Abstract
Herpes simplex virus 1 (HSV-1) is a well-adapted human pathogen that can invade the peripheral nervous system and persist there as a lifelong latent infection. Despite their ubiquity, only one natural isolate of HSV-1 (strain 17) has been sequenced. Using Illumina high-throughput sequencing of viral DNA, we obtained the genome sequences of both a laboratory strain (F) and a low-passage clinical isolate (H129). These data demonstrated the extent of interstrain variation across the entire genome of HSV-1 in both coding and noncoding regions. We found many amino acid differences distributed across the proteome of the new strain F sequence and the previously known strain 17, demonstrating the spectrum of variability among wild-type HSV-1 proteins. The clinical isolate, strain H129, displays a unique anterograde spread phenotype for which the causal mutations were completely unknown. We have defined the sequence differences in H129 and propose a number of potentially causal genes, including the neurovirulence protein ICP34.5 (RL1). Further studies will be required to demonstrate which change(s) is sufficient to recapitulate the spread defect of strain H129. Unexpectedly, these data also revealed a frameshift mutation in the UL13 kinase in our strain F isolate, demonstrating how deep genome sequencing can reveal the full complement of background mutations in any given strain, particularly those passaged or plaque purified in a laboratory setting. These data increase our knowledge of sequence variation in large DNA viruses and demonstrate the potential of deep sequencing to yield insight into DNA genome evolution and the variation among different pathogen isolates.
Herpes simplex virus 1 (HSV-1) is among the most widespread pathogens of the herpesvirus family, with about 60% seroprevalence, indicating exposure or ongoing infection, among adults in the United States (83). HSV-1 infection begins at epithelial surfaces but can progress to the peripheral nervous system, where a lifelong latency is established in neurons (60). HSV-2 is closely related and presents a major public health concern in developing nations, where it is a risk factor for the acquisition of HIV/AIDS (10, 23). Despite the clinical importance of these viruses, only one wild-type genome sequence is available for HSV-1, that of strain 17, which was completed over 20 years ago (41, 42). Remarkably, most of our understanding of HSV-1 biology comes from experiments utilizing just a few common laboratory strains or recent clinical isolates. The only other HSV-1 genome sequence published in the last 2 decades is that of HF10, an oncolytic mutant strain harboring several large genomic deletions and rearrangement relative to the reference strain 17 (78). Since HF10 was itself derived from the nonneuroinvasive and highly attenuated strain HF, the HF10 genome is informative for mutation-based variation but provides little insight into the sequence variation of virulent strains (45, 70). Several studies of specific genes or genomic regions cloned in Escherichia coli have shed more light on interstrain variation in HSV-1, but these studies cannot address variation on a genome-wide scale encompassing every protein in the HSV genome (48, 57, 74, 75). High-throughput sequencing techniques have the potential to address the entire genome of a population without resorting to recombinant DNA techniques and have already enabled substantial inroads into novel pathogen discovery and the genetic characterization of other viruses and pathogens (13, 34, 54, 79, 81).
The genome of HSV-1 is a large double-stranded DNA molecule of 152 kb, with a G/C content of 68%. The HSV genome contains 77 annotated protein-coding sequences, arranged into two unique regions, each of which are flanked by long terminal repeats (9.2 kb and 6.6 kb) (genome diagram in Fig. 1A). In addition to these large repeats, the genome also contains small microsatellite repeats (<100 bp each) and short tandemly reiterated sequences (<500 bp each), also known as variable-number tandem repeats (VNTRs) (14, 41, 42). The large terminal repeats contain a higher concentration of VNTRs and a lower percentage of coding regions than elsewhere in the genome. The VNTRs are highly variable, with the number of repeated units varying both between strains and during replication and repeated passages of the same strain (48, 74-76). The large number of mononucleotide repeats in the HSV-1 reference genome suggested that Illumina's deep sequencing technology, which detects single bases at a time by using reversible chain termination chemistry, would be a useful technology for sequencing these genomes (14, 36).
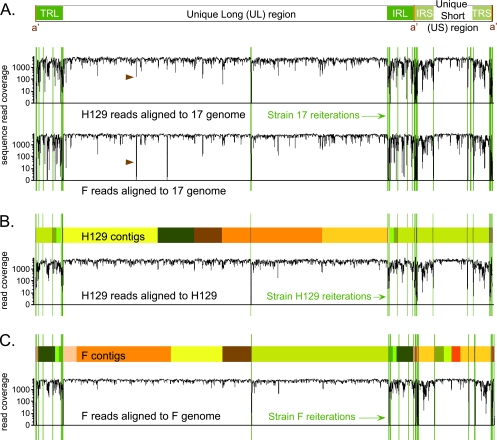
FIG. 1.
Overview of genome coverage for HSV-1 strains F and H129, relative to strain 17 or to their own newly assembled genomes. (A) Diagram of the HSV-1 genome structure, with two unique regions (long and short), each flanked by a pair of terminal repeats. The line graph depicts the depth of sequence read coverage for the newly sequenced strains when each one is aligned to the reference, strain 17. The VNTRs, or reiterations, of strain 17 are overlaid as green lines onto the coverage graphs. Reductions in coverage correlate with the reiterated sequences and also occur more frequently in the terminal repeats that flank each unique region. Brown arrowheads highlight a dip in both new genomes when aligned against strain 17; this is the location of an insertional frameshift in the UL17 gene of strain 17 that leads to alignment mismatch in the nonframeshifted strains F and H129. (B and C) Colored boxes depict the blocks of continuous sequence, or contigs, assembled for each strain. Boxes of the same color represent the same contig; these appear twice when they include sequence in the repeat regions. Sites of reiterations (VNTRs) are overlaid in green. In some cases contigs assembled through a reiteration; in others the contigs terminated at the reiterations. The line graph below the contigs for each strain depicts the improvement in depth of sequence read coverage when data from strain H129 were aligned to the newly assembled H129 genome (B). A similar graph is depicted for strain F (C). Sharp drops in alignment coverage remain at the VNTR sequences inserted from strain 17.
