Abstract
Recently, mutations in the connection subdomain (CN) and RNase H domain of HIV-1 reverse transcriptase (RT) were observed to exhibit dual resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors (NRTIs and NNRTIs). To elucidate the mechanism by which CN and RH mutations confer resistance to NNRTIs, we hypothesized that these mutations reduce RNase H cleavage and provide more time for the NNRTI to dissociate from the RT, resulting in the resumption of DNA synthesis and enhanced NNRTI resistance. We observed that the effect of the reduction in RNase H cleavage on NNRTI resistance is dependent upon the affinity of each NNRTI to the RT and further influenced by the presence of NNRTI-binding pocket (BP) mutants. D549N, Q475A, and Y501A mutants, which reduce RNase H cleavage, enhance resistance to nevirapine (NVP) and delavirdine (DLV), but not to efavirenz (EFV) and etravirine (ETR), consistent with their increase in affinity for RT. Combining the D549N mutant with NNRTI BP mutants further increases NNRTI resistance from 3- to 30-fold, supporting the role of NNRTI-RT affinity in our NNRTI resistance model. We also demonstrated that CNs from treatment-experienced patients, previously reported to enhance NRTI resistance, also reduce RNase H cleavage and enhance NNRTI resistance in the context of the patient RT pol domain or a wild-type pol domain. Together, these results confirm key predictions of our NNRTI resistance model and provide support for a unifying mechanism by which CN and RH mutations can exhibit dual NRTI and NNRTI resistance.
Reverse transcriptase (RT) of HIV-1 was the first target for development of drugs against HIV-1 infection and remains a major target for the exploration of new therapeutic strategies. Out of more than 30 drugs approved by the U.S. Food and Drug Administration for the treatment of HIV-1 infection, 17 comprise nucleoside and nonnucleoside reverse transcriptase inhibitors (NRTIs and NNRTIs, respectively) (http://www.fda.gov/ForConsumers/byAudience/ForPatientAdvocates/HIVandAIDSActivities/ucm118915.htm). To block viral replication more efficiently, three-drug regimens are currently used in standard HIV-1 therapies that include combinations of two NRTIs plus an NNRTI or a protease inhibitor (http://aidsinfo.nih.gov). Selection of drug resistance mutations in response to treatment is a major barrier to successful control of HIV-1 infection, since drug-resistant variants of HIV-1 are selected in response to all approved drugs. An improved understanding of the mechanism of action of antiviral drugs and the mechanisms by which the drug-resistant viruses evade these drugs will facilitate the management of antiviral therapy and facilitate new drug designs.
NNRTIs are a class of very specific and potent anti-HIV-1 drugs that predominantly inhibit reverse transcription (51). In addition, NNRTIs nevirapine (NVP), efavirenz (EFV), and etravirine (ETR) have also been shown to enhance RT dimerization (55). Biochemical and structural analysis of the inhibition of reverse transcription by NNRTIs reveals that their binding induces conformational changes in RT that distort the precise geometry of the DNA polymerase catalytic site; these conformational changes affect the alignment of the primer terminus and slow down phosphodiester bond formation, as well as restrict domain motions and DNA translocation (48, 51, 52, 54). Some NNRTIs can modulate RNase H activity through long-range interactions and, depending upon the structure of the RNA-DNA hybrid substrate, can lead to the inhibition or stimulation of RNase H activity (21, 25, 37, 45, 50). These NNRTIs alter the RNase H cleavage site specificity and rates of the reaction (21), resulting in the accumulation of secondary cleavage products (37, 45), but do not affect the activity of the isolated RNase H domain (25). In addition, RNase H activity can also be affected by the NNRTI binding pocket (NNRTI BP) mutations that confer NNRTI resistance (2, 3, 19) as well as mutations in the polymerase primer grip (20) and the connection subdomain (CN) (11, 27, 38, 46).
Selection of drug-resistant viruses during NNRTI treatment decreases the potency of this class of drugs. For the narrow-spectrum and expanded-spectrum NNRTIs (NVP, DLV, and EFV), a single mutation was frequently sufficient to cause high levels of drug resistance. These mutations generally affect interactions between the inhibitor and the RT, and the affinity of the NNRTI to the RT is a critical factor in determining NNRTI resistance (26, 31, 53). NNRTI resistance mutations located in the NNRTI BP can inhibit drug binding by at least three mechanisms (14, 48); they can cause the loss or alteration of key hydrophobic interactions with the NNRTIs (located in the hydrophobic core of NNRTI BP), they can induce steric hindrance (located in the central region of the NNRTI BP), or they can reduce the entry of the inhibitor to the NNRTI BP (located at the rim of entrance to the NNRTI BP) (26, 31, 48).
We recently proposed a new mechanism of resistance for NRTIs, demonstrating that mutations in the C-terminal portion of RT can contribute to NRTI resistance. We identified mutations in the CN from treatment-experienced patients that enhanced 3′-azido-3′-deoxythymidine (AZT) resistance (34, 35), and closer examination of the CN and RH by us and others has identified additional mutations in this region that are associated with increased drug resistance (6, 11, 13, 23, 29, 43, 56, 62). Several mutations in the CN of RT (N348I, T369I, and E399D) were identified by us and others to be associated with increases in both NRTI and NNRTI resistance (23, 24, 33, 59). Interestingly, some of these CN mutations were previously shown to affect RT heterodimer stability (23). It is typical that mutations selected after treatment with NRTIs or NNRTIs are resistant to several compounds within the same class of drugs; however, it is quite rare for mutations to confer resistance to different classes of compounds. Among the described dual NRTI and NNRTI resistance mutations located in the NNRTI BP and deoxynucleoside triphosphate (dNTP) BP, it has been reported that Y181I/C is cross-resistant to NVP and 2,3-didehydro-2,3-dideoxythymidine (d4T) (4, 5), G190S/A/E is cross-resistant to NVP, d4T, and AZT (39), and Q145M, a very rare mutation in the dNTP BP that is selected in highly treated patients, is cross-resistant to both NRTIs and NNRTIs (41).
Here, we propose a novel mechanism for NNRTI resistance adding to the structural and biochemical approaches one more functional component—the effect of RNase H activity. We propose that the interplay between RNase H cleavage and NRTI excision/NNRTI dissociation is important for resistance to both classes of drugs. Our model provides insights into NNRTI resistance mechanisms and a plausible explanation for the phenomenon of dual resistance to NRTIs and NNRTIs.
MATERIALS AND METHODS
Plasmids, cloning, and mutagenesis.
pHCMV-G expresses the G glycoprotein of vesicular stomatitis virus (VSV-G) (60). pHL[WT] expresses the firefly luciferase reporter gene and all of the HIV-1 proteins except Nef and Env; the vector also contains a natural MscI site at the beginning of pol (amino acids 25/26), a unique Eco47III (amino acids 288/289), a unique SpeI site (amino acids 423/424), and a unique ClaI site (flanking integrase amino acids 4/5) (34). These restriction enzyme sites were used to subclone polymerase (MscI to Eco47III), connection (Eco47III to SpeI), and/or RNase H (SpeI to ClaI site) domain combinations into pHL[WT]. pCMVΔR8.2 expresses all HIV-1 proteins, except envelope, under the control of the human cytomegalovirus (hCMV) immediate early promoter and has the packaging signal (Ψ) and adjacent sequences deleted (32). Site-directed mutagenesis was carried out using the QuikChange XL site-directed mutagenesis kit (Stratagene), and the presence or absence of each mutation was verified by DNA sequencing. The details of the cloning steps are available upon request. Patient samples were as previously described (34).
Antiviral drugs.
NNRTI inhibitors NVP, EFV, and ETR were obtained from the NIH AIDS Research & Reference Reagent Program. Delavirdine (DLV) was purchased from Movarek Biochemicals.
Cells, transfection, and virus production.
