Abstract
It has previously been demonstrated that, during human cytomegalovirus infection, the viral IE2 86 and IE2 40 proteins are both important for the expression of an early-late viral protein, UL84. Here, we show that expression of the UL84 protein is enhanced upon cotransfection with either IE2 86 or IE2 40, although IE2 40 appears to play a more important role. The UL84 protein levels are tightly linked to the amount of IE2 40 present, but this does not appear to be true for IE2 86. RNA remains constant for all corresponding proteins, indicating posttranscriptional regulation of UL84. The first 105 amino acids of UL84 are necessary and sufficient for this phenotype, and this region is also required for an interaction with IE2 86 and IE2 40. Treatment with proteasome inhibitors shows that UL84 exhibits some proteasome-dependent degradation, and UL84 is not protected against this degradation when coexpressed with IE2 86 or IE2 40. UL84 also exhibits an inhibitory effect on IE2 86 and IE2 40 protein levels in these cotransfection assays. Further, we show that the amino acid sequence of UL84 is important for the enhancement governed by IE2 40. These results indicate that IE2 86, IE2 40, and UL84 serve to regulate protein expression in a posttranscriptional fashion and that this regulation is independent of other viral proteins.
Human cytomegalovirus (HCMV), a betaherpesvirus, is the leading viral cause of birth defects and poses a severe threat to immunocompromised individuals. It has a 240-kbp genome that includes at least 150 different open reading frames (ORFs), only a small portion of which have been well characterized. Gene expression occurs in a temporally controlled fashion, and genes are divided into three major classes: immediate-early (IE), early, and late genes (reviewed in reference 34). The IE genes serve to shut down host cell defenses and activate expression of early viral genes, while early and late genes serve primarily in viral replication and structure and assembly of the virus, respectively. Two major IE (MIE) gene products, IE1 72 and IE2 86, are encoded by the UL122-123 coding region. The IE2 86 protein is essential for viral replication and plays a role in transactivating viral early promoters, facilitating repression of its own promoter and regulating expression of many host cellular genes in order to allow proper progression of viral infection. IE1 72, while nonessential for infection at high multiplicity, has also been shown to be important for activating viral and cellular protein expression. Both proteins arise from the same primary transcript, which is alternatively spliced to produce exons 1 to 4 for IE1 72 and exons 1 to 3 and 5 for IE2 86.
Several functional roles have been ascribed to specific domains within IE2 86. DNA binding associated with transcriptional autorepression has been shown to involve amino acids (aa) 290 to 579, while the regions from aa 1 to 98 and 170 to 579 appear to be required for some transcriptional activation functions (7, 29, 33, 38, 47-49, 52, 67). In addition, IE2 86 has been shown to interact with many cellular factors in vitro, but most of these have not been confirmed in the context of infection (for a review, see reference 12).
At late times in the infection, smaller RNAs arise from exon 5 of the IE2 86 gene and encode proteins IE2 60 and IE2 40. The IE2 60 (60-kDA) protein begins translation at methionine 170 of IE2 86, while the IE2 40 (40-kDA) protein initiates at methionine 242. The amino acid numbering system corresponds to the Towne strain of HCMV. A putative TATAA promoter sequence for the RNA specifying IE2 60 lies in the intron between exons 4 and 5, while the predicted promoter for the transcript yielding the IE2 40 protein (and some IE2 60 protein) is located just upstream of the IE2 60 translation initiation site. These proteins have previously been shown to have a role in the transactivation of late genes, as well as repression of the MIE promoter (24, 43, 62). Our lab has extensively studied the roles of these two proteins, particularly at the later stages of infection. Loss of the IE2 60 and IE2 40 proteins was shown to alter the expression of both IE1 72 and IE2 86. We have also identified a role for the IE2 60 and IE2 40 proteins in the expression of two early-late viral proteins, UL83 (pp65) and UL84, and have documented that the loss of these smaller IE2 proteins results in a greater-than-10-fold reduction in viral titer (62).
The regulation of UL84 by the IE2 family of proteins has been of great interest, as UL84 is the only viral protein that has been found to interact with IE2 86 and is the major protein in complex with IE2 86 during infection (10, 23, 41, 43, 50). Low levels of UL84 are present at early times, and the abundance increases significantly after the onset of viral DNA replication (20). UL84 has been shown to have a role in ori-Lyt-dependent replication and downregulation of IE2 86-dependent transactivation functions in transient transfection assays (16) and it is able to interact with the RNA stem-loop sequence within the RNA/DNA hybrid region of ori-Lyt (9). UL84 has also been shown to have homology to the DExD/H box family of proteins and exhibits UTPase activity (11). We have previously shown that UL84 interacts with IE2 86 throughout infection and that IE2 60 and IE2 40 can individually interact with UL84 (43). Further, it has also been proposed that UL84 interacts with a number of other viral and cellular proteins during the course of infection (14-15, 54).
Using IE2 mutant viruses, we determined that loss of the IE2 60 and IE2 40 proteins resulted in a significant loss of UL84 protein expression, and this loss was shown to be posttranscriptional (43, 62). Furthermore, a mutant virus containing a deletion of aa 136 to 290 of IE2 86 (termed IE2 ΔSX), which also does not express IE2 60 or IE2 40, showed similar results (43). In these studies, it was determined that IE2 40 played a more important role in governing UL84 expression at the later stages of infection, given that loss of IE2 40 alone resulted in a significant loss of UL84 (62). Further characterization of the mechanism governing UL84 expression revealed that UL84 RNA could be exported to the cytoplasm and loaded onto the polyribosomes appropriately in IE2 ΔSX mutant-infected cells. Analysis using proteasome inhibitors revealed that this loss of UL84 protein expression was proteasome independent, and the stability of the expressed protein was found to be similar to that of the protein expressed during wild-type (wt) HCMV infection (43).
In this report, we have defined further the mechanisms governing UL84 and IE2 expression. The levels of the UL84 protein, but not RNA, are slightly upregulated when the protein is coexpressed with IE2 86 and much more significantly upregulated when it is coexpressed with IE2 40 in cotransfection assays. We show that the effect of the IE2 proteins on UL84 protein expression does not require other factors that are specific for the infection. The amount of UL84 protein that is expressed is directly dependent on the amount of IE2 40 protein present, although this is not true for IE2 86. Additionally, the expression of UL84 results in a decrease in the levels of the IE2 86 and IE2 40 proteins in these assays. The N-terminal region of UL84 (aa 1 to 105) is necessary and sufficient for the upregulation of UL84 protein expression mediated by the IE2 proteins. We find that UL84 stability is partially governed by the proteasome, but this is independent of the upregulation governed by the IE2 proteins, and the first 105 aa of UL84 are not sufficient. In addition, a protein-protein interaction is likely necessary for the observed effects of the IE2 proteins, given that the amino acid sequence of the N-terminal domain of UL84 is necessary for its binding to IE2 86 and IE2 40. Overall, we have identified important and novel roles for the IE2 family of proteins and have further characterized the unique mechanism by which UL84 expression is governed.
MATERIALS AND METHODS
Cell culture and 293FT transfections.
293FT cells were purchased and cultured according to the manufacturer's instructions (Invitrogen). Media were supplemented with 10% fetal bovine serum (Invitrogen), 2 mM l-glutamine, 50 mg/ml Geneticin (G418), 1 mM sodium pyruvate, and 10 mM nonessential amino acids (all supplements from Invitrogen). Cells were incubated at 37°C in 7% CO2. Twenty-four hours before transfection, cells were seeded in a 12-well plate at a density of approximately 80% confluence in media containing no G418. Immediately prior to transfection, 293FT cells were switched into Opti-MEM (800 μl/well; Invitrogen). Cells were transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. DNA (1.6 μg DNA/well; 0.8 μg of each plasmid) was transfected into each well. Appropriate plasmid DNAs were premixed with Opti-MEM, and then Lipofectamine diluted in Opti-MEM was added. Samples were kept at room temperature for 20 min, and the entire reaction mixture was then added dropwise to the preseeded 293FT cells in Opti-MEM. The Lipofectamine-DNA mix (200 μl) was allowed to incubate with the cells for 24 h, and the medium was then removed and replaced with Dulbecco's modified Eagle medium (DMEM) with no drugs. Twenty-four hours later (48 h posttransfection), cells were rinsed in phosphate-buffered saline (PBS), harvested, snap-frozen in liquid nitrogen, and maintained at −80°C until analysis. For the IE2 40 DNA titration experiment, the same protocol as described above was used, except the samples contained 0.8 μg, 0.4 μg, 0.2 μg, or 0.05 μg of the DNA encoding IE2 40. UL84 DNA concentrations were kept constant, and a control plasmid was added to ensure that the same amount of total DNA was added to each well.