Historically, comparisons of phenotypic and genotypic variations among strains or species of related organisms have provided significant insights to the field of genetics. Similarly, comparison of complete herpesviral genome sequences of clinical and laboratory isolates would greatly facilitate studies of sequence variation and conservation. Significant progress has already been demonstrated for varicella-zoster virus (VZV), Marek's disease virus (MDV), and human cytomegalovirus (HCMV) (8, 12, 53, 58, 66, 67, 72, 85). Sequence analysis can be used to highlight the most conserved, and thus functionally important, domains of proteins, as well as to identify likely regulatory regions in intergenic areas, based on their sequence conservation in the absence of coding pressure. Sequencing the entire genomes of HSV-1 strains with interesting phenotypes will also allow identification of putative causative mutations more comprehensively than single-gene cloning approaches. The unique HSV-1 H129 strain presents one such opportunity; it is the only virus known to transit neural circuits exclusively in an anterograde or forward direction, a finding that has been confirmed in both rodent and primate models (69, 86). H129 was isolated from the brain of an encephalitic patient in 1977, and the limited molecular characterizations thus far have not shed light on any mutations to explain its unique phenotype (17, 30, 33). The distinctive spread characteristics of this strain makes it of great interest to the neuroscience community, where it is used as a directional neural circuit tracer whose spread is complementary to retrograde-limited tracing viruses, such as the attenuated pseudorabies virus (PRV) strain Bartha and various rhabdoviruses (3, 19, 24, 59, 73).
We demonstrate here the successful use of Illumina deep sequencing technology and subsequent analyses to determine the genome sequences of both the unique clinical isolate HSV-1 H129 and a widely used laboratory isolate (strain F). These strains differ in pathogenicity from the previously sequenced strain 17. After peripheral inoculation into mice, strain 17 has a 50% lethal dose (LD50) of 103 PFU, while the LD50 of strain H129 is 105 PFU and for strain F it is >107 PFU (17, 55). Our data demonstrate the extent of variation between these strains across the entire genome of HSV-1, in both coding and noncoding regions. We found many protein-coding variations between strain F and the current genome reference strain 17 by which we can begin to define the spectrum of variability among wild-type HSV-1 isolates. We have fully defined the sequence differences in the unique anterograde spread mutant strain H129 and propose a number of potentially causal genes, including the neurovirulence protein ICP34.5 (RL1). Unexpectedly, our data also revealed a frameshift mutation in the UL13 kinase in our isolate of HSV-1 strain F. This protein is dispensable in cell culture but is required for virulence and spread of infection in animal models (11, 51, 71).
MATERIALS AND METHODS
Virus stocks.
HSV-1 strain F was originally isolated from a facial lesion and maintained as a low-passage stock by B. Roizman and colleagues (18). We received an aliquot from B. Roizman, which was passaged once in Vero cells and then subjected to three rounds of plaque purification. HSV-1 strain H129 is a low-passage clinical isolate received from Richard Dix (17); it is maintained as a low-passage stock. All viral stocks were grown on monolayers of confluent Vero (monkey kidney) cells (ATCC cell line CCL-81).
Nucleocapsid DNA preparation.
Viral nucleocapsid DNA was isolated as previously described (63). Briefly, confluent monolayers of Vero cells were infected at a multiplicity of infection of 5 and harvested by scraping at 24 h postinfection. Cell pellets were rinsed, resuspended, subjected to two rounds of Freon extraction, and pelleted through a glycerol step gradient. Viral nucleocapsids were then lysed using SDS and proteinase K, extracted twice with phenol-chloroform, and ethanol precipitated. Viral DNA was collected by a glass hook, blotted dry, and resuspended in Tris-EDTA (10 mM Tris, pH 7.6; 1 mM EDTA).
Illumina sequencing.
Five-microgram aliquots of HSV-1 strain F and H129 nucleocapsid DNA were processed for sequencing by the Microarray Core Facility at Princeton University's Lewis-Sigler Institute for Integrative Genomics. Two independent sequence libraries were generated by following the manufacturer's protocol for sequencing of genomic DNA (Illumina genomic DNA sample prep kit; protocol part 1003806, revision A), with the slight modification that the column for gel purification was not heated (56). Sequencing was carried out using two lanes of a standard flow cell, using Illumina's standard cluster generation and 36-cycle sequencing kits. The Illumina genome analyzer 2, with SCS 2.3 software, was run for either 36 (one H129 run) or 75 (all other runs) cycles of data acquisition. Image analysis and base calling were performed using the Illumina Pipeline v1.3 under default settings.
De novo and reference-guided assembly.
De novo assembly of the short reads was performed to generate new HSV-1 genomes from the sequence data, followed by a reference-guided assembly of the resulting blocks of contiguous sequences, or contigs. The short sequence reads were first passed through a series of computational filters that removed (i) mononucleotide sequences, (ii) host sequence contamination, and (iii) low-quality sequence. For step i, sequences that consisted of a single nucleotide or a single nucleotide with some N (noncalled) bases were removed. (Step ii) Since virus stocks were prepared on Vero cells, it was critical to identify and remove host DNA sequences. Because the vervet monkey (Vero cell parent) genome sequence is not known, the sequence data were mapped to the human genome (version 36) using the Mapping and Alignment with Qualities (MAQ) software package (32). Sequences homologous to human DNA varied from 0.2 to 15% of the data (see Table S1 in the supplemental material); these were considered host contamination and removed from the analysis. (Step iii) The sequences were then quality trimmed using a modified version of the quality-trimming script supplied with the SSAKE assembler (80). The process of quality trimming removed terminal bases below a quality of 10 and then removed any sequences whose overall resulting length was less than 20 bases. The 36-bp sequencing run for strain H129 (versus all others, of 75-bp length) thus resulted in a net smaller number of sequences for de novo assembly of strain H129 versus strain F. After these filtering procedures, the SSAKE short read assembler was used to assemble the short sequences into contigs, using default parameters.