Human 293T cells (American Type Culture Collection) and a 293T-based cell line, GN-HIV-GFFP (36), were maintained at 5% CO2 and 37°C in Dulbecco's modified Eagle's medium (CellGro) supplemented with 10% fetal calf serum (FCS; HyClone), penicillin (50 U/ml; Gibco), and streptomycin (50 μg/ml; Gibco). Hygromycin (Calbiochem) selection was performed at a final concentration of 270 μg/ml. To produce pHL[WT]-based virus containing the desired mutations, 293T cells were plated at 5 × 106 cells per 100-mm-diameter dishes and transfected by calcium phosphate precipitation in the presence of pHCMV-G. Forty-eight hours later, virus was harvested, filtered through a Millex GS 0.45-μm-pore-size filter (Nalgene), concentrated 20-fold by centrifugation at 25,000 rpm for 90 min (Surespin; Sorvall), and stored at −80°C. Viruses used for the in vitro RNase H cleavage assay (12, 42) were clarified by centrifugation, filtered through a Millex GS 0.45-μm-pore-size filter (Nalgene), centrifuged through a 30% sucrose cushion, and concentrated 100-fold in phosphate-buffered saline (PBS). Viruses for the direct-repeat deletion assays were produced by transiently cotransfecting the cell line GN-HIV1-GFFP with pCMVΔR8.2 (or the mutated variant of RT) and pHCMV-G by calcium phosphate precipitation. Supernatant was collected 24 h after transfection, filtered through a Millex GS 0.45-μm-pore-size filter (Nalgene), and stored at −80°C.
Single-replication-cycle drug susceptibility assay.
Drug susceptibility assays were performed as previously described (36). Briefly, fresh medium containing serial dilutions of a drug or no drugs was added to 293T target cells plated the previous day at 4,000 cells/well onto a 96-well plate. Four hours later, normalized virus was added to each well, and 48 h postinfection, luciferase activity was measured using a 96-well luminometer (LUMIstar Galaxy; BMG Labtech). Data were plotted as the percentage of inhibition of luciferase activity versus log10 drug concentration, and the percentage of inhibition was calculated as follows: [1 − (luciferase activity in the presence of drug/luciferase activity in the absence of drug)] × 100%. The drug concentration at which virus replication was inhibited by 50% (IC50) was calculated using inhibition curves defined by the four-parameter sigmoidal function y = y0 + a/[1 + (x/x0)b] (SIGMAPLOT 8.0 software).
Statistical analysis.
Statistically significant differences in drug resistance and template-switching frequencies were determined using a two-sample t test (SIGMAPLOT 8.0).
RESULTS
CN mutations from treatment-experienced patients possess dual NRTI and NNRTI resistance in the context of a wild-type pol.
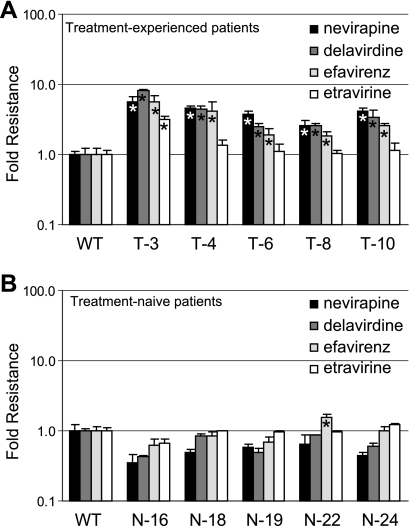
We previously characterized the C-terminal domains (CN plus RH) from subtype B treatment-experienced patients and localized the associated NRTI resistance to mutations in the CN (12, 34). To determine if the patient's CNs also exhibited dual resistance to NNRTIs, we first subcloned the CN from each treatment-experienced patient into an HIV-1 luciferase expression vector containing a wild-type pol (palm, fingers and thumb subdomains) and a wild-type RH. Phenotypic testing for NNRTI resistance of the resulting viruses (Fig. 1A) was then performed, and they were compared to the CNs from treatment-naïve patients (Fig. 1B). All five viruses from the treatment-experienced group (T-3, T-4, T-6, T-8, and T-10) exhibited significantly higher levels of resistance to NVP (up to 6-fold), DLV (up to 8-fold), and EFV (up to 5-fold). Only patient T-3 exhibited a significant 3-fold increase in ETR resistance. As expected, most of the viruses from the treatment-naïve group (N-16, N-18, N-19, N-22, and N-24) did not enhance NNRTI resistance greatly to any of the four drugs tested (Fig. 1B), with the exception of N-22, which exhibited a 1.5-fold increase in EFV resistance. Thus, the CNs from treatment-experienced patients exhibited dual resistance to NRTIs (12, 34) and NNRTIs in the context of a wild-type pol.
FIG. 1.
Effect of CN mutations from patients' RTs on NNRTI resistance. Fold changes in the resistance level to NVP, DLV, EFV, and ETR are shown as vertical bars (mean of two or more independent experiments ± standard error) for viruses containing wild-type pol and CN from treatment-experienced patients' RT T-3, T-4, T-6, T-8, and T-10 (A) or CN from treatment-naïve patients' RT N-16, N-18, N-19, N-22, and N-24 (B). An asterisk within the bar indicates a statistically significant increase in resistance versus wild-type (WT) control (t test, P < 0.05).
CN mutations from treatment-experienced patients possess dual NRTI and NNRTI resistance in the context of the patient-derived RT pol.
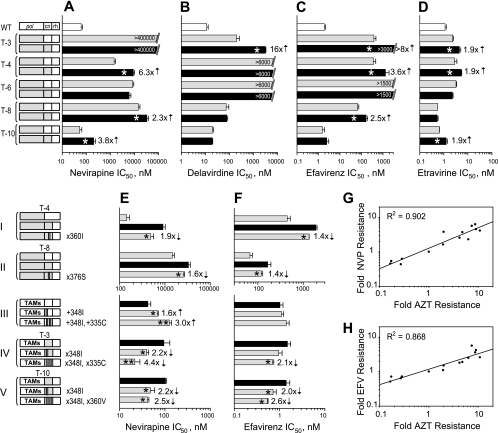
Treatment history of the patients selected for this study included exposure to both NRTI and NNRTI as reported previously and shown in Table 1 (34). Each patient's pol contained different combinations of drug resistance mutations, which included NNRTI resistance mutations. Because interactions between mutations in the pol could potentially modulate the effects of the CN mutations on NNRTI resistance, we analyzed whether mutations in the CNs contributed to NNRTI resistance in the context of the patient-derived RT pol (Fig. 2). The results showed that the level of resistance to all four NNRTIs was elevated when we combined the CN from treatment-experienced patients with the patient-derived RT pol, compared to the resistance level observed with the patient-derived RT pol and wild-type CN and RH domains (Fig. 2A to D). The effect on NNRTI resistance with the pol-CN combination derived from different treatment-experienced patients varied depending upon the NNRTI tested. For example, in Fig. 2A, NVP resistance was increased 6.3-, 2.3-, and 3.8-fold for patients T-4, T-8, and T-10, respectively. The T-3 pol domain already displayed an extremely high-resistance phenotype (>400 μM) due to the presence of V106I and Y188L NVP resistance mutations; therefore, it was not possible to detect the influence of CN mutations on NVP resistance for the pol-CN combination from patient T-3 (Fig. 2A).
TABLE 1.
Patient treatment history with RT inhibitors and mutations in the pol domain and connection subdomaina
| Patient | RTI treatment |
Mutation(s) in pol domain (1 to 296 aa) |
Mutations in CN (297 to 423 aa) | |||
|---|---|---|---|---|---|---|
| NRTI | NNRTI | NRTI-R | NNRTI-R | Other | ||
| T-3 | AZT, 3TC, d4T, ddI | NVP, EFV | D67N, K70R, T215F, K219Q | V106I, Y188L | V60I, K64H, T69N, K122E, I135T, D218E, K233Q, L228H | E312Q, G335C, N348I, R356K, M357T, K358R, K390R, A400T |
| T-4 | AZT, 3TC, ddI, d4T, ABC | NVP, EFV | M41L, D67N, M184V, L210W, T215Y | L100I, K103N | K43E, E44A, K102R, V118I, K122E, D123N, T139K, D177N, G196E, R211T, K219N | E297K, K311R, K358R, A360I, A371V, A400T |
| T-6 | AZT, 3TC, d4T, ABC | EFV | M41L, D67N, L74V, M184V, T215Y | L100I, K103N | V35E, E44D, R83K, K122P, A158S, Q174K, D177E, E194D | E297R, G335D, R356K, M357R, K358R, V365I, T386I, K390R, A400T |
| T-8 | ABC, ddI, TDF | DLV | M41L, L210W, T215Y | K103N, V108I | L74I, V118I | E297K, K311R, I326V, A371V, I375V, A376S, T377M, T386I, K390R |
| T-10 | AZT, 3TC, d4T | None | M184V | None | K73R, Q102K, D123E, I142T, R211T, F214L | E297R, I341F, N348I, K358R, A360V, A376T, A390R, A400T |
aa, amino acids; RTI, reverse transcriptase inhibitor; 3TC, 2′,3′-dideoxy-3′thiacytidine; ddI, 2′,3′-dideoxyinosine; ABC, abacavir; TDF, tenofovir. NRTI-R and NNRTI-R, standard resistance mutations updated in December 2008 by the International AIDS Society—USA. This table is adapted with permission from a report by Nikolenko et al. (34).