Plasmid DNA construction.
All primers and probes used in these cloning studies can be found in Table 1. Plasmids pCDNA3-86F and pCDNA3-40HA have been previously described (42). The original UL84 plasmid, pTARGET-UL84HA, was obtained from G. Pari (University of Nevada, Reno, NV). The UL84 gene coding region was removed from this vector using EcoRI sites that flanked either side of the gene, including the region encoding the hemagglutinin (HA) tag at the UL84 3′ end, and then ligated into the vector pCDNA3 (Invitrogen) with the same sites. To make deletions and amino acid changes, QuikChange mutagenesis (Stratagene) was performed to create the following mutants in UL84: Δ68, Δ105, Δ135, and Δ200. The UL84 frameshift mutant was made by creating 4 different sequential amino acid changes that first shifted the ORF to the second possible ORF, followed by a change to the third ORF. Next, a newly created in-frame stop codon was changed to an alanine codon to allow continued read-through of the current ORF. Following this QuikChange mutagenesis, the ORF was shifted back into the original ORF to allow read-through of the rest of the UL84 protein. This mutant now encodes a new amino acid sequence for the first 175 aa while maintaining most of the RNA sequence for wt UL84. The primers used to create this mutant were designated FS-1, FS-2, FS-3, and FS-4.
TABLE 1.
Primers used to create UL84 mutants
| Mutation | Sequence (5′-3′) of: |
|
|---|---|---|
| Forward primer | Reverse primer | |
| Δ1-68 | ATATGAATTCAGGATGGACAGTGTCCTCCTGAA | GAGCGGCCGCCAGTGTGATGGAT |
| Δ1-105 | ATATGAATTCAGGATGGGCACCTACCATCTGAT | GAGCGGCCGCCAGTGTGATGGAT |
| Δ1-135 | CCTCGAGACGCGTGATTAAACATGGGGGAAGCGAACGACGAGTCTCAAACCG | CGGTTTGAGACTCGTCGTTCGCTTCCCCCATGTTTAATCACGCGTCTCGAGG |
| Δ1-200 | CCTCGAGACGCGTGATTAAACATGTCTCTCTTTCCCGCACGCCCAGGC | GCCTGGGCGTGCGGGAAAGAGAGACATGTTTAATCACGCGTCTCGAGG |
| FS-1 | GACGCGTGATTAAACATGTGCCACGCGTCGACCC | GGGTCGACGCGTGGCACATGTTTAATCACGCGTC |
| FS-2 | CTACCACCACGACCAGATGGGCGTCGCCAG | CTGGCGACGCCCATCTGGTCGTGGTGGTAG |
| FS-3 | CCTCACACTGACGGAGCAACACGACATCCGTC | GACGGATGTCGTGTTGCTCCGTCAGTGTGAGG |
| FS-4 | GAAGAGGACGAGGAGACCGCAACGACGATCGTG | CACGATCGTCGTTGCGGTCTCCTCGTCCTCTTC |
| Hybrid | GGGAGACCCAAGCTTGCCACCATGCCACGCGTCGACCCCAAC | CCGAGAGGCGCGTCTTGCGATCGCCGCGGTGCACCCGAGGCTG |
The hybrid (Hyb) mutant was created using the primers described in Table 1, and the technique was based on the protocol described in reference 17. The templates used in these studies included pCDNA3-UL84 and pCDNA3-UL44. pCDNA3-UL44 contains the region encoding UL44.
Quantitative real-time PCR and real-time RT-PCR.
For the quantitative real-time PCR and real-time reverse transcription-PCR (RT-PCR) analyses, an aliquot of the same cells as those used for Western blot analysis was assayed for DNA and RNA, respectively. Following transfection and freezing of the cells, the DNA was prepared using the Invitrogen Miniprep kit or the Norgen RNA/DNA/protein kit according to the manufacturer's instructions. Concentrations were determined by UV spectrophotometry. Quantitative real-time PCR analyses were performed as previously described (61, 63) using the TaqMan universal PCR master mix (Applied Biosystems), except that the primers and probes used in these analyses were different. In this case, primers and probes were directed against IE2 86 and IE2 40, UL84 (N terminus and C terminus), UL44, and the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) promoter (GAPDHP). The sequences for the primers and probes are listed in Table 2.
TABLE 2.
Primers and probes used for quantitative real-time PCR and RT-PCR studies
| Transcript | Sequence (5′-3′) of: |
||
|---|---|---|---|
| Forward primer | Reverse primer | TaqMan probea | |
| UL84 C terminus | AGACATTGGGACCCTCCGTC | GCGGTGATTCGTTCGGG | FAM-TGGACGATTGGAGCTAG-BHQ |
| UL84 N terminus | TCACTTTCCTCGCCACCACTA | GGAGACTGTCGTCGTCGGTACT | FAM-CACCACGACCATGATGGGCGTC-BHQ |
| UL44 | GCTTTCGCGACAATGTCT | GCCCGATTTCAATATGGAGTTC | FAM-CGTGCACGCAGGCCGAGC-BHQ |
| IE2 C terminus | GCGCAATATCATGAAAGATAAGAACA | GATTGGTGTTGCGGAACATG | FAM-TCGGCGGGGTCGC-BHQ |
| G6PD | TCTACCGCATCGACCACTACC | GCGATGTTGTCCCGGTTC | FAM-ATGGTGCTGAGATTTGCCAACAGGA-BHQ |
| GAPDH-P | TTTCATCCAAGCGTGTAAGGG | CAGGACTGGACTGTGGGCA | FAM-CCCCGTCCTTGACTCCCTAGTGTC-BHQ |
FAM, 6-carboxyfluorescein; BHQ, black hole quencher.
For the real-time RT-PCR analyses, RNA was prepared from the same cells as those used for Western blot analyses at 48 h posttransfection (p.t.) using either the Norgen RNA/DNA/protein purification kit or the Ambion Paris kit according to the manufacturer's instructions. RNA was treated with DNase using the Ambion Turbo DNA-free kit according to the manufacturer's instructions and then quantified using UV spectrophotometry. Samples were diluted to 12.5 ng/μl and analyzed using the same machine as used for DNA quantification (Applied Biosystems; ABI Prism 7000). The real-time RT-PCRs were carried out using the TaqMan one-step RT-PCR master mix reagents (Applied Biosystems) as previously described (61, 63) and primers and probes against the UL84 (5′ and 3′ ends), UL44, IE2, and glucose-6-phosphate dehydrogenase (G6PD) genes (listed in Table 2).
Western blot analyses.
Transfected cells that had been stored at −80°C were lysed in reducing sample buffer as previously described (43), or protein was prepared using the Norgen RNA/DNA/protein purification kit according to the manufacturer's instructions. Lysates were run on an 8 or 10% acrylamide gel and then transferred to a nitrocellulose membrane. Membranes were blocked in 5% milk in Tris-buffered saline containing 0.05% Tween 20 (TBS-T) and probed with the following antibodies: IE2 monoclonal antibody (MAb) 8140 (Chemicon), UL84 MAb (Virusys), HA F-7 MAb (Santa Cruz Biotechnology), UL44 MAb (Virusys), and actin MAb (Sigma-Aldrich). Horseradish peroxidase-coupled anti-mouse IgG antibody was obtained from Calbiochem. Following secondary antibody incubation and washes, proteins were detected with SuperSignal chemiluminescent substrate (Pierce Biotechnology) according to the manufacturer's instructions.