Reference-guided assembly of the best contigs yielded the final reference sequence. Those that were at least 100 bp long and had an average sequence depth, or coverage, of at least 100 sequence reads were passed to the long read assembler MINIMUS (65). All blocks of assembled sequence were surveyed by BLAST to check for erroneous ends, and the most parsimonious and best-supported sequence was accepted when there was disagreement at the ends of joined segments (2). The resulting blocks of sequence, along with any contigs that MINIMUS was unable to assemble further, were aligned to the strain 17 genome using BLAST. The BLAST alignments provided guidance to position blocks of sequence along the genome. Rarely, short mononucleotide runs caused BLAST to place a contig at discontinuous locations. These anomalous breaks were examined and accepted if supported by data from adjacent blocks of sequence. The light orange and light green contigs on the right end of the strain H129 genome are one such example (Fig. 1; labeled minimus2_1 in GenBank and Genome Browser). BLAST also allowed us to place data from assembled blocks of sequence into both repeats when relevant (TRL/IRL and TRS/IRS); this can be seen in Fig. 1 where contig colors match in the repeats.
Short reiterations, or VNTRs, are highly variable in length in both genomic DNA preparations and in cloned DNA, making their assembly a challenge (37, 46, 72, 76, 77). In Illumina sequencing, the average number of repeats in a population of DNA can be accurately estimated by de novo assembly only if the short reads contain unique flanking sequence on one or both ends. This ability is limited by the read length (75 bp in this case). The SSAKE program defaults to assembling the shortest possible number of repeating units supported by the sequence data and may thus underestimate the VNTR lengths for those exceeding 75 bp. As was done for the currently available HSV-1 strain 17 transgenic bacterial artificial chromosomes (BAC) sequence (accession number FJ59328), we marked reiterations of uncertain lengths as such and expanded them to match the published length of the original strain 17 reference sequence. This was done for the following VNTRs: the a′ reiterations, reiterations 1 and 4 in the long repeats, reiterations 1 to 3 in the short repeats, the UL reiteration in UL36, and the US reiteration 1. The exact boundaries of these VNTRs are annotated in the GenBank nucleotide sequences for the corresponding accession numbers for these genomes and are also visible at our genome browser, http://viro-genome.princeton.edu.
Coverage analysis by alignment to reference and new genomes.
MAQ was used to align short Illumina reads against the NCBI HSV-1 genome of strain 17 (RefSeq NC_001806) (44). The default parameters were used to produce an alignment file as well as a consensus sequence. From the consensus, the SNPfilter command was used with default parameters to filter out false-positive single-nucleotide polymorphisms. Once a new genome was assembled for strains H129 and F, the reads were realigned to the new self-genome by using MAQ and analyzed as above.
Determining DNA and amino acid variation.
To determine overall DNA sequence variation, we aligned each pair of genomes using BLAST and compiled a list of differences using the MUMmer sequence analysis package (15). For amino acid variation, we used BLAST to align each piece of coding sequence from the strain 17 reference to the new genome. These coding sequence locations (see GenBank accession nos. GU734771 and GU734772 for exact positions) were used to generate amino acid translations from the new genome. Each new amino acid sequence was aligned to the corresponding strain 17 protein sequence by using BLAST, and differences were compiled as described above (see Table S2 in the supplemental material). Finally, DNA sequence differences in each coding region were tallied as above; these included both silent mutations and nonsynonymous changes that led to protein-level differences (see Table S3). For both DNA and amino acid comparisons, we counted both the total number of changes (e.g., three changes in a row were counted as three) and the number of noncontiguous change events (e.g., three changes in a row were counted as one change event).
PCR analysis of UL13 mutations.
PCR for UL13 used the following primers: forward, CTTACCGAGGTCCATGTCGT, and reverse, CTTTCTAACCGCACACCGAC. PCR products were not cloned but were directly sequenced using internal primers, either CAGTTGGACTTCGCCGTATC in the forward direction or CTGGTCATGTGGCAGCTAAC in the reverse. This technique allowed detection of a mixed population when present.
Nucleotide sequence accession numbers and online data repositories.
Genome sequence data and all annotations described in the manuscript have been deposited at GenBank under accession numbers GU734771 for strain F and GU734772 for strain H129. Annotations include the locations of genes, coding sequences (CDS), repeats, and reiterations. Boundaries of the contiguous sequence blocks (contigs) used to assemble each genome are also included so that the boundaries can be reviewed by future users. Raw sequence reads have been deposited at the NCBI Sequence Read Archive (SRA) under accession numbers SRA010802.1 for strain F and SRA010966.2 for strain H129. These data are all linked under NCBI Genome Project ID 43419. These data can also be viewed at an interactive genome browser at http://viro-genome.princeton.edu. This site includes data from this paper that were not incorporated by GenBank, such as sequence coverage depth maps for each genome (Fig. 1), histograms of sequence differences per 100 bp (see Fig. 2, below), and the location of insertions, deletions, and single-nucleotide changes on each sequence relative to the reference strain 17. Users can view data at the whole-genome scale or investigate the same features at the level of individual genes (see, for example, Fig. S2 in the supplemental material).
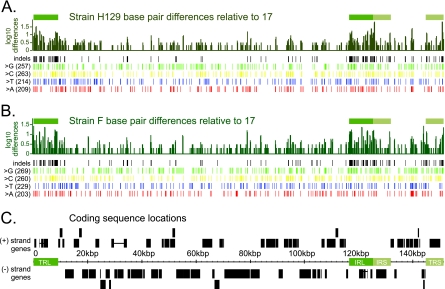
FIG. 2.
DNA-level variation across the HSV-1 genome. (A) Bar graph summarizing the number of base pair differences for strain H129 relative to reference strain 17, with differences averaged over a window of 100 bp. The x axis corresponds to the location along the HSV-1 genome (marked in kbp in panel C). The y axis is plotted on a log scale to make small amounts of difference visible on the graph. Green boxes above the bar graph show the location of the terminal repeats, to highlight the increased DNA sequence variation in these areas. Rows of vertical lines depict base pair variations across the genome, with variation separated out by indels (insertions or deletions), and single nucleotide changes. Single base changes are further subdivided by the identity of the base in the new strain: G, C, T, or A. (B) Similar bar graph and statistics for strain F. (C) Location of coding sequences along the genome of HSV-1, which encodes 77 proteins. Plus- and minus-strand-encoded genes are drawn above and below the genome position line. Repeats are denoted by green boxes. Horizontal lines connect the coding sequence boxes of the only spliced transcripts in HSV-1: that of ICP0 (RL2), which is found in the long repeats TRL and IRL, and UL15, which is found around 30 kbp.
RESULTS
High-throughput sequencing of two viral genomes.