FIG. 2.
Effect of CN mutations from treatment-experienced patients' RTs on NNRTI resistance. (A to D) IC50 determinations for the viruses containing CN mutations from treatment-experienced patients' RTs in the context of their own pol to NVP (A), DLV (B), EFV (C), ETR (D). White bars represent the average IC50 for the wild-type RT control; gray bars represent RTs containing pol domains originated from the patients' virus; black bars represent both pol and CN originated from the patients' virus. Numbers in front of black bars represent fold changes, and asterisks within the black bars indicate a statistically significant difference in IC50 within each pair of RTs connected by a bracket (t test, P < 0.05). Numbers within bars represent the maximum drug concentration used for the testing that was not sufficient to inhibit viral replication. (E and F) IC50 determinations for the viruses containing specific CN mutation(s) from treatment-experienced patients' RTs to NVP (E) and EFV (F). Numbers in front of gray bars represent fold changes, and asterisks within the gray bars indicate a statistically significant difference in IC50 versus the black bar for each group connected by a bracket (t test, P < 0.05). Direction of arrow located near the number indicates a decrease or increase in resistance caused by mutation. An additional asterisk within the bar indicates a statistically significant change in resistance for the double mutant versus the single mutant within the group (t test, P < 0.05). Schematics of the corresponding RT structures are depicted on the left-hand side of panels A, B, C, D (top part), and E and F (bottom part). Groups of constructs are connected by a bracket. White boxes in schematics represent the wild-type sequences, and gray boxes represent the patients' RT sequences; white strips within the gray box indicate the reversion of the mutant amino acid to the wild type; gray strips within the white box indicate a replacement of the wild-type amino acid to the mutant amino acid. Specific mutations are shown near each construct where applicable, designating either the reversion of this mutation to the wild-type amino acid (e.g., ×360I) or the reversion of the wild-type amino-acid to the mutant amino acid (e.g., +348I). Linear regression analysis of resistance changes to AZT versus NVP (G), and to AZT versus EFV (H) caused by CN mutations from treatment-experienced patients. Resistance data for NVP and EFV are collected from Fig. 1A and panels A, C, E, and F of this figure and, for AZT, from reports by Delviks-Frankenberry et al. (12) and Nikolenko et al. (34). The correlation coefficients (R) were determined using SigmaPlot 8.0 software. Fold resistance was calculated as a ratio between IC50s for virus containing specific mutation(s) and correspondent control virus without this mutation(s).
DLV resistance was elevated 16-fold by the presence of the T-3 CN in combination with the patient-derived RT pol (Fig. 2B). It was not possible to determine the exact IC50 due to a high level of resistance to DLV (>6 μM) that resulted from the combination of L100I and K103N mutations in both T-4 and T-6 pols. The addition of the CN from patients T-8 and T-10 to the patient-derived RT pol did not alter the level of resistance to DLV.
EFV resistance was also increased when the CNs from patients T-3, T-4, and T-8 were combined with the patient-derived RT pol (Fig. 2C). The increase in resistance to EFV was more than 8-fold for T-3, 3.6-fold for T-4, and 2.5-fold for T-8; the combination of L100I and K103N in T-6 pol elevated EFV IC50 to >1.5 μM, making it impossible to evaluate the effect of T-6 CN in the pol-CN combination.
Finally, it is noteworthy that the patient's CN affected ETR resistance, which has been reported to have a very high genetic barrier for drug resistance mutation selection (56). As shown in Fig. 2D, the addition of the patients' CN to the patient-derived RT pol increased ETR IC50 1.9-fold for three out of five tested patients (T-3, T-4, and T-10) compared to the addition of the patient-derived RT pol alone.
Overall, the results show that four out of the five CNs from the treatment-experienced patient group (T-3, T-4, T-8, and T-10) significantly enhanced NNRTI resistance when they were tested for resistance in combination with the patient-derived RT pol. Thus, the CNs in the context of their own patient-derived RT pol also exhibit dual NRTI (12, 34) and NNRTI resistance.
To evaluate whether the same mutations in CN (N348I, G335C, A360I/V, and T376S) that were previously shown to enhance resistance to AZT (34), contribute to the observed increase in NNRTI resistance, we determined the resistance level to NVP and EFV for viruses with these selected CN amino acid substitutions. These amino acid substitutions were either added to the wild-type CN with the expectation that they would increase NNRTI resistance or reverted to the wild-type amino acids in the context of patients' RT CN with the expectation that they would reduce NNRTI resistance. The results of this analysis are shown in Fig. 2E and F. Reversion of isoleucine at position 360 in the T-4 CN to an alanine, which is present in wild-type RT, in the presence of the T-4 pol domain, resulted in a 1.9-fold decrease in resistance to NVP (Fig. 2E, group I). Thus, the A360I mutation was associated with NNRTI resistance as well as NRTI resistance (34).
Reversion of serine at position 376 in T-8 CN to an alanine, which is present in wild-type RT, in the presence of the T-8 CN pol domain, resulted in a 1.6-fold decrease in resistance to NVP (Fig. 2E, group II), confirming the contribution of A376S substitution to the NVP resistance.
The N348I substitution alone and in combination with G335C increased NVP resistance 1.6- and 3-fold, respectively, in the context of a pol domain containing thymidine analog mutations (TAMs) (Fig. 2E, group III). We reverted isoleucine at position 348 to the wild-type asparagine, or both isoleucine 348 and cysteine 335 to the wild-type amino acids asparagine and glycine, respectively; in the context of the T-3 CN and a pol domain containing TAMs, these mutants reduced NVP resistance 2.2- and 4.4-fold, respectively (Fig. 2E, group IV). Reversion of isoleucine at position 348 to the wild-type asparagine alone, or in combination with reversion of valine 360 to the wild-type alanine, in the context of T-10 CN and a pol domain containing TAMs reduced NVP resistance 2.2- and 2.5-fold, respectively (Fig. 2E, group V). Overall, these observations confirm that N348I, G335C, and A360V enhance NVP resistance.
Some of the CN mutations were also associated with EFV resistance (Fig. 2F). Reversion of the isoleucine at position 360 to the wild-type alanine in the T-4 CN resulted in a small but statistically significant 1.4-fold reduction in EFV resistance (Fig. 2F, group I); reversion of the serine at position 376 to the wild-type alanine in the T-8 CN resulted in a 1.4-fold reduction in EFV resistance (Fig. 2F, group II); reversion of both isoleucine at position 348 and cysteine at position 335 to the wild-type amino acids asparagine and glycine, respectively, in the T-3 CN resulted in a 2.1-fold reduction in EFV resistance (Fig. 2F, group IV); and reversion of isoleucine 348 to wild-type asparagine alone or in combination with reversion of the valine at position 360 to the wild-type alanine in the T-10 CN resulted in 2.0-fold and 2.6-fold reductions in EFV resistance, respectively (Fig. 2F, group V).
Overall, these results show that mutations N348I, A376S, G335C, and A360V, which were previously shown to increase AZT resistance (34), also increase NVP and EFV resistance. To determine the relationships between increased AZT and NNRTI resistance caused by CN mutations in treatment-experienced patients, we performed linear regression analysis of the fold changes in resistance to AZT versus NVP (Fig. 2G) and to AZT versus EFV (Fig. 2H) that were associated with the presence or absence of CN mutations. We found a very strong correlation between fold changes in resistance for AZT versus NVP (R2 = 0.902; t test, P < 0.0001) as well as for AZT versus EFV (R2 = 0.868; t test, P < 0.0001).
Proposed mechanism for NNRTI resistance conferred by mutations affecting RNase H cleavage.
We previously described a mechanism by which reduced RNase H activity increased resistance to NRTIs by providing more time for excision of the incorporated NRTI (12, 34, 35). Some of the CN mutations identified in our studies also exhibited dual NRTI/NNRTI resistance (33). Similar to our previous report describing a mechanism of NRTI resistance (35), we now propose a new mechanism for NNRTI resistance (Fig. 3).