Immunoprecipitations.
Transfected 293FT cells were lysed in a modified immunoprecipitation buffer containing 10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM dithiothreitol (DTT), 150 mM NaCl, 1% Triton X-100, and a protease inhibitor cocktail (Sigma-Aldrich) for 30 min on ice. Lysed cells were centrifuged at 10,000 × g for 10 min at 4°C to remove cellular debris, and the lysate was allowed to incubate and rotate overnight at 4°C with Protein G-Plus agarose beads (Santa Cruz Biotechnology) coupled with the IE2 antibody that recognizes IE2 86 and IE2 40 or an antibody that recognizes an HA tag (clone F-7; Santa Cruz Biotechnology). The following day, the agarose beads were washed twice with the same buffer that was used for lysis, resuspended in the same reducing sample buffer used for Western blotting, and heated to 95°C. Following centrifugation, the supernatant containing the immunoprecipitated proteins was collected. An aliquot of the sample was taken before immunoprecipitation. This fraction was assessed to determine the amount of protein present before the immunoprecipitation and represents 10% of the sample that was used for immunoprecipitation. All immunoprecipitations were analyzed by Western blotting to determine that the appropriate complexes had been pulled down and to assess which proteins were capable of interaction.
Proteasome inhibitor treatment.
293FT cells were cultured as described above and seeded as for transfection. At 36 h p.t., cells were treated with 10 μM MG132 (Calbiochem) for 12 h and then harvested at 48 h p.t. Cells were processed for Western blot analysis as described above. Two other proteasome inhibitors were tested, salinosporamide (Sal A) and lactacystin, in order to confirm the results with MG132. Sal A was used at 100 nM and was a gift from Bradley Moore (Scripps Institute of Oceanography, University of California, San Diego), while lactacystin was used at 10 μM and was obtained from Calbiochem. Dimethyl sulfoxide (DMSO) was used as a vehicle-only control.
RESULTS
UL84 protein expression is enhanced when the protein is coexpressed with IE2 86 or IE2 40.
Studies with IE2 mutant viruses previously revealed that the expression of UL84 is tightly linked to the presence of the IE2 86, IE2 60, and, most predominantly, IE2 40 proteins (43, 62). Enhancement of UL84 expression during infection was shown to occur at the posttranscriptional level and was not due to inefficient loading of the UL84 RNA onto polyribosomes (43). These data directed our studies to ascertain whether this type of regulation could occur independent of infection and to further delineate the roles of the IE2 proteins in regard to UL84 expression.
To determine whether the IE2 proteins were capable of upregulating the expression of UL84 in the absence of any other viral proteins or infection-specific cellular modifications, 293FT cells were transfected with plasmids encoding the IE2 86, IE2 40, and UL84 proteins. These plasmids were either singly transfected or cotransfected (IE2 86 or IE2 40 plus UL84), and then at 48 h posttransfection (p.t.) cells were harvested and assayed for DNA, RNA, and protein expression. In the case of the singly transfected samples, the quantity of transfected DNA was normalized to the cotransfected samples by adding empty plasmid DNA so that each of the cell cultures received the same amount of DNA. All of the analyses were also repeated in COS-7, U373, and HEK-293 cells to assure that expression patterns were not cell type specific. In all cases, each cell type showed expression patterns similar to those seen in the 293FT cells. Only the experiments in the 293FT cells are shown.
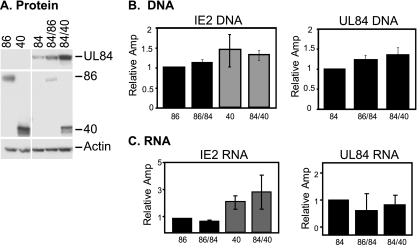
When protein expression was analyzed by Western blotting, significant differences were observed in cotransfected cells (Fig. 1A). The levels of UL84 protein were very low when UL84 was expressed alone and increased slightly when it was coexpressed with IE2 86. However, the levels were greatly enhanced when it was coexpressed with IE2 40. These results are consistent with previous data showing that loss of IE2 40 during infection results in significantly reduced levels of UL84 protein (62). These experiments clearly demonstrate that UL84 expression is somewhat influenced by the presence of IE2 86, but coexpression with IE2 40 appears to have a greater effect.
FIG. 1.
UL84 and IE2 protein, RNA, and DNA expression in transfected 293FT cells. (A) 293FT cells were transfected with either the IE2 86 (86), IE2 40 (40), or UL84 (84) plasmids alone or in combination. Forty-eight hours later, cells were harvested, and UL84, IE2 86, and IE2 40 protein levels were assayed by Western blotting by loading equal amounts of protein per lane. Actin serves as a loading control. (B) Transfected cells with the same constructs as for panel A were assayed for DNA levels by quantitative real-time PCR. IE2 DNAs were assayed using a primer and probe directed against the 3′ end of the IE2 86 gene, thus recognizing both forms of the IE2 constructs. UL84 DNA levels were also assayed alone or in combination with the IE2 constructs using primers and a probe that recognize the N terminus of UL84. Samples were tested in duplicate in each experiment, and the graphs shown are averages of 3 experiments. DNA for each sample was normalized to the cellular GAPDH promoter as a control for the amount of input DNA in each reaction. Graphs are represented as relative amplification (Amp), with the value of the first sample in each graph set to 1. (C) RNA expression for each of the samples was measured by quantitative real-time RT-PCR. Each of the samples was measured using the same primer and probes as for panel B and are normalized to the housekeeping G6PD gene. Graphs represent at least 2 experiments.
The levels of the IE2 proteins were also assayed. Each migrated at the appropriate size, although the IE2 40 protein accumulated to slightly higher levels than IE2 86. Interestingly, the levels of each of these proteins were found to be significantly reduced when the proteins were coexpressed with UL84 (Fig. 1A). Despite the fact that the UL84 protein levels were different, both IE2 proteins were reduced to similar degrees, indicating that the amount of UL84 protein present does not significantly influence this phenotype.
To rule out the possibility that the effects on protein expression were due to DNA transfection efficiency or different levels of mRNA, the amounts of transfected DNA and RNA present in each transfected sample were analyzed by either quantitative real-time PCR or quantitative real-time RT-PCR, respectively. In each case, whether the DNA was singly transfected or cotransfected, the amounts of IE2 86 and IE2 40 DNA were found to be comparable across samples (Fig. 1B). This was true for the multiple assays that were performed during these studies. Similarly, UL84 DNA in these samples remained constant, whether transfected with IE2 86 or IE2 40 DNA or the control plasmid. When the levels of the transfected RNA were assessed, each of these samples was also found to have relatively equivalent amounts of IE2 86 and IE2 40 RNA, although the levels of the IE2 40 RNA were slightly higher than IE2 86 RNA levels. Similar to what was observed during infection, despite the increase in protein levels for UL84, RNA levels remained relatively unaffected when coexpressed with either of the IE2 plasmids (Fig. 1C).
To determine whether cellular processes were necessary to facilitate this regulation, in vitro transcription-translation reactions were conducted in which UL84 and either IE2 40 or IE2 86 were transcribed and translated in the same reaction or IE2 40 or IE2 86 protein was synthesized first and then added to the UL84 in vitro transcription-translation reaction. In no sample was enhancement of UL84 protein expression observed. This suggests that some cellular modifications or processes are necessary for the regulation seen during the cotransfection and infection experiments (data not shown) or that this system is simply not sufficient to recapitulate the phenotypes observed in transfected cells.
UL84 protein expression is dependent on the amount of IE2 40 present.