Nucleocapsid DNA was used as the source material for high-throughput deep sequencing of two new HSV-1 genomes. Two separate sequencing runs were carried out for each strain, providing a total of 17.7 million short sequence reads for H129 and 14.1 million for F (see Table S1 in the supplemental material). To provide a general outline of genome coverage, we used MAQ software to align these reads against the only currently available wild-type HSV-1 genome of strain 17 (NCBI record NC_001806) (32). This technique revealed an average coverage depth of over 1,000 sequence reads per base pair in the unique regions of the genome and revealed much lower and more variable coverage depth in the terminal repeats that flank each unique region (Fig. 1A). This variable coverage reflects more base changes, insertions, and/or deletions in the repeat regions of the new strains, relative to the reference sequence. Since alignment approaches are not well equipped computationally to handle insertions, deletions, and repetitive sequences (22, 32), we used de novo sequence assembly as a productive alternative approach.
De novo assembly and reiterated sequences.
In de novo assembly, short sequence reads are assembled into larger blocks by using overlapping stretches of homology between the reads. This technique produces longer stretches of continuous sequence, termed contigs. To improve the de novo assembly process, we identified and removed host DNA sequences that always contaminate viral DNA preparations. Host sequences amounted to 0.2 to 15% of the data (see Table S1 in the supplemental material). We used BLAST to order the assembled contigs along the reference genome. Many of these sequence blocks terminated at the VNTRs, or reiterations, found throughout the HSV-1 genome (Fig. 1B and C) (2). We note that all currently available high-throughput methods for sequence determination are unable to identify the length of a VNTR unless the VNTR is within the actual sequence read length (12, 22, 36). Among these data, only imperfect reiterations or those less than the sequence read length of 75 bp could be accurately sized by the presence of unique flanking sequence. Despite this, we were able to assemble the entire genome as follows: we verified that the longer reiterations contained sequence of the same repeating units, and then we extended the VNTR length to match the number in the currently published reference strain 17 (see Materials and Methods for a list of expanded VNTRs). This method provides as much consistency as possible in overall gene positions and genome length. In summary, the new genome sequence assembled for strain F is 152,151 bp, while that of strain H129 is 152,066 bp, both of which are similar to the length of strain 17 at 152,261 bp (see Fig. S1 in the supplemental material).
To confirm the accuracy of these new genome assemblies, we realigned all of the sequence reads for each strain, and this time we used the appropriate self-genome as an alignment guide. This method revealed a more consistent, high level of coverage across the genome, with significant reduction in coverage only at the VNTRs where proxy sequence was inserted from strain 17 (Fig. 1B and C). For strain F, 97.6% of the nonreiteration portions of the genome have 100-fold or greater sequence coverage and 95.6% have 1,000-fold or greater depth of coverage. For strain H129, 97.5% of the nonreiteration portions of the genome have 100-fold or greater sequence coverage, with 93.4% of that at a coverage of 1,000-fold or greater. The slightly lower coverage depth of strain H129 is because one of the two sequencing runs had a shorter read length, 36 bp, instead of the 75-bp length used for all other runs. Even data from this short read data set could be assembled into high-quality sequence. These newly assembled genomes were next used to assess DNA-level sequence variation across the genome.
DNA-level sequence variation.
Variation among viral genomes reflects the processes of mutation and recombination. Subsequent selection pressures fix these changes in populations, and these pressures vary during replication in vivo and in vitro. High-throughput sequencing is especially well suited to reveal the full extent of overall genome variation between strains, because it comprehensively surveys the entire genome sequence in a given population of DNA. In pairwise alignments of each new genome against the reference, we found that strain F had 961 bp changes relative to strain 17, while H129 had 943 bp changes relative to strain 17 (Fig. 2A and B; the figure shows changes by base type, A, C, G, and T) (see also the summary in Fig. S1 of the supplemental material). Gaps are created in the alignment whenever one strain has an insertion or deletion relative to the reference strain. For strain F, there were 332 bp of insertions and 431 bp of deletions relative to strain 17. Strain H129 had 298 bp of insertions and 496 bp of deletions relative to strain 17. Overall, these nucleotide differences are dispersed throughout the genome, with a slightly greater concentration of differences in the repeats relative to unique regions (Fig. 2A and B).
We also examined the number of evolutionary change events in the DNA sequences, where contiguous variations, such as deletions of several bases in a row, are considered one event. These change events were examined for intergenic regions, coding sequences, and untranslated regions (UTRs). Not surprisingly, the lowest rate of change from the strain 17 reference was found in coding regions, where evolutionary pressure is likely highest: six changes per kb in strain F or five per kb in H129. In contrast, both new strains had three times more changes (15/kb) in intergenic regions and a similarly high rate in the UTRs (17/kb in F and 18/kb in H129). If we analyze the large terminal repeats separately from the rest of the genome, the most noticeable changes emerge in the intergenic repeat regions, where the differences from the reference strain are at 17 per kb for both F and H129, versus just 10 (H129) or 11 (F) per kb for intergenic, nonrepeat regions. However, all of these changes together represent a <1% deviation from the HSV-1 strain 17 genome sequence, indicating a high degree of overall DNA sequence conservation among these three strains. We likewise found that the relative positions of the open reading frames are similar in all three genomes (Fig. 2C). Although the positions of these coding sequences are largely conserved, we next addressed the conservation and variation of the resulting protein sequences.
Amino acid coding-level changes.
To a first approximation, selection for function leads to maintenance of sequence fidelity. Therefore, we determined which of the base pair changes, insertions, and deletions affected the coding sequence. Overall, we found 310 amino acid differences between wild-type strains F and 17 and 281 amino acid differences between H129 and the reference strain 17 (summarized in Fig. S1 of the supplemental material). Strains F and H129 have fewer overall amino acid differences with each other, totaling 231 across the proteome. These amino acid differences occur throughout the complement of 77 proteins encoded by HSV-1 (Fig. 3) and can be categorized as changes where strains F and H129 share the same amino acid residue with each other, but differ from strain 17, versus those positions where only strain H129 or strain F has a unique amino acid relative to the other two strains (see Table S2 for a full list of all amino acid differences for each protein). In a prior analysis using a limited number of genes, Norberg and colleagues found that strain 17 and strain F were divergent enough to fall into distinct clades (47, 48). Since these clades are distinguishable based on restriction digest patterns in the US4 and US7 genes (48), we applied this approach and found that strain F and strain H129 fall into the same clade, while strain 17 does not (data not shown). This similarity in clade may reflect the fact that strains F and H129 were both isolated from patients in the United States, while strain 17 was isolated from a Scottish patient (17, 18, 42).