FIG. 3.
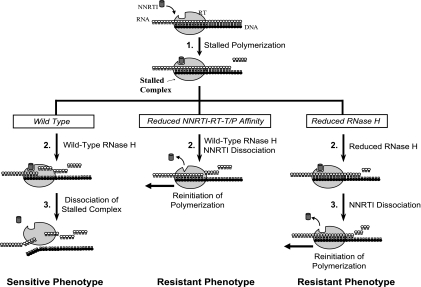
A model for NNRTI-mediated abrogation of HIV-1 replication. DNA primer strand and RNA template strand (stretches of black and white circles, respectively), RT (large gray oval), and NNRTI (dark gray cylinder) are shown schematically. The thick arrow reflects sequential events during HIV-1 replication in the presence of NNRTI. Once initiated, polymerization continues until NNRTI binds to RT-template-primer complex forming a PI complex. The left panel represents events for wild-type RT leading to NNRTI-sensitive phenotype; the middle and right panels represent events for RT with reduced affinity to NNRTI and RNase H-defective RT, which lead to NNRTI-resistant phenotype. The sequential events in the reverse transcription during NNRTI exposure include RNase H cleavage, dissociation of NNRTI, and either formation of a PI complex for NNRTI-susceptible phenotype or continuation of polymerization for NNRTI-resistant phenotype.
A schematic representation of sequential steps in reverse transcription during NNRTI exposure is outlined in Fig. 3, with the left panel representing the sensitive (wild-type) phenotype. NNRTIs abrogate DNA synthesis by binding to the RT and forming a polymerization-incompetent NNRTI-RT-template-primer (T/P) complex (PI complex) (Fig. 3, top panel, step 1). RNase H degradation of the template continues (Fig. 3, left panel, step 2) until the PI complex dissociates and reverse transcription is terminated (step 3). To become resistant to NNRTIs, RT could potentially evolve using two main pathways—reduced NNRTI-RT affinity (Fig. 3, middle panel) and reduced RNase H activity (Fig. 3, right panel)—which can also exist in combination. First, selection of mutations in the NNRTI BP that reduce NNRTI binding affinity to RT can lead to the resistance phenotype (Fig. 3, middle panel, step 2). Due to the reduced affinity and faster dissociation of NNRTI from RT, the PI complex exists for a shorter period of time, allowing polymerization to resume before significant template degradation occurs. Second, selection of mutations in RT that reduce RNase H cleavage can also lead to the resistance phenotype (Fig. 3, right panel). Due to reduced RNase H cleavage, the NNRTI-RT-T/P complex can remain intact for a longer period of time (Fig. 3, right panel, step 2), allowing more time for dissociation of NNRTI from the PI complex and subsequent reinitiation of reverse transcription (Fig. 3, right panel, step 3) and leading to the resistance phenotype.
Effect of D549N RNase H mutation on resistance to NVP, DLV, EFV, and ETR.
To test the predictions of our model, we analyzed the relationships between RNA template degradation and NNRTI resistance by using the D549N RNase H mutant, which is known to reduce RNase H activity (9). The D549N mutation increased resistance to NVP 5-fold and to DLV 1.8-fold in the context of a wild-type pol (Fig. 4A and B, WT). The observed increases in NVP and DLV resistance were consistent with the hypothesis that decreasing RNase H cleavage by the D549N mutation allowed sufficient time for NVP and DLV to dissociate from the PI complex before the template was significantly degraded, leading to the reinitiation of polymerization and the resistance phenotype. In contrast to the results obtained for NVP and DLV, the level of resistance to EFV and ETR was not increased by the D549N mutation in the context of a wild-type pol (Fig. 4C and D, WT). The NNRTI-RT-T/P dissociation constants (Kd) are available from the literature for NVP, DLV, and EFV (58) (Fig. 4E), but not ETR. We assume that the Kd for the ETR-RT complex is comparable to or lower than that for the EFV-RT complex, because the IC50s for NVP, DLV, and EFV correlate with their Kd values, and the IC50 for ETR (1.1 nM) is lower than that for EFV (1.9 nM) (Fig. 4E). We reasoned that because EFV and ETR have a much higher affinity for the RT (Kd for EFV, 2.6 nM) than do NVP and DLV (Kd, 25 nM and 16.6 nM, respectively), the EFV- or ETR-containing PI complexes are more stable than are the NVP- and DLV-containing PI complexes. Thus, we hypothesized that reducing RNase H activity by D549N mutation does not provide sufficient time for EFV or ETR to dissociate from the RT and resume DNA synthesis before the template is significantly degraded.
FIG. 4.
Effect of reduced RNase H activity on the resistance level to NNRTIs NVP (A), DLV (B), EFV (C), and ETR (D). Vertical bars represent IC50s (nM) for each drug; gray bars correspond to the IC50s by viruses bearing the wild-type RNase H domain; black bars represent viruses with the D549N mutation. Fold changes in IC50s versus wild-type control are indicated above each bar. Error bars represent the standard error for the results from 2 to 11 experiments. (E) Summary table for the Kd values for the NNRTI-RT complex from the work by Xia et al. (58) and IC50s for WT viruses. (F) Linear regression analysis of fold NNRTI resistance associated with NNRTI BP mutations in the presence (x axis) or absence of the D549N substitution (y axis). Fold resistance was calculated as a ratio between IC50s for virus containing specific mutation(s) versus WT control using resistance data from panels A, B, C, and D. The correlation coefficient (R) was determined using SigmaPlot 8.0 software.
Affinity of NNRTIs to the RT is a critical parameter in determining the influence of RNase H defective mutants on NNRTI resistance.
To test the hypothesis that the affinity of the NNRTI to the RT is a key component in determining whether reducing RNase H activity affects NNRTI resistance, we created NNRTI-resistant RT mutants with amino acid substitutions L100I, K103N, V106A, V179F, and Y181C in the NNRTI BP that alter the binding characteristics of the mutant RTs for the NNRTIs (8, 31, 47, 53, 61). The effects of most of these NNRTI resistance mutations, as well as other mutations, on NVP and EFV affinity to the RT are summarized in Table 2. The results from these previously published studies clearly demonstrate that the NNRTI BP mutations decrease the affinity of the mutant RTs to the NNRTIs.
TABLE 2.
Decreases in affinities of NVP and EFV to common NNRTI-resistant mutants of RTa
| NNRTI-resistant mutant | Fold change in affinities (Kimut/Kiwt) for: |
Reference | |
|---|---|---|---|
| EFV | NVP | ||
| L100I mutant | ND | 22.5 | 31 |
| 5 | 40 | 61 | |
| 5 | 17.5 | 47 | |
| 4 | 22.5 | 8 | |
| K103N mutant | ND | 17.5 | 31 |
| 20 | 22 | 61 | |
| 50 | 12.5 | 47 | |
| 5.3 | 17.5 | 8 | |
| V106A mutant | ND | 25 | 31 |
| 1.3 | 25 | 8 | |
| Y181C mutant | ND | 132b | 53 |
| Y181I mutant | ND | 90 | 31 |
| 30 | 50 | 61 | |
| 30 | 90 | 47 | |
| 5 | 90 | 8 | |
| V179D mutant | 5 | 50 | 61 |
| 3.3 | 5 | 8 | |
| Y188L mutant | ND | 45 | 31 |
| 12.6 | 45 | 8 | |
The fold changes in affinities (Kimut/Kiwt) for NVP and EFV were determined through measurements of the equilibrium dissociation constant (Ki = Koff/Kon) for ternary complexes.
These Kimut and Kiwt values were determined for the NVP-E-DNA ternary complex without magnesium.
These mutants were also created in combination with D549N to assay for the combined effects of both reducing NNRTI affinity and reducing RNase H cleavage. L100I, V106A, and K103N mutants alone exhibited NVP resistance increased 2-, 60-, and 116-fold, respectively (Fig. 4A); when these mutations were combined with D549N, NVP resistance was even further enhanced 6-, 950-, and 718-fold, respectively. Similar to the results obtained with NVP, the L100I, V106A, and K103N mutations alone increase resistance to DLV 37-, 15-, and 8-fold, respectively (Fig. 4B); when combined with D549N, these mutations further increased DLV resistance 61-, 65-, and 242-fold, respectively. Overall, the D549N mutation enhanced resistance to NVP and DLV in the context of a wild-type RT or in combination with NNRTI BP mutations.