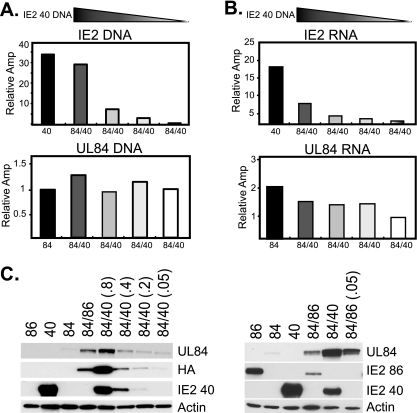
To determine how the regulation of UL84 protein expression might be related to the amount of IE2 40 protein present in the cells, the amount of transfected IE2 40 plasmid was incrementally decreased while the UL84 plasmid concentration remained constant. Starting with 0.8 μg of IE2 40 plasmid, DNA was decreased to 0.4, 0.2, and 0.05 μg, while 0.8 μg of UL84 DNA was used for all samples. Analysis of both the DNA and RNA levels by quantitative real-time PCR and quantitative real-time RT-PCR, respectively, revealed appropriate reduction of the IE2 40 plasmid (Fig. 2A and B). The amount of IE2 40 RNA expressed was also proportional to the amount of input DNA (Fig. 2B). In all cases, however, the amounts of UL84 DNA and RNA remained relatively unaffected by the presence of the varying levels of IE2 40 (Fig. 2A and B).
FIG. 2.
Titration of IE2 40 DNA affects UL84 protein expression. (A) Decreasing amounts of IE2 40 (40) DNA were transfected (0.8, 0.4, 0.2, and 0.05 μg), while a constant amount of UL84 (84) DNA was transfected into cells. The levels of transfected DNA were analyzed by quantitative real-time PCR and normalized to the GAPDH promoter as a loading control. Values are shown as relative amplification (Amp), with the lowest value set to 1. Both IE2 and UL84 DNAs were assessed. (B) IE2 40 and UL84 RNAs from the same samples as in panel A were measured by quantitative real-time RT-PCR and normalized to the cellular housekeeping G6PD gene. (C) UL84 and IE2 40 protein expression was analyzed from the same samples as in panels A and B by Western blotting (left). UL84 expression was also measured with an antibody that recognizes the C-terminal tag HA to confirm UL84 antibody results (left). UL84 expression was assessed after cotransfection with 20-fold-less IE2 86 plasmid [right; 84/86 (0.05)]. Actin serves as a loading control.
In contrast, expression of UL84 protein was directly correlated with the amount of IE2 40 protein that was present. Samples that contained the most IE2 40 protein also expressed the most UL84 protein, and the levels of the UL84 protein were coordinately reduced as the levels of IE2 40 declined, as measured with an antibody recognizing UL84 or the HA tag on the UL84 protein (Fig. 2C, left). Similar experiments were conducted with decreasing amounts of IE2 86 plasmid; however, these data indicated that UL84 expression is not directly linked to the amount of IE2 86 protein present in the sample. In this case, UL84 expression remained slightly increased compared to that in the sample with UL84 transfected alone, even when the amount of IE2 86 was decreased over 20-fold (Fig. 2C, right). The IE2 86 protein levels dropped below the level of detection on the exposure shown but can be seen upon very long exposure of the blot (data not shown). These data indicate that IE2 86 likely regulates UL84 protein expression in a different manner than IE2 40 and that the levels of UL84 protein are more directly linked to the expression of the IE2 40 protein than to that of the IE2 86 protein.
Deletions within the UL84 sequence affect expression of the protein and its enhancement by the IE2 proteins.
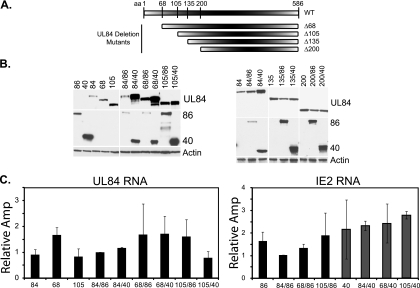
To determine which domains of UL84 were important for the enhanced protein levels, deletions throughout UL84 were created. Specific domains were chosen based on previous studies suggesting that the first 105 aa of UL84 were important for an interaction with IE2 86 (16). Based on these data, we constructed UL84 mutants that lacked either the first 68 (Δ68), 105 (Δ105), 135 (Δ135), or 200 (Δ200) aa (Fig. 3A). Each of the mutant constructs was confirmed by sequencing and tested for expression in 293FT cells to ensure that the protein was produced and that the size of each protein was as expected (data not shown; Fig. 3B).
FIG. 3.
The N-terminal 105 aa of UL84 are important for the enhancement of protein expression governed by the IE2 proteins. (A) Deletions throughout the N terminus of UL84 were created by PCR mutagenesis and included the first 68 (Δ68), 105 (Δ105), 135 (Δ135), and 200 (Δ200) aa. The schematic is not to scale. (B) Protein expression of UL84 (wt and mutants), IE2 86, and IE2 40 was measured using Western blot analyses. Actin serves as a loading control. Mutants are represented by the corresponding amino acid deletions (68, 105, 135, and 200). (C) RNA for each sample was analyzed as for Fig. 1B. Only the Δ68 and Δ105 mutants are shown. Values are shown as relative amplification (Amp) and are relative to the 84/86 sample, with a value set to 1.
The relative expression of each of the UL84 mutants transfected singly and in combination with either IE2 86 or IE2 40 was analyzed by Western blotting. Interestingly, when the UL84 mutants were expressed alone, we observed an increase in the accumulation of the protein as the N-terminal region was deleted (Fig. 3B). Removal of the first 68 aa resulted in a slight enhancement of protein expression, while deletion of the first 105 aa was associated with a significant increase in mutant protein level. Larger N-terminal deletions of 135 or 200 aa did not lead to an additional increase in UL84 protein above that observed with the UL84 mutant missing the first 105 aa. However, the increased level of the mutant UL84 proteins when they were expressed alone was still lower than the level observed when wt UL84 was cotransfected with IE2 40 (Fig. 3B). Expression of the UL84 mutant protein with the 68-aa N-terminal deletion was also significantly enhanced when the mutant was coexpressed with IE2 86 or IE2 40, but cotransfection of IE2 86 or IE2 40 did not increase the level of the UL84 mutant protein missing the first 105 aa above that observed when the mutant was singly transfected. As noted above, the levels of the IE2 86 and IE2 40 proteins were lower when the proteins were coexpressed with wt UL84. Interestingly, the mutant with a deletion of the first 105 aa was unable to inhibit IE2 86 or IE2 40 protein expression, indicating that this N-terminal domain is also important for the inhibitory effect on the IE2 proteins. Deletion of the first 135 or 200 aa showed results similar to those for deletion of the first 105 aa (Fig. 3B).
To determine whether the RNAs for each of these samples remained constant when coexpressed, RNA was quantified by real-time RT-PCR analyses (Fig. 3C). As before, no significant difference in the amounts of UL84 RNA in the samples was observed. Coexpression with either IE2 86 or IE2 40 did not affect the levels of UL84 RNA for either the Δ68 or the Δ105 mutant. The RNA levels for the mutants Δ135 and Δ200 remained unchanged as well (data not shown). The expression of the IE2 86 or IE2 40 RNAs also was not affected by the presence of either the Δ68 or Δ105 mutant, indicating that regulation of the protein levels occurred posttranscriptionally.
Interaction of UL84 with IE2 86 and IE2 40 is dependent on the first 105 aa of UL84.
Since UL84 has previously been shown to bind to IE2 86 and IE2 40 (10, 43, 50) and since the above deletion analysis revealed differences in the relative enhancement of the UL84 mutant protein levels when the proteins were coexpressed with IE2 86 or IE2 40, the interaction between the UL84 mutant proteins and IE2 proteins was measured. 293FT cells were singly transfected or cotransfected with plasmids, and the UL84 mutant proteins were then assayed for formation of complexes with the IE2 proteins 48 h later. Cells were harvested, snap-frozen in liquid nitrogen, and then lysed and subjected to immunoprecipitation using an antibody that recognizes IE2 86 and IE2 40 coupled to agarose beads. Following immunoprecipitation, Western blot analyses were conducted for each of the transfected proteins. Relative expression of the UL84 protein before immunoprecipitation can be observed (Fig. 4, Pre lanes). Similar to previous results, the amount of UL84 protein increased as the N-terminal domain was removed. The deletion of aa 1 to 68 of UL84 resulted in a slight reduction in its interaction with IE2 86 or IE2 40, but the efficiency of complex formation was still significant. The deletion of aa 1 to 105 of UL84, however, resulted in complete loss of detectable binding to both IE2 86 and IE2 40 (Fig. 4, IP lanes). These data indicate that the first 105 aa of UL84 are involved in the interaction with both IE2 proteins.
