, early in differentiation, HCMV primarily induces caspase-dependent cell death, and pUL36/vICA is required to prevent caspase-dependent death that otherwise inhibits replication, a function anticipated from the cell-death suppressor mechanism reported for this viral protein and now reported here for a specific stage of monocyte-macrophage differentiation. In contrast, the UL36 gene primarily prevents caspase-independent cell death at late stages of differentiation to macrophages, a result that reveals an unrecognized function for the gene.
DISCUSSION
Cytomegaloviruses encode multiple genes to evade intrinsic and immune-controlled defenses that activate cell death pathways. The antiapoptotic protein vICA, identified through an unbiased, cell-based screen for viral proteins that prevent Fas- or TNF-mediated cell death, binds procaspase-8 to prevent activation (73). Adventitious mutations (73) and transient complementation of targeted mutations (current studies) indicate that vICA/pUL36 also prevents extrinsic apoptosis in infected HFs. Current studies evaluated HCMV replication in monocytes differentiating to macrophages and suggest that vICA/pUL36 is critical throughout differentiation. Previous sequence and functional analyses of the MCMV homolog supported the expectation for a critical role because M36 prevents virus-induced, caspase-dependent death in differentiated macrophages (18, 54, 57). Importantly, those observations utilized a novel approach, replacing the M36 gene with a dominant negative FADD protein, that suggested that extrinsic apoptosis is the primary mechanism restricting growth of M36-deficient virus. Unexpectedly, the current studies revealed that vICA/pUL36 controls caspase-independent cell death in differentiated macrophages. M36-deficient MCMV induces caspase-dependent apoptosis in either primary macrophages or macrophage cell lines (17, 18, 57). Thus, the macrophage response to HCMV may indicate general differences in the host response to these species-restricted viruses, may be the result of other, nonconserved viral cell death suppressor functions, or may indicate differences specific to THP-1 cells. Each of these possibilities requires further evaluation; however, none detracts from the novel observation that vICA/pUL36 controls caspase-independent cell death. In contrast, HCMV required vICA/pUL36 to control caspase-dependent death in cells early in differentiation to macrophages. Whether MCMV has a similar requirement for M36 during monocyte-to-macrophage differentiation is unknown. Thus, our results have highlighted differentiation-dependent cellular responses to the virus as well as distinct roles for vICA/pUL36 at different stages of macrophage differentiation.
Monocyte-macrophage differentiation and cell death control by vICA/pUL36.
The process of monocyte-to-macrophage differentiation occurs over several days. Gene expression profiles have indicated both transient and stable changes that occur within the first 3 days and remain altered throughout 7 days, respectively (35, 44, 49). Here, HCMV infections have highlighted functional differences in antiviral responses in comparisons of monocytic THP-1 cells at early and late stages of macrophage differentiation. THP-1 cells are considered a model for macrophage differentiation as the result of functional, morphological, and membrane antigen expression (2, 85, 86) and share with primary cells a transcriptional program of many commonly regulated genes (35). Donor-to-donor variability inherent in primary monocyte-derived macrophages contrasts with more uniform responses of THP-1 cells, making this cell line useful in virus-host cell interaction studies (12). Previous evaluations of HCMV infections of THP-1 cells have explored the nonpermissive state of undifferentiated cells (5, 30, 31, 42, 56, 60, 87, 91, 92). In contrast, our current studies have focused on cells permissive to viral replication. However, infections in undifferentiated controls indicated that the control viruses used in our study did not induce apoptosis of nonpermissive cells, as might be anticipated from an earlier report (60). More specifically, Moon et al. reported that HCMV strain Towne induces apoptosis in undifferentiated THP-1 but not HL-60 cells (60). Although an apparent contradiction, our previous studies have, in fact, revealed that laboratory strains certainly include variants with adventitious mutations impacting cell death suppressor genes (73). Importantly, our current studies employed both viruses with specific deletions and complementation by exogenous expression to evaluate cell death. Thus, future efforts will be needed to clarify viral components that promote or prevent apoptosis in infections of undifferentiated, nonpermissive THP-1 cells. These comparisons and future studies to determine the role of vICA/pUL36-controlled caspase-independent responses in primary macrophages will be guided by the results here and will provide a more complete understanding of the role of the UL36 gene. Further, these studies will also guide future efforts to determine the impact of the UL36 gene in the context of viruses more closely related to clinical isolates that include additional genes (13). Regardless of the outcome, our studies indicate that the THP-1 monocyte-macrophage differentiation model is useful for studies of HCMV gene function during permissive replication and, importantly, indicate that the function of the UL36 gene is incompletely understood.