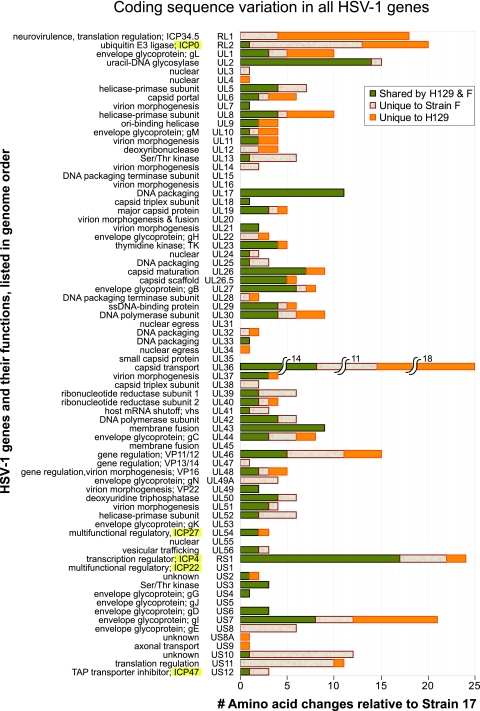
FIG. 3.
Coding sequence variation across the HSV-1 proteome. Histogram depicting the total number of amino acid differences observed in strains F and H129, relative to the reference strain 17, for every protein in the HSV-1 genome. Changes are color coded to depict the proportion of changes that are different from strain 17 but common between strains F and H129, versus those that are unique to strain F or H129. Proteins are listed in spatial order of occurrence along the HSV-1 genome. Names of the five immediate-early genes, expressed first upon herpesvirus infection of a host cell, are highlighted in yellow (ICP0, ICP27, ICP4, ICP22, and ICP47). Genes in the repeats, ICP34.5 (RL1), ICP0 (RL2), and ICP4 (RS1), are each listed only once, at their first position of occurrence. Ten genes have no amino acid changes at all. Protein functions are summarized to the left of each gene name.
Complete coding sequence conservation.
The analysis of amino acid differences across the HSV-1 proteome revealed 10 genes with complete conservation across strains F, H129, and 17: the capsid protein UL35; tegument protein UL16; the envelope protein UL20 and glycoproteins gK (UL53) and gJ (Us5); and the nonstructural proteins UL15, UL31, UL45, UL55, and ICP22 (Us1). These proteins vary in coding sequence length, from 92 amino acids for glycoprotein J (US5) to 735 amino acids for the DNA terminase subunit protein UL15, indicating that sequence length is not the primary criterion for complete amino acid conservation. Several of the genes in this group are known to be dispensable for growth in cell culture, such as UL20, gK (UL53), ICP22 (Us1), gJ (Us5), UL45, and UL55, but their conservation suggests an evolutionary advantage to preserving their functions.
Amino acid differences unique to the mutant strain H129.
Although the complete conservation of coding sequences across these strains is noteworthy, we were particularly interested in deducing the likely mutations behind the unique anterograde spread phenotype of the clinical isolate strain H129. Rather than the typical HSV-1 bidirectional spread from infected neurons, H129 appears to only exit via axonal connections from the presynaptic to postsynaptic cell, producing an overall phenotype of exclusively anterograde-directed spread along neural circuits in vivo. We searched for amino acid changes unique to H129 relative to both reference strain 17 and to the newly sequenced strain F, to uncover the mutations responsible for this directional spread phenotype. We first examined genes with the largest number of amino acid changes overall and highlighted those with many changes unique to strain H129 (Fig. 4A). These included the large tegument protein UL36, the neurovirulence protein ICP34.5 (RL1), the ubiquitin E3 ligase ICP0 (RL2), and the envelope glycoproteins gI (US7) and gL (UL1). This analysis revealed that some genes with large numbers of amino acid changes, such as the transcriptional regulator ICP4 (RS1; see Fig. S2 in the supplemental material) and the uracil-DNA glycosylase UL2, have changes that are largely shared with wild-type strain F, suggesting that these are less likely candidates to explain the unique phenotype of strain H129. Since gene length reflects the target size for mutations accumulated over time, we also normalized the number of amino acid changes observed for gene length (Fig. 4B). Several of the same genes are highlighted again, including ICP34.5 (RL1), gI (US7), and gL (UL1), while the short tegument protein UL11 now arises as another potential candidate. Strain F has many amino acid differences in several of the same genes: UL36, ICP0 (RL2), and gI (US7) (see Fig. S3 in the supplemental material). The genes that have large numbers of amino acid changes, both overall and with respect to gene length, are likely candidates to explain all or part of the H129 phenotype.
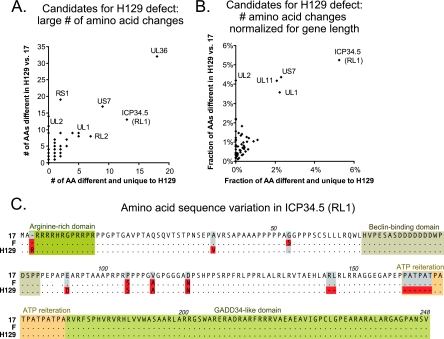
FIG. 4.
Potential causative mutations of the H129 spread defect. (A) Scatter plot of the strain H129 proteins, showing the largest overall number of amino acid differences from the wild-type reference strain 17 (y axis) versus the subset of these changes that are also different from the wild-type strain F (x axis) and are thus unique to H129. The most extreme points are labeled to identify the proteins. (B) Because gene length affects the total possible number of observed mutations, the same data were normalized for gene length. The scatter plot presents H129 proteins with the largest overall percentage of amino acids differing from the reference strain 17 (y axis), versus the subset of these changes that are also different from wild-type strain F (x axis). (C) Amino acid alignment of the ICP34.5 (RL1) protein in strains 17, F, and H129. Previously described functional domains are boxed, while amino acid residues that differ from reference strain 17 are highlighted in red. Amino acids that match the reference are represented by dots for clarity; dashes represent deletions in the respective strains.