FIG. 4.
The first 105 aa of UL84 are important for interaction with IE2 86 and IE2 40. wt UL84 and each of the N-terminal mutants, Δ68 (68) and Δ105 (105), were assayed for their interaction with both IE2 86 and IE2 40 using immunoprecipitation assays as described in Materials and Methods. Expression of each of the proteins was analyzed before immunoprecipitation (Pre) in order to assess the relative expression levels. Following immunoprecipitation, the complexes were eluted from the agarose beads and subjected to Western blot analysis. Results are shown in the immunoprecipitation (IP) lanes. Antibodies to UL84 (wt and mutants), IE2 86, and IE2 40 were used in the analysis. The Pre samples are 10% of the sample used for IP.
UL84 protein is sensitive to proteasomal degradation, independent of the presence of the IE2 proteins.
To assess whether the lack of UL84 expression when it was transfected alone in 293FT cells was due to proteasomal degradation, transfected cells were treated with the proteasome inhibitors lactacystin, MG132, and Sal A. Relative expression patterns described above (Fig. 3 and 4) were maintained in all experiments. All proteasome inhibitors showed the same results, and therefore only the results with MG132 are shown here. Treatment of singly transfected cells with MG132 for 12 h prior to harvest resulted in increased levels of UL84 protein (Fig. 5). However, the UL84 levels in the presence of the proteasome inhibitor were still significantly lower than the levels of UL84 coexpressed with IE2 40 in the absence of the inhibitor. This was also true for the deletion mutants Δ68 and Δ105. Interestingly, the levels of the UL84 protein also increased when it was coexpressed with either of the IE2 proteins in the presence of MG132, although the relative increase was slightly less. These data indicate that the IE2 proteins provide some protection from degradation, but they are not sufficient to prevent this degradation entirely. In contrast to that of UL84, the levels of IE2 86 and IE2 40, either alone or in combination with UL84, were not affected by the presence of the proteasome inhibitor, indicating that the IE2 proteins were not subject to proteasome degradation during this period of time.
FIG. 5.
Proteasome inhibitor treatment affects the level of UL84 expression, but coexpression with the IE2 proteins does not protect UL84 from proteasomal degradation. Thirty-six hours p.t., cells were incubated with a proteasome inhibitor (MG132; 10 μM; +) or mock treated with DMSO (−) and then assayed for protein expression 12 h later. UL84 expression was assayed by Western blotting with or without IE2 86 or IE2 40. Actin serves as a loading control.
The first 105 aa of UL84 are sufficient for enhanced expression from the IE2 proteins.
Given that the first 105 aa of UL84 were important for the binding of UL84 to the IE2 proteins as well as the enhanced levels of UL84 when coexpressed with the IE2 proteins, we next tested whether this UL84 domain was sufficient for enhanced expression when the protein was coexpressed with IE2 86 or IE2 40. To assess this, the first 105 aa were cloned onto the N terminus of another HCMV viral protein, UL44 (Fig. 6A). This protein was termed UL44-84 Hyb (Hyb). As before, transfections were conducted in 293FT cells, and the samples were analyzed for protein expression. As a control, IE2 86 and IE2 40 were cotransfected with UL44 to determine if they could enhance the expression of UL44 in the same way as UL84. Coexpression of IE2 86 or IE2 40 with UL44 had no effect on UL44 protein levels (Fig. 6A). However, when IE2 40 or IE2 86 was cotransfected with the UL44-84 Hyb plasmid, significant increases in hybrid protein expression were observed compared to results for the singly transfected plasmid (Fig. 6A). This protein can be detected with either an antibody recognizing the first 105 aa of UL84 or an antibody recognizing UL44. Expression of the hybrid protein did not inhibit expression of IE2 86 or IE2 40, indicating that although the first 105 aa of UL84 are sufficient for enhanced expression by the IE2 proteins, they are not sufficient for mediating the decrease in IE2 protein expression.
FIG. 6.
The first 105 aa of UL84 are sufficient for enhancement of expression governed by the IE2 proteins but not for susceptibility to proteasomal degradation. (A) The first 105 aa were added to the amino terminus of another viral protein, UL44 (Hyb). Coexpression of this protein with IE2 86 and IE2 40 was assessed, as was the expression of UL44. The expression of the Hyb protein was assayed using an antibody directed against UL84 and UL44. (B) Proteasome treatment was conducted as in Fig. 5, except that amount of Hyb protein was analyzed in combination with IE2 86 (86) and IE2 40 (40). Cells were either treated with the proteasome inhibitor MG132 (+) or mock treated with DMSO (−). UL84 and UL44 protein expression was assayed as a control. Actin serves as a loading control in both panels A and B. (C) RNA was analyzed for IE2 86 (86), IE2 40 (40), UL44 (44), and Hyb samples. All samples were normalized to G6PD. Values are shown as relative amplification (Amp), with the first sample in the graph having a value set to 1.
To test whether the Hyb protein was also subject to proteasomal degradation, as observed for the full-length UL84 protein, the levels of the Hyb protein were measured after treatment of the transfected cells with the proteasome inhibitors MG132 and lactacystin (only MG132 is shown). Cells were maintained in media with the inhibitor for 12 h prior to harvest (36 to 48 h p.t.), and the samples were assessed for protein content by Western blot analysis. As a control, wt UL84 was analyzed to ensure that the proteasome treatment was effective (Fig. 6B). Expression of UL44 transfected alone was also assessed with or without proteasome inhibition, and no difference in expression between the two samples was seen. In contrast to results for the full-length UL84 protein, changes in the levels of the Hyb protein were not detected in the presence of the proteasome inhibitors, suggesting that the first 105 aa of UL84 alone are likely not sufficient for targeting the protein to the proteasome for degradation in the transfected cells. Again, expression of IE2 86 and IE2 40 remained unaffected by inhibition of the proteasome.
The RNA for each of the above samples was analyzed in multiple experiments using quantitative real-time RT-PCR (Fig. 6C). The IE2 86 and IE2 40 RNAs showed no difference in expression when cotransfected with the UL44 or UL44-84 Hyb plasmid. Similarly, the amounts of Hyb and UL44 RNA were analyzed. In each case, no differences in the levels of RNA were observed when either Hyb or UL44 was cotransfected with IE2 86 or IE2 40, further confirming the previous results that the enhancement provided by the IE2 proteins is posttranscriptional.
We did note in the above studies, however, that the levels of the Hyb protein were significantly lower than the levels of the UL44 protein in the transfected cells. To determine whether this was due to differential RNA expression, we compared the levels of the Hyb RNA, UL84 RNA, and UL44 RNA. This was accomplished using 2 sets of primers and probes, one specific for the UL84 sequence and the other specific for the UL44 sequence present in the hybrid (Fig. 7). Interestingly, we observed a 6- to 8-fold decrease in the amount of Hyb RNA produced in the transfected cells compared to UL44 RNA. In contrast, the amount of Hyb RNA was comparable to that of wt UL84 RNA. This difference in RNA expression likely explains the difference in UL44 and Hyb protein levels.
FIG. 7.
Hyb RNA is expressed to the same level as UL84 RNA but to a significantly lower level than UL44 RNA. RNA was prepared from the same samples as in Fig. 6A and analyzed by quantitative real-time RT-PCR. Primers and probes directed near the 5′ end of the UL84 gene were used to compare the amounts of RNA in the Hyb and UL84 samples (UL84 RNA). Primers and probes to UL44 were used to compare the expression levels of the Hyb RNA and UL44 RNA (UL44 RNA). Values are shown as relative amplification (Amp), with the lowest value set to 1.