Caspase-dependent cell death at early stages of differentiation.
Currently, the pathway to caspase-dependent death of THP-1 cells infected early in differentiation is incompletely understood; however, results thus far suggest that activation of caspase-8 occurs following responses that initiate from within the infected cell. Previous analyses have shown that HCMV-infected THP-1 cultures produce TNF (87), a known activator of caspase-8 that could bind death receptors on infected cells. These early analyses also suggested that the TNF was largely produced by uninfected cells within the THP-1 cultures, consistent with other reports of HCMV-induced bystander effects (22, 87). Other investigators have shown that HCMV downregulates TNF receptor 1 (TNFR1) on infected THP-1 cells (3), and this mechanism may be sufficient to evade the consequences of TNF produced by bystander cells. In the current study, the impact of multiplicity of infection on vICA-deficient virus replication was evaluated in order to establish appropriate conditions for cell death analyses. The vICA-deficient viruses could not be rescued by high-multiplicity infections, as might be expected from apoptosis induced by extrinsic factors released from uninfected, bystander cells. In contrast, the results were more consistent with control mechanisms that initiated from within the cell although this conclusion relies on infection conditions that included some uninfected cells at all multiplicities. Thus, additional studies are required to determine whether extrinsic factors or death receptors play any role in the events required to induce caspase-dependent death of THP-1 cells infected at early stages of differentiation.
Caspase-independent cell death pathways controlled by CMV.
Multiple caspase-independent cell death programs have been characterized (43, 45). Consequences of caspase-independent cell death include increased inflammation and pathology while apoptosis is an intrinsic pathogen clearance process that alarms the immune system to initiate cellular responses (39). Knowledge of CMV-encoded gene products that control caspase-independent prodeath responses is emerging, as is the evidence for single gene products controlling multiple pathways. Caspase-independent programs that control cytomegaloviruses include a serine protease pathway controlled by the mitochondrial protease HtrA2/Omi and receptor interacting protein (RIP) kinase-dependent cell death (46, 53, 89, 90). Viral proteins that control these pathways include vMIA/pUL37x1 and M45. Similar to vICA/pUL36, vMIA/pUL37x1 also controls caspase-dependent apoptosis if it is induced during replication (51). Thus far, the timing of activation correlates with the consequence of the specific caspase-dependent or -independent pathway on viral replication. The HtrA2/Omi-dependent death pathway, controlled by vMIA/pUL37x1, is activated late in replication and has a minimal impact on viral yield (51, 53). However, infected cells die 2 to 3 days earlier than with infection by parental virus, and it is anticipated that premature HtrA2-dependent death may play a more important role in vivo. Initiated soon after infection begins, the RIP-dependent death pathway, controlled by M45, effectively controls replication (10, 46, 89, 90). Our studies here have revealed that vICA/pUL36 also falls within this group of gene products that control caspase-independent death and that the caspase-independent death induced within differentiated THP-1 cells effectively controls viral replication.
vICA/pUL36-mediated suppression of caspase-independent death has not been reported prior to this study. The observation that specific amino acids critical to caspase-8 binding may also be critical to vICA/pUL36-mediated protection from caspase-independent death may indicate a nonproteolytic role for the cellular protein in the HCMV-induced death pathway. However, significant concerns raised by reports of infected-HF-specific degradation of UL36mut prevent any such conclusions at this time (64). Thus, additional effort will be necessary to determine the mechanism of vICA/pUL36 protection, events of the caspase-independent death process, and consequences of caspase-dependent or -independent cell death.