We tested the same NNRTI BP mutants alone or in combination with D549N mutation for their resistance to EFV (Fig. 4C). We found that the L100I, V106A, and K103N increased resistance to EFV 9-, 1.4-, and 32-fold, respectively. In contrast to the results obtained with NVP and DLV, the L100I-plus-D549N and V106A-plus-D549N combinations of mutations did not further increase resistance to EFV; however, the combination of the K103N and D549N mutations resulted in an increase in EFV resistance from 32- to 161-fold (Fig. 4C). The L100I and V106A mutations reduced the affinity of EFV for the RT-T/P-dTTP 5-fold, whereas the K103N mutation reduced the EFV affinity to the RT-T/P-dTTP 20- to 50-fold in the same studies (Table 2) (47, 61). We hypothesize that as a consequence of the reduced affinity, the D549N mutation, when combined with the K103N mutation, provided sufficient time for EFV to dissociate from the RT-T/P before significant template degradation, allowing resumption of DNA synthesis. On the other hand, the smaller changes in EFV affinity induced by the L100I and V106A mutations, in combination with D549N, did not provide sufficient time for EFV to dissociate from the RT-T/P complex before significant template degradation; as a result, DNA synthesis could not be reinitiated, and EFV resistance was not increased.
ETR is a new NNRTI recently approved for the treatment of HIV infection. It is a potent inhibitor of viral replication, and RT mutants containing standard NNRTI resistance mutations remain sensitive to inhibition by ETR (1, 56). As reported previously, the K103N mutation does not increase resistance to ETR (Fig. 4D). Because single NNRTI resistance mutations do not significantly increase resistance to ETR (56), we generated mutants containing a combination of L100I plus K103N plus Y181C and V179F plus Y181C. Consistent with a previous report (56), the L100I plus K103N plus Y181C combination increased ETR resistance 41-fold, and the V179F plus Y181C combination increased ETR resistance 87-fold (Fig. 4D); combining these mutations with the D549N mutation further increased ETR resistance to 309- and 300-fold, respectively.
Analysis of relationships between the increases in NNRTI resistance caused by the NNRTI BP mutations in the context of wild-type RNase H or in the presence of D549N mutation revealed a very strong correlation between the fold changes in resistance when comparing the absence and presence of the D549N mutation (R2 = 0.858; t test, P < 0.0001).
To rule out the possibility that the results shown in Fig. 4 were not due to a structural alteration specific to the D549N mutation, we analyzed the effects of Q475A and Y501A, which are RNase H primer grip mutations that also reduce RNase H activity (11), on NNRTI resistance in the context of a wild-type pol domain. We found that Q475A and Y501A increased resistance to NNRTIs NVP (9- to 15-fold), DLV (3- to 6-fold), and EFV (1.5- to 1.7-fold), but not ETR, which correlated with the affinities of the NNRTIs to the RT. Overall, these results support the predictions of our NNRTI resistance model and demonstrate a correlation between the affinity of the NNRTIs to the RT and the ability of the mutations D549N, Q475A, and Y501A to further enhance NNRTI resistance (data not shown).
To determine whether the PI complex can resume DNA synthesis after dissociation from the NNRTI, we analyzed the effects of NNRTIs on RT switching frequency by using an in vivo assay (36). We observed that ETR and EFV increased the wild-type RT template-switching frequency only 1.4- to 1.5-fold, whereas DLV and NVP, which bind to RT with a lower affinity, increased the RT template-switching frequency 2.0- and 2.3-fold. Interestingly, EFV increased the RT template-switching frequency of the K103N mutant RT to a greater extent than it did WT RT (2.5-fold) (data not shown). These observations suggested that the reduced affinity of EFV to the K103N mutant RT allows the template-primer complex to resume DNA synthesis more efficiently (data not shown).
Mutations in RNase H, CNs isolated from patient-derived RTs, and mutations in the NNRTI BP reduce in vitro RNase H cleavage activity.
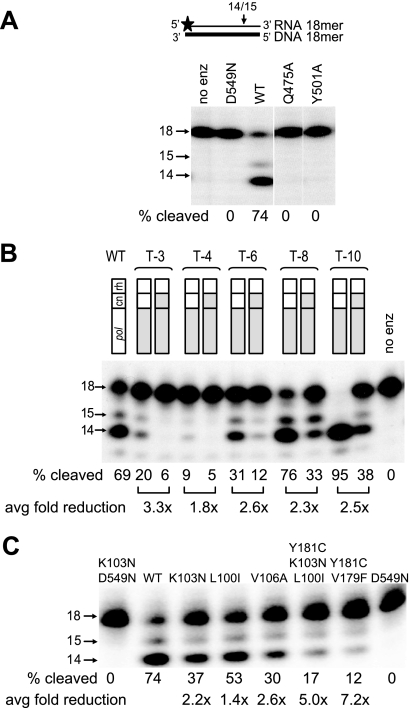
To confirm that mutations D549N, Q475A, and Y501A, which increased NNRTI resistance, also decreased RNase H cleavage, we performed in vitro RNase H cleavage assays (Fig. 5A). RNase H primary cleavages were analyzed using an 18-mer RNA-DNA hybrid and virion-associated RTs as previously described (12). Cleavage of the RNA in the RNA-DNA hybrid resulted in the formation of 14- and 15-nucleotide (nt) RNA fragments. The results showed that compared to the WT RT, the D549N, Q475A, and Y501A mutants were all severely defective in their ability to carry out RNase H primary cleavage.
FIG. 5.
RNase H cleavage of NNRTI-resistant mutants. (A) RNase H primary cleavages for D549N, Q475A, and Y501A mutants. The assay is schematically depicted above the gel autoradiogram; horizontal arrows show the size of the RNA product. The average percentages of cleaved product (proportion of 15- and 14-nt products) are shown for the representative gel from two independent experiments. (B) RNase H primary cleavages for the patients' RT pol alone and in combination with patient's CN. Fold reduction in RNase H cleavages is calculated for each pair of RTs (connected by a bracket) and presented below as an average of the results from five independent experiments. (C) RNase H cleavages by mutant RTs containing NNRTI BP resistant mutations. The percentages of cleaved product (proportion of 15- and 14-nt products) are shown for the representative gel. Fold reduction in cleavages is calculated versus WT control. Average fold reduction was calculated from the results for five independent experiments.
We previously observed that the CNs obtained from patient-derived RTs reduced RNase H cleavage in combination with TAMs (12). To determine whether the CNs obtained from patient-derived RTs reduced RNase H cleavage in combination with the patient-derived RT pol domains, we performed in vitro RNase H cleavage assays (Fig. 5B). We found that the patient-derived RT pol domains in combination with wild-type CN and RH exhibited reduced RNase H cleavage activity for patients T3, T-4, and T-6, but not for patients T-8 or T-10. Combining the patient-derived CN domains with the patient-derived RT pol resulted in a further reduction in RNase H cleavage from 1.8- to 3.3-fold for all five patients. The results confirmed that mutations in the CNs isolated from patient-derived RTs further reduced RNase H cleavage activity.
The results obtained with the RT pol domains from patients T-3, T-4, and T-6, in the context of wild-type CN and RH domains, showed that mutations in these pol domains reduced RNase H cleavage activity (Fig. 5B). It has been previously observed that mutations in the NNRTI BP and elsewhere in pol can reduce RNase H cleavage activity (2, 3, 14, 19, 20, 27, 38, 46, 57). To determine whether the mutations in the NNRTI BP in these patients' pol domains reduced RNase H cleavage activity, we tested the effects of several NNRTI BP mutations (Fig. 5C). The results showed that NNRTI BP mutations reduced primary RNase H cleavage activity by 2- to 7-fold. These results showed that NNRTI BP mutations can directly reduce RNase H activity and may confer resistance to NNRTIs by reducing RNase H cleavage activity as well as by reducing the affinity of the NNRTIs to the RT.
DISCUSSION
Based on the growing evidence for the existence of dual NRTI and NNRTI resistance (4, 23, 24, 33, 40, 59), we have explored the mechanism by which mutations in RT can confer dual resistance. Here, we propose a new unifying mechanism for NNRTI resistance that explains the dual NRTI and NNRTI resistance conferred by mutations in the CN and RNase H of RT. We suggest that there are analogous steps in reverse transcription that are affected by both NRTI and NNRTI exposure, and both classes of drugs block reverse transcription by making PI complexes. To reinitiate polymerization, the incorporated NRTI or the bound NNRTI is either excised from the blocked primer or dissociated from the RT-T/P complex, respectively. We previously demonstrated that reducing RNase H cleavages substantially increases resistance to some NRTIs, such as AZT and d4T, by providing more time for excision of the incorporated NRTI, resulting in more efficient reinitiation of DNA synthesis (35). In our NNRTI resistance model, reducing RNase H cleavages allows more time for dissociation of the NNRTI from the PI complex, leading to more efficient reinitiation of DNA synthesis. These similar mechanisms can explain how some mutations that reduce RNase H activity can confer dual NRTI and NNRTI resistance.