The UL84 amino acid sequence is important for the enhancement governed by IE2 40.
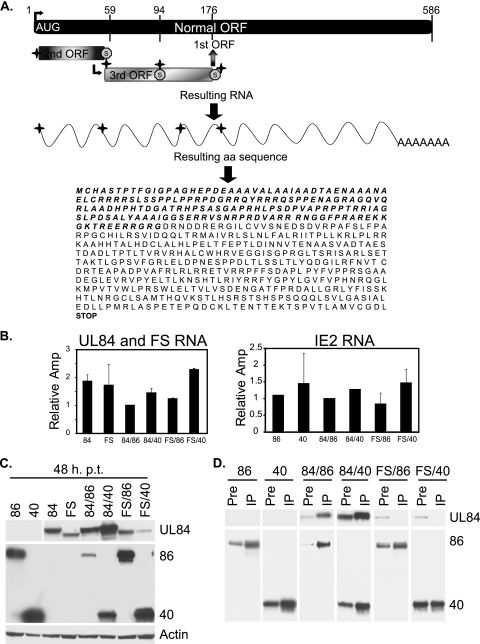
Since the enhancement by IE2 40 appeared to be at the level of protein synthesis, it was possible that the structure of the RNA encoding the amino-terminal 105 aa of UL84 rather than the amino acid sequence itself was involved. To address this question, we constructed a UL84 mutant that maintained the same RNA sequence (except for the minimal number of base changes required to create the mutant) but encoded a protein with a different amino acid sequence for the N-terminal 175 residues (Fig. 8A shows a schematic). This new mutant is referred to as UL84-Frameshift (FS).
FIG. 8.
The UL84 amino acid sequence is important for regulation governed by IE2 86 and IE2 40 and for the interaction of UL84 with the IE2 proteins. (A) Schematic representation of the mutations (crosses) made in the RNA to shift the amino acid sequence to that encoded by the second and third open reading frames (ORFs). The original AUG is depicted and is the site of the first mutation, and all subsequent mutations are shown. The resulting RNA has four mutations, while the resulting protein is the product of a new ORF for the first 175 aa and is the product of the original ORF for the remainder of the protein. The resulting amino acid sequence is shown, with the new amino acid sequence indicated in boldface italics. (B) The RNA for each of the samples was measured by quantitative real-time RT-PCR. Analyses were conducted as for previous figures, and each of the relative RNA values is shown. Values are shown as relative amplification (Amp), with the sample 84/86 value set to 1. (C) The frameshift mutant (FS) was assayed by Western blotting in combination with IE2 86 and IE2 40. Actin serves as a loading control. (D) Interactions between each of the proteins assessed in panels B and C were measured using immunoprecipitation assays. Antibody specific for IE2 86 and IE2 40 was coupled to agarose beads, and then transfected samples were subjected to immunoprecipitation with the IE2-coupled beads. The amount of protein present before the immunoprecipitation is shown (Pre), followed by the amount of protein that was pulled down in the immunoprecipitation (IP).
293FT cells were transfected with the UL84 (wt), IE2 40, IE2 86, and FS plasmids as before. At 48 h p.t., cells were harvested and analyzed for the quantity of DNA, RNA, and protein. DNA was measured by quantitative real-time PCR, and results showed that equal amounts of the UL84 plasmid and FS plasmid had been transfected in all samples (data not shown). Further, the amounts of IE2 40 and IE2 86 DNA in each of the samples transfected with these DNAs were comparable (data not shown). The amount of RNA was also assessed using quantitative real-time RT-PCR, and this revealed that, similar to wt UL84, the FS mutant was not affected transcriptionally in the presence of IE2 86 or IE2 40. Moreover, the amounts of IE2 86 and IE2 40 RNA remained constant when samples were cotransfected with UL84 or FS DNA (Fig. 8B).
Next, the protein level for each sample was analyzed (Fig. 8C). The UL84 and FS protein levels were measured by Western blotting with an antibody that was specific for a region downstream of aa 176, thus recognizing both proteins. The amounts of IE2 86 and IE2 40 protein were measured with an antibody that was directed against the common C-terminal region. As before, UL84 protein expression was enhanced slightly when the protein was cotransfected with IE2 86, although this effect again was minimal compared to the significant enhancement governed by IE2 40. The levels of the FS mutant protein alone were slightly lower than that of wt UL84. However, when the FS mutant was cotransfected with either IE2 86 or IE2 40, no enhancement of the protein was detected. These data indicate that the low levels of UL84 protein when expressed alone are not solely due to the first 175 aa of UL84, but this protein sequence is important for IE2 40-mediated enhancement of UL84. The amounts of IE2 86 and IE2 40 protein were also assessed. As before, IE2 86 and IE2 40 protein expression was repressed when they were coexpressed with UL84 (Fig. 8C). However, expression of IE2 86 and IE2 40 when they were cotransfected with FS did not show any inhibition.
To determine whether IE2 86 and IE2 40 could form a complex with the FS mutant, we performed coimmunoprecipitation experiments as described for Fig. 4 (Fig. 8D). As expected, UL84 bound efficiently to IE2 86 and IE2 40 (see IP lanes). However, the FS mutant was unable to form a complex with either IE2 86 or IE2 40. Although we could not rule out that the small number of base changes made in the FS mutant RNA influenced the structure of the RNA, it is highly unlikely that these changes were responsible for the differences seen in the protein expression and binding experiments. Taken together, these studies strongly support the conclusion that the N-terminal amino acid sequence of UL84 is important for enhanced expression of UL84 by the IE2 proteins, which is likely mediated by direct binding of this region to the IE2 proteins.
DISCUSSION
Many functions have been ascribed to the MIE proteins during HCMV infection. It is known that IE1 72 and IE2 86 serve in shutting down host defenses immediately upon infection and play a role in dysregulating the cell cycle and interfering with apoptosis (5, 30, 36, 65, 66, 69). IE2 86 also regulates the expression of many cellular and viral transcripts throughout infection (for reviews, see references 12, 34, and 53). Much of the regulation governed by IE2 86 is thought to be controlled by protein-protein interactions, given the vast number of proteins that IE2 86 has been shown to bind to (2, 4, 6-8, 13, 18, 19, 21, 22, 25, 26, 28, 31, 32, 46, 48-51, 60, 68). Although IE2 86 has been extensively studied, the functions of the two early-late proteins (IE2 60 and IE2 40) that arise from the C-terminal half of IE2 86 remain less well understood. In previous studies, our lab has shown that IE2 60 and IE2 40 are important for the late stages of viral infection, impacting the expression of several viral early-late proteins. While regulation of other early-late proteins (for instance, UL83) was shown be transcriptional, the regulation of one particular early-late protein, UL84, was shown to be posttranscriptional (42, 43, 62).
UL84 has been shown to be important for ori-Lyt-dependent DNA replication and can downregulate the ability of IE2 86 to activate some early promoters in transient assays (16, 37). In addition, UL84 is the only other viral protein that IE2 86 has been shown to interact with to date, and our previous studies demonstrated that IE2 60 and IE2 40 have the ability to interact with UL84 individually, independent of IE2 86. Infection with recombinant mutant viruses that did not express the IE2 60 and IE2 40 proteins resulted in a significant decrease in UL84 protein, but not RNA, expression. In these studies, IE2 40 was found to be more important for regulating UL84 protein expression (43, 62). The effects of the IE2 proteins on both viral and cellular transcription have been studied extensively, but it is important to understand the other potential roles these proteins may play in regard to regulating protein expression.