Summary.
Monocyte-macrophages are important cells for HCMV, having proposed roles in dissemination during primary infection and in reactivation of latent virus. Accumulating evidence indicates that caspase-8, the target of vICA/pUL36, is critical to monocyte-to-macrophage differentiation (33, 34, 68). The impact of differentiation on viral replication has also been well established. Efforts to determine how the virus counteracts antiviral defenses mediated by caspase-8 without interfering with caspase-8-dependent differentiation are ongoing. The current studies indicate that the process of differentiation reduces the role of caspase-8 as an initiator of a caspase-dependent death pathway activated in response to HCMV, leaving open the questions of how the cell controls caspase-8-dependent death and which antiviral pathway is activated in cells at late stages of differentiation to macrophages.
Acknowledgments
We are grateful for the expert cell culture assistance of William Kaiser and to Tim Sparer, Ritesh Tandon, Jenny Ahlqvist, Lisa Daley, and Edward Mocarski for critical reading of the manuscript. Additionally, we acknowledge Lisa Daley for technical advice on flow cytometry.
This work was supported by National Blood Foundation funds and PHS grant RO3 AI076640, both awarded to A.L.M. Additional support was provided by funds from PHS grant RO1 AI020211 and Georgia Cancer Coalition funds awarded to Edward Mocarski.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Abbate, J., J. C. Lacayo, M. Prichard, G. Pari, and M. A. McVoy. 2001. Bifunctional protein conferring enhanced green fluorescence and puromycin resistance. Biotechniques 31:336-340. [DOI] [PubMed] [Google Scholar]
- 2.Auwerx, J. 1991. The human leukemia cell line, THP-1: a multifaceted model for the study of monocyte-macrophage differentiation. Experientia 47:22-31. [DOI] [PubMed] [Google Scholar]
- 3.Baillie, J., D. A. Sahlender, and J. H. Sinclair. 2003. Human cytomegalovirus infection inhibits tumor necrosis factor alpha (TNF-α) signaling by targeting the 55-kilodalton TNF-alpha receptor. J. Virol. 77:7007-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber, G. N. 2001. Host defense, viruses and apoptosis. Cell Death Differ. 8:113-126. [DOI] [PubMed] [Google Scholar]
- 5.Beisser, P. S., L. Laurent, J. L. Virelizier, and S. Michelson. 2001. Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes. J. Virol. 75:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bevan, I. S., M. R. Walker, and R. A. Daw. 1993. Detection of human cytomegalovirus DNA in peripheral blood leukocytes by the polymerase chain reaction. Transfusion 33:783-784. [DOI] [PubMed] [Google Scholar]
- 7.Blink, E., N. A. Maianski, E. S. Alnemri, A. S. Zervos, D. Roos, and T. W. Kuijpers. 2004. Intramitochondrial serine protease activity of Omi/HtrA2 is required for caspase-independent cell death of human neutrophils. Cell Death Differ. 11:937-939. [DOI] [PubMed] [Google Scholar]
- 8.Britt, W. 2007. Virus entry into host, establishment of infection, spread in host, mechanisms of tissue damage, p. 737-764. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 9.Brocchieri, L., T. N. Kledal, S. Karlin, and E. S. Mocarski. 2005. Predicting coding potential from genome sequence: application to betaherpesviruses infecting rats and mice. J. Virol. 79:7570-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 11.Cam, M., W. Handke, M. Picard-Maureau, and W. Brune. 2009. 9 October 2009. Cytomegaloviruses inhibit Bak- and Bax-mediated apoptosis with two separate viral proteins. Cell Death Differ. doi: 10.1038/cdd.2009.147. [DOI] [PubMed]
- 12.Cassol, E., M. Alfano, P. Biswas, and G. Poli. 2006. Monocyte-derived macrophages and myeloid cell lines as targets of HIV-1 replication and persistence. J. Leukoc. Biol. 80:1018-1030. [DOI] [PubMed] [Google Scholar]