Our results strongly suggest that the affinity of the NNRTI is an important factor in determining whether reducing RNase H activity will enhance NNRTI resistance. We propose that EFV and ETR, which have a higher affinity for the RT than do NVP and DLV, form high-affinity “dead-end” complexes that are unable to reinitiate DNA synthesis, even after reducing RNase H cleavage. We reason that the off rates of EFV and ETR may be significantly lower than those of NVP and DLV; the low rates of EFV and ETR dissociation do not allow sufficient time for resumption of DNA synthesis within the time frame in which viral DNA synthesis must be completed. The observation that mutations in the NNRTI BP that increased EFV and ETR resistance were sensitive to the resistance-enhancing effect of RNase H cleavage activity is consistent with this hypothesis. The NNRTI BP mutations analyzed in our studies were previously shown to reduce the affinity of NVP and EFV to the RT and RT-T/P (8, 31, 47, 53, 61). Although there is no biochemical data available at this time, it is likely that NNRTI BP mutations that confer resistance to DLV and ETR also reduce the affinity of these drugs to the mutant RTs (10, 16).
Our studies indicate that there is a balance between NNRTI affinity to the RT and template RNA degradation. We observed that different NNRTI BP mutations, which are expected to reduce the affinity of NNRTIs to the RT, in combination with the D549N RNase H-deficient mutation resulted in an increased level of NNRTI resistance. Therefore, the interplay between RNase H cleavages and affinity of the NNRTI to RT is important in determining the level of NNRTI resistance. The effect of NNRTI exposure on the frequency of template switching provides additional evidence for the existence of this balance.
The impact of RNase H-defective mutants on the levels of resistance was different for the different classes of drugs. In general, reducing RNase H activity had a greater impact on AZT resistance than on NNRTI resistance. Only two of the 10 RNase H primer grip mutations tested, Q475A and Y501A, increased resistance to NNRTIs, while all 10 increased AZT resistance (data not shown). Q475A and Q501A mutants exhibited the highest increases in AZT resistance, suggesting that substantial reductions in RNase H activity are required to increase resistance to NNRTIs. The increases in NNRTI resistance observed by CN mutations are a few fold, which is less than the effects observed with NNRTI BP mutations, such as K103N. The clinical significance of these mutations is not known at present. However, even small differences in NNRTI resistance could provide a selective advantage to the virus, which could facilitate the selection of highly resistant variants.
CN and RNase H mutations could reduce RNase H activity through different mechanisms that ultimately enhance NNRTI resistance. It was recently shown that CN mutations N348I, A360V, and Q509L reduce the positioning of the RT in the RNase H cleavage mode, especially on the short hybrids, leading to the reduction of RNase H activity and stabilizing the binding of RT in the polymerase-dependent mode (7, 15). It is likely that both reduced RNase H and increased binding in the polymerase-dependent mode enhance NRTI and NNRTI resistance.
While our results indicate that the CN mutations enhance dual NRTI/NNRTI resistance by reducing RNase H activity, it is possible that these mutations affect other properties of RT that also contribute to drug resistance. It has been proposed that T369I and N348I mutants confer dual NRTI and NNRTI resistance by affecting the RT heterodimer stability (23). While a recent study did not reveal any obvious correlation between RT dimer stability and NNRTI resistance (18), further studies are needed to determine the relationship between RT dimerization, RNase H activity, the balance between polymerase and RNase H activities, and NRTI/NNRTI drug resistance.
In another study, it was observed that upon binding to an NNRTI, RT is positioned on the template-primer in an orientation that favors RNase H cleavage, rather than the orientation that favors DNA synthesis (22). In addition to the role of CN mutations described in our NNRTI resistance model, these mutations could potentially counteract the ability of NNRTIs to enhance RNase H cleavages. Assuming that NNRTIs inhibit viral replication, in part by enhancing RNase H cleavages, the CN mutations may increase NNRTI resistance by counteracting the RNase H cleavage-enhancing effect of the NNRTIs.
The reduction of RNase H activity by NNRTI BP mutations (2, 3, 17-19, 49, 57) was previously thought to primarily diminish viral fitness, suppressing the selection of the NNRTI BP mutations during therapy. We propose here that the NNRTI BP mutations are selected in part because they reduce RNase H activity, which in turn enhances NNRTI resistance. The NNRTI BP mutations may reduce RNase H activity by affecting the template-primer binding or affecting its positioning at the RNase H active site; NNRTI BP mutations localized in the p51 subunit could potentially also contribute to the observed effect. We speculate that a large number of mutations in the NNRTI BP can reduce NNRTI affinity and enhance resistance, that a subset of these mutations also reduce RNase H activity, and that these mutations are preferentially selected in response to therapy because they affect both NNRTI affinity and RNase H activity.
The prevalence of dual NRTI/NNRTI resistance mutations, such as N348I, T369I, and E399D, in the treatment-experienced patients is 10 to 16% (23, 24, 33, 43, 59), which is comparable to the prevalence of other drug resistance mutations (30). Additionally, mutations that confer low or intermediate levels of resistance to ETR are quite common (30). Our results suggest that CN mutations will have little or no effect on ETR resistance unless they are combined with NNRTI BP mutations that confer ETR resistance. However, clinical observations and in vitro selection experiments highlight the role of CN mutations in ETR resistance. For example, the E399D mutation that reduced sensitivity to AZT and EFV (23) was associated with a 14.4-fold reduction in sensitivity to ETR (43), emphasizing the potential impact for the mutations from CN on ETR susceptibility. CN mutation T386A was also selected in the presence of ETR during in vitro experiments, suggesting a role in ETR resistance (56).
These results from our in vivo assays, in vitro RNase H activity assays, as well as previously published NNRTI-RT affinities, provide strong correlative data that support the NNRTI resistance model. Biochemical studies directly demonstrating that the RT template-primer complex remains intact, allowing resumption of DNA synthesis after the inhibitor has dissociated from the complex, would provide direct evidence to support this key feature of the model. In summary, this proposed mechanism explains how the same mutations in the CN can reduce sensitivity to structurally and functionally distinct NRTIs and NNRTIs, highlighting the importance of interactions between distinct parts of RT that play a significant role in the evolution of the drug resistance.