The current studies demonstrate that the posttranscriptional regulation of UL84 by IE2 40 can occur in the absence of other viral proteins. Similar to what occurs during infection, IE2 86 is able to enhance the expression of UL84 protein to some degree, while the levels of UL84 markedly increase when it is coexpressed with IE2 40. This regulation is associated with the amount of IE2 40 present, given that a decrease in IE2 40 results in a concomitant loss of UL84 protein expression. These data indicate that the increased levels of UL84 protein in the context of infection, particularly at later times, as well as independent of infection, are tightly coupled to the expression of the IE2 40 protein, and this may be one way that HCMV facilitates temporal regulation of protein expression. IE2 60 may have the ability to facilitate the same type of regulation, although previous studies showed that loss of IE2 60 alone did not result in a loss of UL84 protein expression during infection (62). Given that aberrant expression of many HCMV proteins has been shown to be detrimental to infection, this mechanism of control may be advantageous to the virus for achieving finely tuned protein levels. Moreover, it has been demonstrated that IE2 86 has the potential to interact with multiple cellular proteins, and for the majority of these proteins, the interacting domain is also present in IE2 40. Thus, a key question to be addressed is whether the IE2 gene products also regulate the expression of any of these cellular proteins at a translational level.
In this study, we have further defined the domains of UL84 that are necessary for the IE2 40-mediated enhancement of the protein level. We show that the N-terminal 105 aa of UL84 are important for the interaction with IE2 86 and IE2 40, and deletion of the region encoding these amino acids results in loss of the enhancement by IE2 86 and IE2 40. Although the data suggested that a protein-protein interaction was required, it was important to address the issue of whether the UL84 RNA was sufficient to facilitate this enhancement or if the appropriate amino acid sequence was necessary. This question was answered by constructing a mutant UL84 gene that maintained the RNA sequence except for 4 minor changes but encoded a different amino acid sequence at the N terminus. The mutant protein was not enhanced by the presence of IE2 40 and was unable to form a complex with IE2 86 or IE2 40. Based on this result, we conclude that the N-terminal 105-aa protein sequence, and not the UL84 RNA itself, is necessary for the IE2 40-mediated increase in UL84 protein levels. Further support for this conclusion was provided by the experiment showing that attachment of these 105 aa to the N-terminal region of another protein recapitulated regulation by the IE2 proteins.
Based on bioinformatic analysis, it appears that the first 105 aa of UL84 are highly unstructured (D. H. Spector and R. L. Sanders, unpublished results). It is therefore possible that IE2 40 regulates UL84 protein expression by aiding in the correct folding of the protein. In this manner, IE2 40 could potentially act as a chaperone protein, modulating protein structure and stability. Given that the majority of UL84 is in complex with the IE2 proteins (43), it is likely that this interaction is necessary for maintenance of UL84 protein levels. We do not know yet whether IE2 40 first interacts with UL84 as the protein is being synthesized and cannot exclude the possibility that IE2 40 serves to remove or displace a translational repressor or acts as a translational enhancer, possibly to relieve translational pausing. The question of whether the interaction of IE2 40 and UL84 occurs cotranslationally on the ribosome is an important one, and studies are in progress to determine if this is the case.
One difference between the infection and transfection studies was that UL84 appeared to be more sensitive to proteasomal degradation in the transient assays (43). Although there are many explanations for this observation, one possibility is that, during viral infection, the ubiquitin-proteasome degradation pathway that can target UL84 has been compromised or altered. Our lab and others have shown that HCMV replication requires that the proteasome remain active throughout infection (27, 39, 40, 56). Yet, the anaphase-promoting complex (APC) E3 ubiquitin ligase is specifically disabled during infection, allowing stabilization of its target proteins (55, 64). Although we do not have direct evidence that UL84 can be a target of the APC, it does have a consensus APC D-box degradation signal beginning at aa 183 (RGILCVVSN). In the cotransfection studies, the presence of IE2 40 (or IE2 86) was not sufficient to protect UL84 from proteasomal degradation. However, it is important to note that the enhancement of the UL84 levels by IE2 40 was still significantly greater than that provided by inhibition of the proteasome in cells transfected only with UL84. It is possible that IE2 86 and IE2 40 aid in the protection of UL84 from proteasomal degradation to some degree, but these experiments suggest that this is not the sole method by which UL84 protein expression and stability are governed.
UL84 has been shown to interact with a number of other cellular proteins that may have some importance in the mechanisms observed in these studies. For instance, UL84 has recently been shown to interact with the ubiquitin-conjugating E2 enzyme, ribosomal protein P0, casein kinase II, and p32 (14). It is possible that one or more of these proteins facilitate the mechanisms identified in these studies or that one or more cellular proteins aid in the regulation governed by the IE2 proteins.
Many other viruses utilize mechanisms of posttranscriptional regulation to mediate proper expression of both viral and cellular proteins throughout infection (1, 3, 35, 44, 45, 57-59). These mechanisms likely serve to provide multiple levels of control of viral and cellular expression patterns, making the environment most favorable for productive viral infection.
Acknowledgments
We thank E. S. Huang and G. Pari for antibodies directed against UL84. We thank G. Pari for providing the original UL84 vector pTARGET-UL84HA. We also thank the members of the Spector lab for their helpful feedback during the course of this work.
This work was supported by NIH grants CA073490 and CA034729.
Footnotes
Published ahead of print on 3 March 2010.
REFERENCES
- 1.Arias, C., D. Walsh, J. Harbell, A. C. Wilson, and I. Mohr. 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog. 5:e1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berndt, A., H. Hofmann-Winkler, N. Tavalai, G. Hahn, and T. Stamminger. 2009. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J. Virol. 83:12881-12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyne, J. R., and A. Whitehouse. 2006. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. U. S. A. 103:15190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant, L. A., P. Mixon, M. Davidson, A. J. Bannister, T. Kouzarides, and J. H. Sinclair. 2000. The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF. J. Virol. 74:7230-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castillo, J. P., A. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caswell, R., C. Hagemeier, C.-J. Chiou, G. Hayward, T. Kouzarides, and J. Sinclair. 1993. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J. Gen. Virol. 74:2691-2698. [DOI] [PubMed] [Google Scholar]
- 7.Chiou, C.-J., J. Zong, I. Waheed, and G. S. Hayward. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 67:6201-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, K. S., S.-J. Kim, and S. Kim. 1995. The retinoblastoma gene product negatively regulates transcriptional activation mediated by the human cytomegalovirus IE2 protein. Virology 208:450-456. [DOI] [PubMed] [Google Scholar]
- 9.Colletti, K. S., K. E. Smallenburg, Y. Xu, and G. S. Pari. 2007. Human cytomegalovirus UL84 interacts with an RNA stem-loop sequence found within the RNA/DNA hybrid region of oriLyt. J. Virol. 81:7077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colletti, K. S., Y. Xu, S. A. Cei, M. Tarrant, and G. S. Pari. 2004. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J. Virol. 78:9203-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colletti, K. S., Y. Xu, I. Yamboliev, and G. S. Pari. 2005. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 280:11955-11960. [DOI] [PubMed] [Google Scholar]
- 12.Fortunato, E. A., and D. H. Spector. 1999. Regulation of human cytomegalovirus gene expression. Adv. Virus Res. 54:61-128. [DOI] [PubMed] [Google Scholar]
- 13.Furnari, B. A., E. Poma, T. F. Kowalik, S.-M. Huong, and E.-S. Huang. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 67:4981-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, Y., K. Colletti, and G. S. Pari. 2008. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 82:96-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, Y., and G. S. Pari. 2009. Interaction of human cytomegalovirus pUL84 with casein kinase 2 is required for oriLyt-dependent DNA replication. J. Virol. 83:2393-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebert, S., S. Schmolke, G. Sorg, S. Floss, B. Plachter, and T. Stamminger. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 71:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiser, M., R. Cebe, D. Drewello, and R. Schmitz. 2001. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques 31:88-90, 92. [DOI] [PubMed] [Google Scholar]
- 18.Hagemeier, C., R. Caswell, G. Hayhurst, J. Sinclair, and T. Kouzarides. 1994. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 13:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, Y. S., L. Xu, and E. S. Huang. 1992. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J. Virol. 66:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, C., Y. Wang, D. Tsao, S. Tung, Y. Lin, and C. Wu. 2000. Antagonism between members of the CNC-bZIP family and the immediate-early protein IE2 of human cytomegalovirus. J. Biol. Chem. 275:12313-12320. [DOI] [PubMed] [Google Scholar]
- 23.Hwang, E. S., Z. Zhang, H. Cai, D. Y. Huang, S. M. Huong, C. Y. Cha, and E. S. Huang. 2009. Human cytomegalovirus IE1-72 protein interacts with p53 and inhibits p53-dependent transactivation by a mechanism different from that of IE2-86 protein. J. Virol. 83:12388-12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins, D. E., C. L. Martens, and E. S. Mocarski. 1994. Human cytomegalovirus late protein encoded by ie2: a transactivator as well as a repressor of gene expression. J. Gen. Virol. 75:2337-2348. [DOI] [PubMed] [Google Scholar]
- 25.Jupp, R., S. Hoffmann, A. Depto, R. M. Stenberg, P. Ghazal, and J. A. Nelson. 1993. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J. Virol. 67:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 67:7539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaspari, M., A. Zimmermann, R. Schilf, and E. Bogner. 2008. Proteasome inhibitor MG132 blocks viral DNA replication and assembly of human cytomegalovirus. FEBS Lett. 582:666-672. [DOI] [PubMed] [Google Scholar]
- 28.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang, D., and T. Stamminger. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J. Virol. 67:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukac, D. M., and J. C. Alwine. 1999. Effects of human cytomegalovirus major immediate-early proteins in controlling the cell cycle and inhibiting apoptosis: studies with ts13 cells. J. Virol. 73:2825-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses. In D. M. Knipe and P. M. Howley (ed.), Fields' virology, 5th ed., vol. 2. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA.