ICP34.5 and other candidates that could account for the H129 spread defect.
Substantial amino acid changes may affect protein structure and function. ICP34.5 (RL1) is a well-known neurovirulence gene previously demonstrated to affect the spread of HSV-1 strains in vivo (6, 82). The H129 strain has one extra arginine in an N-terminal arginine-rich domain of ICP34.5 (38) and two unique amino acid changes that fall on either side of the Beclin-binding domain mediating ICP34.5's effect on autophagy (Fig. 4C) (50). The other H129-specific changes in ICP34.5 are two small deletions, one of which is in the Ala-Thr-Pro (ATP) reiteration. Although long reiterated sequences are not determined with accuracy by de novo assembly, H129 has an extremely short ATP reiteration of only 33 bp, which we validated by PCR (data not shown). Short ATP reiterations in ICP34.5 have been previously associated with decreased neurovirulence (7, 38). The C terminus of ICP34.5 has a domain akin to that of the mammalian protein GADD34 (growth arrest and DNA damage), which blocks protein shutoff by host cells and facilitates viral replication. However, this domain is unchanged in both newly sequenced strains (25). ICP34.5's role in neurovirulence and these H129-specific changes in the amino acid sequence suggest ICP34.5 as a prime candidate for further studies of the H129 phenotype.
We also examined the coding sequence differences of a number of other candidate proteins. The short tegument protein UL11 has a total of four amino acid changes in these strains, two of which are specific to H129. UL11 is highly conserved among herpesviruses and plays a role in virion envelopment through its interaction with the tegument protein UL16 (4, 31, 35, 84); however, none of the observed changes lie in the functional interaction domains of this protein. Glycoprotein gI (US7) is another potential candidate because of its dimerization with glycoprotein gE (US8) and its roles in immunoglobulin binding, axonal sorting, and virulence (43, 64). H129 has 17 amino acid changes in gI, of which 8 are shared with the wild-type strain F and another 7 result from a change in length of a VNTR in gI. Although Norberg and colleagues have shown that the amino acids encoded by this reiteration are substrates for O-linked glycosylation, the VNTR varies in length among many clinical isolates, making its change unlikely to be responsible for the unique phenotype of H129 (48, 49). The remaining two mutations in gI that are unique to H129 lie outside its known functional domains. Another glycoprotein, gL (UL1), has five amino acid changes unique to the H129 strain, plus an additional three shared with the F strain. Glycoprotein gL (UL1) is part of the HSV-1 fusion complex that includes glycoproteins gH, gB, and gD (52). Three of the H129-specific changes lie near a region recently suggested to be part of a gL-gH interaction domain (21), which if disabled could make gL an attractive candidate to explain part of the H129 phenotype. The largest number of amino acid changes in the H129 strain was found in the essential tegument protein VP1/2 (UL36) (1, 16, 29, 62). This multifunctional protein is also the largest in HSV-1, at 3,139 amino acids, a length that dwarfs the 18 amino acid changes in the H129 strain when these changes are normalized for length. These additional candidate proteins, either alone or together, may contribute to the anterograde spread phenotype of the H129 strain and warrant further investigation.
Background mutation detection: the UL13 kinase.
In addition to uncovering mutations in the H129 strain, we found an unexpected mutation in the wild-type strain F isolate: a frameshift in the UL13 kinase gene resulting from the deletion of one C in a mononucleotide run of six Cs. This frameshift changes the amino acid sequence of UL13 from amino acid 120 forward and then introduces a stop codon that truncates the protein at residue 150 instead of the normal length of 518 amino acids. To verify this mutation, we PCR amplified this region and directly sequenced the PCR product to assess any variability in the stock population. All plaque-purified strain F stocks in our lab carried this mutation, while isolates of strains NS, RE, and several ICP34.5 mutants of strain F did not (82). The original stock of strain F in our laboratory displayed a mixed population of mutant and wild-type sequence, demonstrating the likely source of the frameshift found in the plaque-purified stock used for sequencing. The sequence of UL13 in all other strains matches that of the original strain 17 reference at this position, indicating that our sequenced strain is indeed a UL13 mutant. All amino acid comparisons between strains in this paper were done with a corrected version of UL13. The strain F genome sequence submitted to NCBI has been corrected to the parental version, with a notation of the location of the frameshift in the sequenced isolate.
DISCUSSION
Genome sequencing of clinical and lab isolates of HSV-1 provides rich data on interstrain variations at both the DNA and amino acid levels. It presents the opportunity to map simple and complex phenotypes of interest to specific genes, as we have done with the unique strain H129, whose anterograde spread phenotype is of crucial interest to the field of neural circuit tracing. Defining the full genetic spectrum of any virus stock also allows one to find previously undetected mutations, as demonstrated by the unexpected UL13 kinase mutation found in our otherwise-wild-type strain F. Further analysis of these data, including complementation testing with the candidate mutations of the H129 strain, will allow us to determine causality in these genotype-phenotype connections.
Future advances in genome sequencing of herpesviruses.
One sequencing run provided extensive coverage (>1,000-fold) of the genome (Fig. 1), far beyond the depth used for most genome sequencing projects (12, 26, 27, 39, 40, 61). Future sequencing will be done by multiplexing four or more strains per run, providing more power for interstrain comparisons. To handle the enormous sequence output of these projects, improvements in de novo assembly will be required for facile analysis. We used a combination of de novo assembly followed by alignment to position large blocks of sequence along the reference genome, but this method cannot fully address the possibility of transpositions or other rearrangements. Standard restriction fragment length polymorphism methods can be used to address these issues, and deep sequencing technologies using longer reads or paired-end sequencing may also assist in assembly. We cannot overemphasize the importance of the source of DNA used for future sequencing projects. Our data demonstrate that plaque-purified viral DNA may fix variations from the original stock into sequence artifacts, as demonstrated by the UL13 kinase mutation. Single genomes that are cloned into BACs reflect cloning of a single genome from a diverse population, and they will likely have similar issues of genetic bottlenecks and unintentionally selected mutations.
Limitations of VNTR sequencing.