- 13.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan, G., E. R. Bivins-Smith, M. S. Smith, P. M. Smith, and A. D. Yurochko. 2008. Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation toward an M1 macrophage. J. Immunol. 181:698-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, H. Y., and X. Yang. 2000. Proteases for cell suicide: functions and regulation of caspases. Microbiol. Mol. Biol. Rev. 64:821-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chipuk, J. E., and D. R. Green. 2005. Do inducers of apoptosis trigger caspase-independent cell death? Nat. Rev. Mol. Cell Biol. 6:268-275. [DOI] [PubMed] [Google Scholar]
- 17.Cicin-Sain, L., J. Podlech, M. Messerle, M. J. Reddehase, and U. H. Koszinowski. 2005. Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J. Virol. 79:9492-9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cicin-Sain, L., Z. Ruzsics, J. Podlech, I. Bubic, C. Menard, S. Jonjic, M. J. Reddehase, and U. H. Koszinowski. 2008. Dominant-negative FADD rescues the in vivo fitness of a cytomegalovirus lacking an antiapoptotic viral gene. J. Virol. 82:2056-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Droin, N., S. Cathelin, A. Jacquel, L. Guery, C. Garrido, M. Fontenay, O. Hermine, and E. Solary. 2008. A role for caspases in the differentiation of erythroid cells and macrophages. Biochimie 90:416-422. [DOI] [PubMed] [Google Scholar]
- 20.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Festjens, N., S. Cornelis, M. Lamkanfi, and P. Vandenabeele. 2006. Caspase-containing complexes in the regulation of cell death and inflammation. Biol. Chem. 387:1005-1016. [DOI] [PubMed] [Google Scholar]
- 22.Gafa, V., O. Manches, A. Pastor, E. Drouet, P. Ambroise-Thomas, R. Grillot, and D. Aldebert. 2005. Human cytomegalovirus downregulates complement receptors (CR3, CR4) and decreases phagocytosis by macrophages. J. Med. Virol. 76:361-366. [DOI] [PubMed] [Google Scholar]
- 23.Garrido, C., and G. Kroemer. 2004. Life's smile, death's grin: vital functions of apoptosis-executing proteins. Curr. Opin. Cell Biol. 16:639-646. [DOI] [PubMed] [Google Scholar]
- 24.Gerna, G., F. Baldanti, and M. G. Revello. 2004. Pathogenesis of human cytomegalovirus infection and cellular targets. Hum. Immunol. 65:381-386. [DOI] [PubMed] [Google Scholar]
- 25.Goldmacher, V. S. 2005. Cell death suppression by cytomegaloviruses. Apoptosis 10:251-265. [DOI] [PubMed] [Google Scholar]
- 26.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J. W. Han, R. J. Lutz, S. Watanabe, E. D. Cahir McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. U. S. A. 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, L. M., R. P. Cherla, C. van den Hoogen, W. C. van Haaften, S. Y. Lee, and V. L. Tesh. 2005. Comparative evaluation of apoptosis induced by Shiga toxin 1 and/or lipopolysaccharides in human monocytic and macrophage-like cells. Microb. Pathog. 38:63-76. [DOI] [PubMed] [Google Scholar]
- 28.Hertel, L., V. G. Lacaille, H. Strobl, E. D. Mellins, and E. S. Mocarski. 2003. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J. Virol. 77:7563-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho, P. K., and C. J. Hawkins. 2005. Mammalian initiator apoptotic caspases. FEBS J. 272:5436-5453. [DOI] [PubMed] [Google Scholar]
- 30.Huang, T. H., T. Oka, T. Asai, T. Okada, B. W. Merrills, P. N. Gertson, R. H. Whitson, and K. Itakura. 1996. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 24:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioudinkova, E., M. C. Arcangeletti, A. Rynditch, F. De Conto, F. Motta, S. Covan, F. Pinardi, S. V. Razin, and C. Chezzi. 2006. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene 384:120-128. [DOI] [PubMed] [Google Scholar]
- 32.Jarvis, M. A., and J. A. Nelson. 2002. Human cytomegalovirus persistence and latency in endothelial cells and macrophages. Curr. Opin. Microbiol. 5:403-407. [DOI] [PubMed] [Google Scholar]