Acknowledgments
We especially thank Wei-Shau Hu for intellectual input throughout the project, John Coffin for valuable discussion of results, and Steve Hughes for critical comments during manuscript preparation.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mentioning of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Andries, K., H. Azijn, T. Thielemans, D. Ludovici, M. Kukla, J. Heeres, P. Janssen, B. De Corte, J. Vingerhoets, R. Pauwels, and M. P. de Bethune. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, R. H., M. Wisniewski, R. A. Bambara, and L. M. Demeter. 2001. The Y181C mutant of HIV-1 reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors alters the size distribution of RNase H cleavages. Biochemistry 40:4087-4095. [DOI] [PubMed] [Google Scholar]
- 4.Baldanti, F., S. Paolucci, G. Maga, N. Labo, U. Hubscher, A. Y. Skoblov, L. Victorova, S. Spadari, L. Minoli, and G. Gerna. 2003. Nevirapine-selected mutations Y181I/C of HIV-1 reverse transcriptase confer cross-resistance to stavudine. AIDS 17:1568-1570. [DOI] [PubMed] [Google Scholar]
- 5.Blanca, G., F. Baldanti, S. Paolucci, A. Y. Skoblov, L. Victorova, U. Hubscher, G. Gerna, S. Spadari, and G. Maga. 2003. Nevirapine resistance mutation at codon 181 of the HIV-1 reverse transcriptase confers stavudine resistance by increasing nucleotide substrate discrimination and phosphorolytic activity. J. Biol. Chem. 278:15469-15472. [DOI] [PubMed] [Google Scholar]
- 6.Brehm, J. H., D. Koontz, J. D. Meteer, V. Pathak, N. Sluis-Cremer, and J. W. Mellors. 2007. Selection of mutations in the connection and RNase H domains of human immunodeficiency virus type 1 reverse transcriptase that increase resistance to 3′-azido-3′-dideoxythymidine. J. Virol. 81:7852-7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehm, J. H., J. W. Mellors, and N. Sluis-Cremer. 2008. Mechanism by which a glutamine to leucine substitution at residue 509 in the ribonuclease H domain of HIV-1 reverse transcriptase confers zidovudine resistance. Biochemistry 47:14020-14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butini, S., M. Brindisi, S. Cosconati, L. Marinelli, G. Borrelli, S. S. Coccone, A. Ramunno, G. Campiani, E. Novellino, S. Zanoli, A. Samuele, G. Giorgi, A. Bergamini, M. Di Mattia, S. Lalli, B. Galletti, S. Gemma, and G. Maga. 2009. Specific targeting of highly conserved residues in the HIV-1 reverse transcriptase primer grip region. 2. Stereoselective interaction to overcome the effects of drug resistant mutations. J. Med. Chem. 52:1224-1228. [DOI] [PubMed] [Google Scholar]
- 9.Cristofaro, J. V., J. W. Rausch, S. F. Le Grice, and J. J. DeStefano. 2002. Mutations in the ribonuclease H active site of HIV-RT reveal a role for this site in stabilizing enzyme-primer-template binding. Biochemistry 41:10968-10975. [DOI] [PubMed] [Google Scholar]
- 10.Das, K., A. D. Clark, Jr., P. J. Lewi, J. Heeres, M. R. De Jonge, L. M. Koymans, H. M. Vinkers, F. Daeyaert, D. W. Ludovici, M. J. Kukla, B. De Corte, R. W. Kavash, C. Y. Ho, H. Ye, M. A. Lichtenstein, K. Andries, R. Pauwels, M. P. De Bethune, P. L. Boyer, P. Clark, S. H. Hughes, P. A. Janssen, and E. Arnold. 2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 47:2550-2560. [DOI] [PubMed] [Google Scholar]
- 11.Delviks-Frankenberry, K. A., G. N. Nikolenko, R. Barr, and V. K. Pathak. 2007. Mutations in human immunodeficiency virus type 1 RNase H primer grip enhance 3′-azido-3′-deoxythymidine resistance. J. Virol. 81:6837-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delviks-Frankenberry, K. A., G. N. Nikolenko, P. L. Boyer, S. H. Hughes, J. M. Coffin, A. Jere, and V. K. Pathak. 2008. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc. Natl. Acad. Sci. U. S. A. 105:10943-10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delviks-Frankenberry, K. A., G. N. Nikolenko, F. Maldarelli, S. Hase, Y. Takebe, and V. K. Pathak. 2009. Subtype-specific differences in the human immunodeficiency virus type 1 reverse transcriptase connection subdomain of CRF01_AE are associated with higher levels of resistance to 3′-azido-3′-deoxythymidine. J. Virol. 83:8502-8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domaoal, R. A., and L. M. Demeter. 2004. Structural and biochemical effects of human immunodeficiency virus mutants resistant to non-nucleoside reverse transcriptase inhibitors. Int. J. Biochem. Cell Biol. 36:1735-1751. [DOI] [PubMed] [Google Scholar]
- 15.Ehteshami, M., G. L. Beilhartz, B. J. Scarth, E. P. Tchesnokov, S. McCormick, B. Wynhoven, P. R. Harrigan, and M. Gotte. 2008. Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3′-azido-3′-deoxythymidine through both RNase H-dependent and -independent mechanisms. J. Biol. Chem. 283:22222-22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esnouf, R. M., J. Ren, A. L. Hopkins, C. K. Ross, E. Y. Jones, D. K. Stammers, and D. I. Stuart. 1997. Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor. Proc. Natl. Acad. Sci. U. S. A. 94:3984-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan, N., K. B. Rank, D. E. Slade, S. M. Poppe, D. B. Evans, L. A. Kopta, R. A. Olmsted, R. C. Thomas, W. G. Tarpley, and S. K. Sharma. 1996. A drug resistance mutation in the inhibitor binding pocket of human immunodeficiency virus type 1 reverse transcriptase impairs DNA synthesis and RNA degradation. Biochemistry 35:9737-9745. [DOI] [PubMed] [Google Scholar]
- 18.Figueiredo, A., S. Zelina, N. Sluis-Cremer, and G. Tachedjian. 2008. Impact of residues in the nonnucleoside reverse transcriptase inhibitor binding pocket on HIV-1 reverse transcriptase heterodimer stability. Curr. HIV Res. 6:130-137. [DOI] [PubMed] [Google Scholar]
- 19.Gerondelis, P., R. H. Archer, C. Palaniappan, R. C. Reichman, P. J. Fay, R. A. Bambara, and L. M. Demeter. 1999. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J. Virol. 73:5803-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh, M., P. S. Jacques, D. W. Rodgers, M. Ottman, J. L. Darlix, and S. F. Le Grice. 1996. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 35:8553-8562. [DOI] [PubMed] [Google Scholar]
- 21.Gopalakrishnan, V., and S. Benkovic. 1994. Effect of a thiobenzimidazolone derivative on DNA strand transfer catalyzed by HIV-1 reverse transcriptase. J. Biol. Chem. 269:4110-4115. [PubMed] [Google Scholar]
- 22.Grobler, J. A., G. Dornadula, M. R. Rice, A. L. Simcoe, D. J. Hazuda, and M. D. Miller. 2007. HIV-1 reverse transcriptase plus-strand initiation exhibits preferential sensitivity to non-nucleoside reverse transcriptase inhibitors in vitro. J. Biol. Chem. 282:8005-8010. [DOI] [PubMed] [Google Scholar]
- 23.Gupta, S., S. Fransen, E. E. Paxinos, W. Huang, E. Stawiski, C. J. Petropoulos, and N. T. Parkin. 2006. Infrequent occurrence of mutations in the C-terminal region of reverse transcriptase modulates susceptibility to RT inhibitors. Antiviral Ther. 11:S143-S143. [Google Scholar]
- 24.Hachiya, A., E. N. Kodama, S. G. Sarafianos, M. M. Schuckmann, Y. Sakagami, M. Matsuoka, M. Takiguchi, H. Gatanaga, and S. Oka. 2008. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 82:3261-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hang, J. Q., Y. Li, Y. Yang, N. Cammack, T. Mirzadegan, and K. Klumpp. 2007. Substrate-dependent inhibition or stimulation of HIV RNase H activity by non-nucleoside reverse transcriptase inhibitors (NNRTIs). Biochem. Biophys. Res. Commun. 352:341-350. [DOI] [PubMed] [Google Scholar]
- 26.Hsiou, Y., J. Ding, K. Das, A. D. Clark, Jr., P. L. Boyer, P. Lewi, P. A. Janssen, J. P. Kleim, M. Rosner, S. H. Hughes, and E. Arnold. 2001. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J. Mol. Biol. 309:437-445. [DOI] [PubMed] [Google Scholar]
- 27.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2003. Mutation of amino acids in the connection domain of human immunodeficiency virus type 1 reverse transcriptase that contact the template-primer affects RNase H activity. J. Virol. 77:8548-8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Kemp, S. D., C. Shi, S. Bloor, P. R. Harrigan, J. W. Mellors, and B. A. Larder. 1998. A novel polymorphism at codon 333 of human immunodeficiency virus type 1 reverse transcriptase can facilitate dual resistance to zidovudine and L-2′,3′-dideoxy-3′-thiacytidine. J. Virol. 72:5093-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llibre, J. M., J. R. Santos, T. Puig, J. Molto, L. Ruiz, R. Paredes, and B. Clotet. 2008. Prevalence of etravirine-associated mutations in clinical samples with resistance to nevirapine and efavirenz. J. Antimicrob. Chemother. 62:909-913. [DOI] [PubMed] [Google Scholar]