The HSV-1 genome contains 24 documented VNTRs or reiterations (41, 42). In both HSV and the related alphaherpesvirus varicella-zoster virus, VNTR lengths vary between strains and also during multiple passages of the same strain (37, 46, 53, 72, 74-77). Precision in defining the length of reiterated sequences is impossible for most sequencing technologies, and even paired-end reads do not offer precise length determinations because of variations in the insert size of the sequencing libraries. Thus, many published genome studies across a wide range of species either do not report data for reiterated sequences or exclude data mapping to repetitive regions from any further analyses (26, 27, 39, 40, 61). While the approach used here yielded sequence reads covering all HSV-1 reiterations, currently available assembly methods precluded accurate length determinations for about half of the HSV-1 reiterations (22). According to current genome finishing standards recommended for all species from viral to eukaryotic, these HSV-1 genomes would be considered noncontiguous finished because of the imprecision of VNTR length (9). Future efforts to determine VNTR length by traditional sequencing methods may allow for further understanding of variations in these regions.
Background mutation detection.
The loss of a functional UL13 kinase protein in our plaque-purified isolate of HSV-1 strain F provides a cautionary note to our confidence in purportedly wild-type laboratory strains and also to the genetic background of strains used for directed mutagenesis. It is common to assume that DNA genomes are inherently stable and exhibit almost no variation in sequence during laboratory passage. However, until now, we have been unable to comprehensively analyze the entire genome complement, or background, of any given strain, and thus our knowledge of genetic drift in culture has been limited at best. The UL13 kinase, like many HSV-1 proteins, is not required for growth in vitro and only marginally affects virus fitness in cell culture (11, 51, 71), allowing its mutation to pass unnoticed. Surprisingly, the homologous UL13 kinase of MDV is also frequently deleted during laboratory passage, suggesting that mutation of UL13 may provide some as-yet-unknown adaptive advantage to growth in cultured cells (5, 28, 68). There are at least two other examples of nonessential genes found to be truncated in passaged HSV lab strains: a terminal truncation of gI (Us7) in the KOS321 strain (48), and a truncated vhs protein (UL41) in the HSV-2 HG52 strain (20). As high-throughput sequencing technologies become more facile and widespread, it may be feasible to routinely sequence lab isolates and mutagenized strains, in order to screen for unexpected, unnoticed mutations. In this regard, a powerful use of whole genome sequencing will be the analysis of suppressor mutations, which is a useful method to detect genetic interactions.
Completely conserved proteins: potential therapeutic targets.
The complete conservation of 10 coding sequences across all three strains suggests that this group includes proteins vital to viral function in vivo and in vitro and less tolerant of sequence variation. In comparing these 10 genes to other sequences available in GenBank and to the published genome of the mutant HF10 strain, only four of these, UL31, UL35, UL45, and gJ (US5), were still invariant (78). As more genome sequences become available, it will be important to see if these proteins remain unchanged. Further examination on a protein-by-protein level may also reveal that some genes with only one or two coding changes are in fact also highly conserved, with minor changes that do not affect their functional domains. The preservation of a coding sequence unit across a large number of divergent HSV strains indicates a promising target for antiviral discovery.
Defining the H129 mutant genotype and phenotype.
The full genotype of the previously uncharacterized strain H129 is of significant interest to the neuroanatomical circuit tracing community, because it is the only strain known whose spread is limited to the forward, or anterograde, direction (3, 17, 24, 59, 69, 86). This phenotype complements the opposing retrograde-only spread of the related alphaherpesvirus PRV strain Bartha, as well the rabies virus-derived tracers (19, 73). Finding all of the sequence differences in the H129 strain is the first step toward defining the causative mutation(s). Because there is as yet no in vitro assay for the H129 directional spread phenotype, testing of complementation and sufficiency will require either the development of such an assay or the use of rodent models. Given the mutations observed in several candidate genes, such as ICP34.5 (RL1), gL (UL1), and UL36, the phenotype of H129 may well be polygenic, adding complexity to future studies. However, the ability of this unique strain to provide insight to neuronal biology and viral infection makes it a worthy goal.
Since the original source of the H129 clinical isolate was an encephalitic patient (17), an interesting question arises: was the unique biology of H129 involved in the disease? It also is possible that the H129 phenotype had nothing to do with the disease. The patient may have had other genetic differences that led to viral encephalitis, and these provided an opportunity for the H129 mutant to thrive. Unfortunately, the lack of patient samples from that time and the inability for further testing in humans preclude our ability to answer these questions. The best insight may come from future studies, where if a case of herpetic encephalitis is observed, both the patient genome and the viral genome can be assayed simultaneously. By correlation, we may then be able to predict whether HSV-induced encephalitis usually results from patient genetics, viral genetics, or a combination of both.
Use of comparative virology and genome sequencing to map complex phenotypes.
These data provided the complete sequence of two new genomes of HSV-1 and demonstrated the large degree of coding sequence variability in a DNA virus of high replication fidelity. The abundance of protein-level variation provides an impetus to continue sequencing projects aimed at discovering the sequence variabilities in clinical isolates of HSV-1. Clearly, these methods provide rich data for comparisons across strains, but they also directly suggest straightforward experiments to map specific genotype differences to known phenotypic differences.
In the case of hard-to-study clinical phenotypes, such as latency, reactivation, and tissue tropism, high-throughput genome sequencing of divergent virus strains will now enable unbiased and comprehensive association of phenotypes to differences at multiple genetic loci. In our proof-of-principle example, we used the new sequence of strain F, in combination with the previously published strain 17, to help identify the likely causative mutations in the mutant strain H129. Similarly, genome sequencing could be used to map complex traits, such as tendency to latency or reactivation frequency, where candidate loci could be found by comparing variations across the genomes of multiple genetically divergent strains that share these phenotypes. Whole-genome assay techniques will provide data and a means to map viral genotype differences to phenotypes previously defined in human patients, particularly those that are difficult to accurately replicate or study in animal and cell culture models.
Supplementary Material
Acknowledgments
We thank J. Buckles, C. Chiriac, Y. Tafuri, and the Lewis-Sigler Institute for Integrative Genomics Microarray Facility for technical support. We thank M. Llinás, M. Lyman, O. Kobiler, members of the Enquist lab, and anonymous reviewers for feedback on these data and the manuscript.