- 33.Kang, T. B., T. Ben-Moshe, E. E. Varfolomeev, Y. Pewzner-Jung, N. Yogev, A. Jurewicz, A. Waisman, O. Brenner, R. Haffner, E. Gustafsson, P. Ramakrishnan, T. Lapidot, and D. Wallach. 2004. Caspase-8 serves both apoptotic and nonapoptotic roles. J. Immunol. 173:2976-2984. [DOI] [PubMed] [Google Scholar]
- 34.Kang, T. B., G. S. Oh, E. Scandella, B. Bolinger, B. Ludewig, A. Kovalenko, and D. Wallach. 2008. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice. J. Immunol. 181:2522-2532. [DOI] [PubMed] [Google Scholar]
- 35.Kohro, T., T. Tanaka, T. Murakami, Y. Wada, H. Aburatani, T. Hamakubo, and T. Kodama. 2004. A comparison of differences in the gene expression profiles of phorbol 12-myristate 13-acetate differentiated THP-1 cells and human monocyte-derived macrophage. J. Atheroscler. Thromb. 11:88-97. [DOI] [PubMed] [Google Scholar]
- 36.Kollert-Jons, A., E. Bogner, and K. Radsak. 1991. A 15-kilobase-pair region of the human cytomegalovirus genome which includes US1 through US13 is dispensable for growth in cell culture. J. Virol. 65:5184-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo, K., H. Kaneshima, and E. S. Mocarski. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc. Natl. Acad. Sci. U. S. A. 91:11879-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo, K., J. Xu, and E. S. Mocarski. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. U. S. A. 93:11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krysko, D. V., K. D'Herde, and P. Vandenabeele. 2006. Clearance of apoptotic and necrotic cells and its immunological consequences. Apoptosis 11:1709-1726. [DOI] [PubMed] [Google Scholar]
- 40.Lamkanfi, M., N. Festjens, W. Declercq, T. Vanden Berghe, and P. Vandenabeele. 2007. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 14:44-55. [DOI] [PubMed] [Google Scholar]
- 41.Launay, S., O. Hermine, M. Fontenay, G. Kroemer, E. Solary, and C. Garrido. 2005. Vital functions for lethal caspases. Oncogene 24:5137-5148. [DOI] [PubMed] [Google Scholar]
- 42.Lee, C. H., G. C. Lee, Y. J. Chan, C. J. Chiou, J. H. Ahn, and G. S. Hayward. 1999. Factors affecting human cytomegalovirus gene expression in human monocyte cell lines. Mol. Cells 9:37-44. [PubMed] [Google Scholar]
- 43.Leist, M., and M. Jaattela. 2001. Four deaths and a funeral: from caspases to alternative mechanisms. Nat. Rev. Mol. Cell Biol. 2:589-598. [DOI] [PubMed] [Google Scholar]
- 44.Liu, H., B. Shi, C. C. Huang, P. Eksarko, and R. M. Pope. 2008. Transcriptional diversity during monocyte to macrophage differentiation. Immunol. Lett. 117:70-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lockshin, R. A., and Z. Zakeri. 2002. Caspase-independent cell deaths. Curr. Opin. Cell Biol. 14:727-733. [DOI] [PubMed] [Google Scholar]
- 46.Mack, C., A. Sickmann, D. Lembo, and W. Brune. 2008. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc. Natl. Acad. Sci. U. S. A. 105:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manzur, M., P. Fleming, D. C. Huang, M. A. Degli-Esposti, and C. E. Andoniou. 2008. Virally mediated inhibition of Bax in leukocytes promotes dissemination of murine cytomegalovirus. Cell Death Differ. [DOI] [PubMed]
- 48.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez, F. O., S. Gordon, M. Locati, and A. Mantovani. 2006. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177:7303-7311. [DOI] [PubMed] [Google Scholar]
- 50.McCormick, A. L. 2008. Control of apoptosis by human cytomegalovirus. Curr. Top. Microbiol. Immunol. 325:281-295. [DOI] [PubMed] [Google Scholar]