- 31.Maga, G., M. Amacker, N. Ruel, U. Hubscher, and S. Spadari. 1997. Resistance to nevirapine of HIV-1 reverse transcriptase mutants: loss of stabilizing interactions and thermodynamic or steric barriers are induced by different single amino acid substitutions. J. Mol. Biol. 274:738-747. [DOI] [PubMed] [Google Scholar]
- 32.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. U. S. A. 93:11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolenko, G. N., K. A. Delviks-Frankenberry, A. Jere, and V. K. Pathak. 2008. Mutations in the reverse transcriptase connection and RNase H domains exhibit dual resistance to nucleoside and non-nucleoside reverse transcriptase inhibitors. Antivir. Ther. 13:A55-A55. [Google Scholar]
- 34.Nikolenko, G. N., K. A. Delviks-Frankenberry, S. Palmer, F. Maldarelli, M. J. Fivash, Jr., J. M. Coffin, and V. K. Pathak. 2007. Mutations in the connection domain of HIV-1 reverse transcriptase increase 3′-azido-3′-deoxythymidine resistance. Proc. Natl. Acad. Sci. U. S. A. 104:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolenko, G. N., S. Palmer, F. Maldarelli, J. W. Mellors, J. M. Coffin, and V. K. Pathak. 2005. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: balance between RNase H activity and nucleotide excision. Proc. Natl. Acad. Sci. U. S. A. 102:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolenko, G. N., E. S. Svarovskaia, K. A. Delviks, and V. K. Pathak. 2004. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J. Virol. 78:8761-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palaniappan, C., P. J. Fay, and R. A. Bambara. 1995. Nevirapine alters the cleavage specificity of ribonuclease H of human immunodeficiency virus 1 reverse transcriptase. J. Biol. Chem. 270:4861-4869. [DOI] [PubMed] [Google Scholar]
- 38.Palaniappan, C., M. Wisniewski, P. S. Jacques, S. F. Le Grice, P. J. Fay, and R. A. Bambara. 1997. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J. Biol. Chem. 272:11157-11164. [DOI] [PubMed] [Google Scholar]
- 39.Paolucci, S., F. Baldanti, G. Campanini, R. Cancio, A. Belfiore, G. Maga, and G. Gerna. 2007. NNRTI-selected mutations at codon 190 of human immunodeficiency virus type 1 reverse transcriptase decrease susceptibility to stavudine and zidovudine. Antiviral Res. 76:99-103. [DOI] [PubMed] [Google Scholar]
- 40.Paolucci, S., F. Baldanti, G. Maga, R. Cancio, M. Zazzi, M. Zavattoni, A. Chiesa, S. Spadari, and G. Gerna. 2004. Gln145Met/Leu changes in human immunodeficiency virus type 1 reverse transcriptase confer resistance to nucleoside and nonnucleoside analogs and impair virus replication. Antimicrob. Agents Chemother. 48:4611-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paolucci, S., F. Baldanti, M. Tinelli, G. Maga, and G. Gerna. 2003. Detection of a new HIV-1 reverse transcriptase mutation (Q145M) conferring resistance to nucleoside and non-nucleoside inhibitors in a patient failing highly active antiretroviral therapy. AIDS 17:924-927. [DOI] [PubMed] [Google Scholar]
- 42.Parniak, M. A., K. L. Min, S. R. Budihas, S. F. Le Grice, and J. A. Beutler. 2003. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal. Biochem. 322:33-39. [DOI] [PubMed] [Google Scholar]
- 43.Poveda, E., C. de Mendoza, T. Pattery, M. Gonzalez Mdel, J. Villacian, and V. Soriano. 2008. Phenotypic impact of resistance mutations on etravirine susceptibility in HIV patients with prior failure to nonnucleoside analogues. AIDS 22:2395-2398. [DOI] [PubMed] [Google Scholar]
- 44.Reference deleted.
- 45.Radzio, J., and N. Sluis-Cremer. 2008. Efavirenz accelerates HIV-1 reverse transcriptase ribonuclease H cleavage, leading to diminished zidovudine excision. Mol. Pharmacol. 73:601-606. [DOI] [PubMed] [Google Scholar]
- 46.Rausch, J. W., D. Lener, J. T. Miller, J. G. Julias, S. H. Hughes, and S. F. Le Grice. 2002. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry 41:4856-4865. [DOI] [PubMed] [Google Scholar]
- 47.Samuele, A., A. Kataropoulou, M. Viola, S. Zanoli, G. La Regina, F. Piscitelli, R. Silvestri, and G. Maga. 2009. Non-nucleoside HIV-1 reverse transcriptase inhibitors di-halo-indolyl aryl sulfones achieve tight binding to drug-resistant mutants by targeting the enzyme-substrate complex. Antiviral Res. 81:47-55. [DOI] [PubMed] [Google Scholar]
- 48.Sarafianos, S. G., B. Marchand, K. Das, D. M. Himmel, M. A. Parniak, S. H. Hughes, and E. Arnold. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma, S. K., N. Fan, K. B. Rank, L. A. Kopta, R. A. Olmsted, S. M. Poppe, D. E. Slade, R. C. Thomas, and W. G. Tarpley. 1995. A drug-resistance mutation (G190E) in the inhibitor binding pocket impairs both RNA-dependent DNA polymerase and ribonuclease H activities of HIV-1 reverse transcriptase. J. Acquir. Immune Defic. Syndr. Hum. Retrovir. 10:2. [Google Scholar]
- 50.Shaw-Reid, C. A., B. Feuston, V. Munshi, K. Getty, J. Krueger, D. J. Hazuda, M. A. Parniak, M. D. Miller, and D. Lewis. 2005. Dissecting the effects of DNA polymerase and ribonuclease H inhibitor combinations on HIV-1 reverse-transcriptase activities. Biochemistry 44:1595-1606. [DOI] [PubMed] [Google Scholar]
- 51.Sluis-Cremer, N., and G. Tachedjian. 2008. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res. 134:147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sluis-Cremer, N., N. A. Temiz, and I. Bahar. 2004. Conformational changes in HIV-1 reverse transcriptase induced by nonnucleoside reverse transcriptase inhibitor binding. Curr. HIV Res. 2:323-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spence, R. A., K. S. Anderson, and K. A. Johnson. 1996. HIV-1 reverse transcriptase resistance to nonnucleoside inhibitors. Biochemistry 35:1054-1063. [DOI] [PubMed] [Google Scholar]
- 54.Spence, R. A., W. M. Kati, K. S. Anderson, and K. A. Johnson. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267:988-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srivastava, S., N. Sluis-Cremer, and G. Tachedjian. 2006. Dimerization of human immunodeficiency virus type 1 reverse transcriptase as an antiviral target. Curr. Pharm. Des. 12:1879-1894. [DOI] [PubMed] [Google Scholar]
- 56.Vingerhoets, J., H. Azijn, E. Fransen, I. De Baere, L. Smeulders, D. Jochmans, K. Andries, R. Pauwels, and M. P. de Bethune. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, J., C. Dykes, R. A. Domaoal, C. E. Koval, R. A. Bambara, and L. M. Demeter. 2006. The HIV-1 reverse transcriptase mutants G190S and G190A, which confer resistance to non-nucleoside reverse transcriptase inhibitors, demonstrate reductions in RNase H activity and DNA synthesis from tRNA(Lys, 3) that correlate with reductions in replication efficiency. Virology 348:462-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia, Q., J. Radzio, K. S. Anderson, and N. Sluis-Cremer. 2007. Probing nonnucleoside inhibitor-induced active-site distortion in HIV-1 reverse transcriptase by transient kinetic analyses. Protein Sci. 16:1728-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yap, S. H., C. W. Sheen, J. Fahey, M. Zanin, D. Tyssen, V. D. Lima, B. Wynhoven, M. Kuiper, N. Sluis-Cremer, P. R. Harrigan, and G. Tachedjian. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yee, J. K., A. Miyanohara, P. LaPorte, K. Bouic, J. C. Burns, and T. Friedmann. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 91:9564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zanoli, S., S. Gemma, S. Butini, M. Brindisi, B. P. Joshi, G. Campiani, C. Fattorusso, M. Persico, E. Crespan, R. Cancio, S. Spadari, U. Hubscher, and G. Maga. 2008. Selective targeting of the HIV-1 reverse transcriptase catalytic complex through interaction with the “primer grip” region by pyrrolobenzoxazepinone non-nucleoside inhibitors correlates with increased activity towards drug-resistant mutants. Biochem. Pharmacol. 76:156-168. [DOI] [PubMed] [Google Scholar]
- 62.Zelina, S., C. W. Sheen, J. Radzio, J. W. Mellors, and N. Sluis-Cremer. 2008. Mechanisms by which the G333D mutation in human immunodeficiency virus type 1 reverse transcriptase facilitates dual resistance to zidovudine and lamivudine. Antimicrob. Agents Chemother. 52:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]