- 35.Mohr, I. 2004. Neutralizing innate host defenses to control viral translation in HSV-1 infected cells. Int. Rev. Immunol. 23:199-220. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of the cell cycle. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pari, G. S. 2008. Nuts and bolts of human cytomegalovirus lytic DNA replication. Curr. Top. Microbiol. Immunol. 325:153-166. [DOI] [PubMed] [Google Scholar]
- 38.Pizzorno, M. C., M.-A. Mullen, Y.-N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prösch, S., C. Priemer, C. Hoflich, C. Liebenthaf, N. Babel, D. H. Kruger, and H. D. Volk. 2003. Proteasome inhibitors: a novel tool to suppress human cytomegalovirus replication and virus-induced immune modulation. Antivir. Ther. 8:555-567. [PubMed] [Google Scholar]
- 40.Sadanari, H., J. Tanaka, Z. Li, R. Yamada, K. Matsubara, and T. Murayama. 2009. Proteasome inhibitor differentially regulates expression of the major immediate early genes of human cytomegalovirus in human central nervous system-derived cell lines. Virus Res. 142:68-77. [DOI] [PubMed] [Google Scholar]
- 41.Samaniego, L. A., M. J. Tevethia, and D. J. Spector. 1994. The human cytomegalovirus 86-kilodalton immediate-early 2 protein: synthesis as a precursor polypeptide and interaction with a 75-kilodalton protein of probably viral origin. J. Virol. 68:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders, R. L., C. L. Clark, C. S. Morello, and D. H. Spector. 7 May 2008. Development of cell lines that provide tightly controlled temporal translation of the human cytomegalovirus IE2 proteins for complementation and functional analyses of growth-impaired and non-viable IE2 mutant viruses. J. Virol. [Epub ahead of print.] doi: 10.1128/JVI.00675-08. [DOI] [PMC free article] [PubMed]
- 43.Sanders, R. L., C. J. D. Rosario, E. A. White, and D. H. Spector. 2008. Internal deletions of IE2 86 and loss of the late IE2 60 and IE2 40 proteins encoded by human cytomegalovirus affect the levels of UL84 protein but not the amount of UL84 mRNA or the loading and distribution of the mRNA on polysomes. J. Virol. 82:11383-11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandri-Goldin, R. M. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241-5256. [DOI] [PubMed] [Google Scholar]
- 45.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz, R., B. Helmich, and D. H. Spector. 1996. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 70:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz, R., M. H. Sommer, A. Scully, and D. H. Spector. 1994. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J. Virol. 68:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86 kDa protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sommer, M. H., A. L. Scully, and D. H. Spector. 1994. Transactivation by the human cytomegalovirus IE2 86-kDa protein requires a domain that binds to both TATA box-binding protein and the retinoblastoma protein. J. Virol. 68:6223-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spector, D. J., and M. J. Tevethia. 1994. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J. Virol. 68:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speir, E., R. Modali, E.-S. Huang, M. B. Leon, F. Sahwl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 52.Stenberg, R. M., A. S. Depto, J. Fortney, and J. A. Nelson. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J. Virol. 63:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stinski, M. F., and D. T. Petrik. 2008. Functional roles of the human cytomegalovirus essential IE86 protein. Curr. Top. Microbiol. Immunol. 325:133-152. [DOI] [PubMed] [Google Scholar]
- 54.Strang, B. L., E. Sinigalia, L. A. Silva, D. M. Coen, and A. Loregian. 2009. Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84. J. Virol. 83:7581-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran, K., J. A. Mahr, J. Choi, J. G. Teodoro, M. R. Green, and D. H. Spector. 2008. Accumulation of substrates of the anaphase-promoting complex (APC) during human cytomegalovirus infection is associated with the phosphorylation of Cdh1 and the dissociation and relocalization of the APC subunits. J. Virol. 82:529-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran, K., J. A. Mahr, and D. H. Spector. 2010. Proteasome subunits relocalize during human cytomegalovirus infection, and proteasome activity is necessary for efficient viral gene transcription. J. Virol. 84:3079-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walsh, D., and I. Mohr. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 20:461-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walsh, D., and I. Mohr. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18:660-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walsh, D., C. Perez, J. Notary, and I. Mohr. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 79:8057-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wara-Aswapati, N., Z. Yang, W. R. Waterman, Y. Koyama, S. Tetradis, B. K. Choy, A. C. Webb, and P. E. Auron. 1999. Cytomegalovirus IE2 protein stimulates interleukin 1β gene transcription via tethering to Spi-1/PU.1. Mol. Cell. Biol. 19:6803-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, E. A., C. J. Del Rosario, R. L. Sanders, and D. H. Spector. 2007. The IE2 60-kilodalton and 40-kilodalton proteins are dispensable for human cytomegalovirus replication but are required for efficient delayed early and late gene expression and production of infectious virus. J. Virol. 81:2573-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, E. A., and D. H. Spector. 2005. Exon 3 of the human cytomegalovirus major immediate-early region is required for efficient viral gene expression and for cellular cyclin modulation. J. Virol. 79:7438-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiebusch, L., M. Bach, R. Uecker, and C. Hagemeier. 2005. Human cytomegalovirus inactivates the G0/G1-APC/C ubiquitin ligase by Cdh1 dissociation. Cell Cycle 4:1435-1439. [DOI] [PubMed] [Google Scholar]
- 65.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wiebusch, L., and C. Hagemeier. 2001. The human cytomegalovirus immediate early 2 protein dissociates cellular DNA synthesis from cyclin dependent kinase activation. EMBO J. 20:1086-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeung, K. C., C. M. Stoltzfus, and M. F. Stinski. 1993. Mutations of the human cytomegalovirus immediate-early 2 protein defines regions and amino acid motifs important in transactivation of transcription from the HIV-1 LTR promoter. Virology 195:786-792. [DOI] [PubMed] [Google Scholar]
- 68.Yoo, Y. D., C.-J. Chiou, K. S. Choi, Y. Yi, S. Michelson, S. Kim, G. S. Hayward, and S.-J. Kim. 1996. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor B1 gene through an Egr-1 binding site. J. Virol. 70:7062-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]