- 51.McCormick, A. L., C. D. Meiering, G. B. Smith, and E. S. Mocarski. 2005. Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J. Virol. 79:12205-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCormick, A. L., and E. S. Mocarski. 2007. Betaherpesvirus modulation of the host response to infection, p. 321-332. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 53.McCormick, A. L., L. Roback, and E. S. Mocarski. 2008. HtrA2/Omi terminates cytomegalovirus infection and is controlled by the viral mitochondrial inhibitor of apoptosis (vMIA). PLoS Pathog. 4:e1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCormick, A. L., A. Skaletskaya, P. A. Barry, E. S. Mocarski, and V. S. Goldmacher. 2003. Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology 316:221-233. [DOI] [PubMed] [Google Scholar]
- 55.McCormick, A. L., V. L. Smith, D. Chow, and E. S. Mocarski. 2003. Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J. Virol. 77:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meier, J. L., and M. F. Stinski. 1997. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J. Virol. 71:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendelson, M., S. Monard, P. Sissons, and J. Sinclair. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J. Gen. Virol. 77:3099-3102. [DOI] [PubMed] [Google Scholar]
- 59.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. U. S. A. 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moon, M. S., G. C. Lee, J. H. Kim, H. A. Yi, Y. S. Bae, and C. H. Lee. 2003. Human cytomegalovirus induces apoptosis in promonocyte THP-1 cells but not in promyeloid HL-60 cells. Virus Res. 94:67-77. [DOI] [PubMed] [Google Scholar]
- 61.Moorman, N. J., I. M. Cristea, S. S. Terhune, M. P. Rout, B. T. Chait, and T. Shenk. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park, S. M., R. Schickel, and M. E. Peter. 2005. Nonapoptotic functions of FADD-binding death receptors and their signaling molecules. Curr. Opin. Cell Biol. 17:610-616. [DOI] [PubMed] [Google Scholar]
- 64.Patterson, C. E., and T. Shenk. 1999. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J. Virol. 73:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perlman, H., L. J. Pagliari, C. Georganas, T. Mano, K. Walsh, and R. M. Pope. 1999. FLICE-inhibitory protein expression during macrophage differentiation confers resistance to Fas-mediated apoptosis. J. Exp. Med. 190:1679-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pop, C., P. Fitzgerald, D. R. Green, and G. S. Salvesen. 2007. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry 46:4398-4407. [DOI] [PubMed] [Google Scholar]
- 67.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rebe, C., S. Cathelin, S. Launay, R. Filomenko, L. Prevotat, C. L'Ollivier, E. Gyan, O. Micheau, S. Grant, A. Dubart-Kupperschmitt, M. Fontenay, and E. Solary. 2007. Caspase-8 prevents sustained activation of NF-κB in monocytes undergoing macrophagic differentiation. Blood 109:1442-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 70.Schotte, P., W. Declercq, S. Van Huffel, P. Vandenabeele, and R. Beyaert. 1999. Non-specific effects of methyl ketone peptide inhibitors of caspases. FEBS Lett. 442:117-121. [DOI] [PubMed] [Google Scholar]
- 71.Schwerk, C., and K. Schulze-Osthoff. 2003. Non-apoptotic functions of caspases in cellular proliferation and differentiation. Biochem. Pharmacol. 66:1453-1458. [DOI] [PubMed] [Google Scholar]
- 72.Sindre, H., G. E. Tjoonnfjord, H. Rollag, T. Ranneberg-Nilsen, O. P. Veiby, S. Beck, M. Degre, and K. Hestdal. 1996. Human cytomegalovirus suppression of and latency in early hematopoietic progenitor cells. Blood 88:4526-4533. [PubMed] [Google Scholar]
- 73.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U. S. A. 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith, M. S., G. L. Bentz, J. S. Alexander, and A. D. Yurochko. 2004. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J. Virol. 78:4444-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sordet, O., C. Rebe, S. Plenchette, Y. Zermati, O. Hermine, W. Vainchenker, C. Garrido, E. Solary, and L. Dubrez-Daloz. 2002. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood 100:4446-4453. [DOI] [PubMed] [Google Scholar]
- 76.Stanier, P., D. L. Taylor, A. D. Kitchen, N. Wales, Y. Tryhorn, and A. S. Tyms. 1989. Persistence of cytomegalovirus in mononuclear cells in peripheral blood from blood donors. Br. Med. J. 299:897-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stern, J. L., J. Z. Cao, J. Xu, E. S. Mocarski, and B. Slobedman. 2008. Repression of human cytomegalovirus major immediate early gene expression by the cellular transcription factor CCAAT displacement protein. Virology 378:214-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Su, Y., R. Adair, C. N. Davis, N. L. DiFronzo, and A. M. Colberg-Poley. 2003. Convergence of RNA cis elements and cellular polyadenylation factors in the regulation of human cytomegalovirus UL37 exon 1 unspliced RNA production. J. Virol. 77:12729-12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taylor-Wiedeman, J., J. G. Sissons, L. K. Borysiewicz, and J. H. Sinclair. 1991. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 72:2059-2064. [DOI] [PubMed] [Google Scholar]
- 80.Taylor-Wiedeman, J., P. Sissons, and J. Sinclair. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 68:1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tenney, D. J., and A. M. Colberg-Poley. 1991. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology 182:199-210. [DOI] [PubMed] [Google Scholar]
- 82.Tenney, D. J., and A. M. Colberg-Poley. 1991. Human cytomegalovirus UL36-38 and US3 immediate-early genes: temporally regulated expression of nuclear, cytoplasmic, and polysome-associated transcripts during infection. J. Virol. 65:6724-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tenney, D. J., and A. M. Colberg-Poley. 1990. RNA analysis and isolation of cDNAs derived from the human cytomegalovirus immediate-early region at 0.24 map units. Intervirology 31:203-214. [DOI] [PubMed] [Google Scholar]
- 84.Terhune, S., E. Torigoi, N. Moorman, M. Silva, Z. Qian, T. Shenk, and D. Yu. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81:3109-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsuchiya, S., Y. Kobayashi, Y. Goto, H. Okumura, S. Nakae, T. Konno, and K. Tada. 1982. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 42:1530-1536. [PubMed] [Google Scholar]
- 86.Tsuchiya, S., M. Yamabe, Y. Yamaguchi, Y. Kobayashi, T. Konno, and K. Tada. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26:171-176. [DOI] [PubMed] [Google Scholar]
- 87.Turtinen, L. W., A. Assimacopoulos, and A. T. Haase. 1989. Increased monokines in cytomegalovirus infected myelomonocytic cell cultures. Microb. Pathog. 7:135-145. [DOI] [PubMed] [Google Scholar]
- 88.Turtinen, L. W., and B. J. Seufzer. 1994. Selective permissiveness of TPA differentiated THP-1 myelomonocytic cells for human cytomegalovirus strains AD169 and Towne. Microb. Pathog. 16:373-378. [DOI] [PubMed] [Google Scholar]
- 89.Upton, J. W., W. J. Kaiser, and E. S. Mocarski. 2008. Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J. Biol. Chem. 283:16966-16970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Upton, J. W., W. J. Kaiser, and E. S. Mocarski. Pathogen subversion of RIP3-dependent necrosis. Cell Host Microbe, in press. [DOI] [PMC free article] [PubMed]
- 91.Weinshenker, B. G., S. Wilton, and G. P. Rice. 1988. Phorbol ester-induced differentiation permits productive human cytomegalovirus infection in a monocytic cell line. J. Immunol. 140:1625-1631. [PubMed] [Google Scholar]
- 92.Yee, L. F., P. L. Lin, and M. F. Stinski. 2007. Ectopic expression of HCMV IE72 and IE86 proteins is sufficient to induce early gene expression but not production of infectious virus in undifferentiated promonocytic THP-1 cells. Virology 363:174-188. [DOI] [PubMed] [Google Scholar]