We acknowledge funding from a Center Grant (NIH/NIGMS P50 GM071508), the New Jersey Commission on Spinal Cord Research (M.L.S.), NIH P40 RR 018604 (L.W.E. and M.L.S.), and a supplement to NIH R01 AI 033063 (M.L.S.).
Footnotes
Published ahead of print on 10 March 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abaitua, F., R. N. Souto, H. Browne, T. Daikoku, and P. O'Hare. 2009. Characterization of the herpes simplex virus (HSV)-1 tegument protein VP1-2 during infection with the HSV temperature-sensitive mutant tsB7. J. Gen. Virol. 90:2353-2363. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Archin, N. M., and S. S. Atherton. 2002. Rapid spread of a neurovirulent strain of HSV-1 through the CNS of BALB/c mice following anterior chamber inoculation. J. Neurovirol. 8:122-135. [DOI] [PubMed] [Google Scholar]
- 4.Baird, N. L., P. C. Yeh, R. J. Courtney, and J. W. Wills. 2008. Sequences in the UL11 tegument protein of herpes simplex virus that control association with detergent-resistant membranes. Virology 374:315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blondeau, C., N. Chbab, C. Beaumont, K. Courvoisier, N. Osterrieder, J. F. Vautherot, and C. Denesvre. 2007. A full UL13 open reading frame in Marek's disease virus (MDV) is dispensable for tumor formation and feather follicle tropism and cannot restore horizontal virus transmission of rRB-1B in vivo. Vet. Res. 38:419-433. [DOI] [PubMed] [Google Scholar]
- 6.Bolovan, C. A., N. M. Sawtell, and R. L. Thompson. 1994. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J. Virol. 68:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bower, J. R., H. Mao, C. Durishin, E. Rozenbom, M. Detwiler, D. Rempinski, T. L. Karban, and K. S. Rosenthal. 1999. Intrastrain variants of herpes simplex virus type 1 isolated from a neonate with fatal disseminated infection differ in the ICP34.5 gene, glycoprotein processing, and neuroinvasiveness. J. Virol. 73:3843-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley, A. J., N. S. Lurain, P. Ghazal, U. Trivedi, C. Cunningham, K. Baluchova, D. Gatherer, G. W. Wilkinson, D. J. Dargan, and A. J. Davison. 2009. High-throughput sequence analysis of variants of human cytomegalovirus strains Towne and AD169. J. Gen. Virol. 90:2375-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chain, P. S., D. V. Grafham, R. S. Fulton, M. G. Fitzgerald, J. Hostetler, D. Muzny, J. Ali, B. Birren, D. C. Bruce, C. Buhay, J. R. Cole, Y. Ding, S. Dugan, D. Field, G. M. Garrity, R. Gibbs, T. Graves, C. S. Han, S. H. Harrison, S. Highlander, P. Hugenholtz, H. M. Khouri, C. D. Kodira, E. Kolker, N. C. Kyrpides, D. Lang, A. Lapidus, S. A. Malfatti, V. Markowitz, T. Metha, K. E. Nelson, J. Parkhill, S. Pitluck, X. Qin, T. D. Read, J. Schmutz, S. Sozhamannan, P. Sterk, R. L. Strausberg, G. Sutton, N. R. Thomson, J. M. Tiedje, G. Weinstock, A. Wollam, and J. C. Detter. 2009. Genomics genome project standards in a new era of sequencing. Science 326:236-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, L., P. Jha, B. Stirling, S. K. Sgaier, T. Daid, R. Kaul, and N. Nagelkerke. 2007. Sexual risk factors for HIV infection in early and advanced HIV epidemics in sub-Saharan Africa: systematic overview of 68 epidemiological studies. PLoS One 2:e1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulter, L. J., H. W. Moss, J. Lang, and D. J. McGeoch. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 74:387-395. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham, C., D. Gatherer, B. Hilfrich, K. Baluchova, D. J. Dargan, M. Thomson, P. D. Griffiths, G. W. Wilkinson, T. F. Schulz, and A. J. Davison. 11 November 2009, posting date. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J. Gen. Virol. 91:605-615. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, B. M., and M. K. Waldor. 2009. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: one fewer porin is enough. Nucleic Acids Res. 37:5757-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deback, C., D. Boutolleau, C. Depienne, C. E. Luyt, P. Bonnafous, A. Gautheret-Dejean, I. Garrigue, and H. Agut. 2009. Utilization of microsatellite polymorphism for differentiating herpes simplex virus type 1 strains. J. Clin. Microbiol. 47:533-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delcher, A. L., S. L. Salzberg, and A. M. Phillippy. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics 10:unit 10.3. [DOI] [PubMed] [Google Scholar]
- 16.Desai, P., G. L. Sexton, E. Huang, and S. Person. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J. Virol. 82:11354-11361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dix, R. D., R. R. McKendall, and J. R. Baringer. 1983. Comparative neurovirulence of herpes simplex virus type 1 strains after peripheral or intracerebral inoculation of BALB/c mice. Infect. Immun. 40:103-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 19.Enquist, L. W. 2002. Exploiting circuit-specific spread of pseudorabies virus in the central nervous system: insights to pathogenesis and circuit tracers. J. Infect. Dis. 186(Suppl. 2):S209-S214. [DOI] [PubMed] [Google Scholar]
- 20.Everett, R. D., and M. L. Fenwick. 1990. Comparative DNA sequence analysis of the host shutoff genes of different strains of herpes simplex virus: type 2 strain HG52 encodes a truncated UL41 product. J. Gen. Virol. 71:1387-1390. [DOI] [PubMed] [Google Scholar]
- 21.Fan, Q., E. Lin, and P. G. Spear. 2009. Insertional mutations in herpes simplex virus type 1 gL identify functional domains for association with gH and for membrane fusion. J. Virol. 83:11607-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flicek, P., and E. Birney. 2009. Sense from sequence reads: methods for alignment and assembly. Nat. Methods 6:S6-S12. [DOI] [PubMed] [Google Scholar]
- 23.Freedman, E., and A. Mindel. 2004. Epidemiology of herpes and HIV co-infection. J. HIV Ther. 9:4-8. [PubMed] [Google Scholar]
- 24.Garner, J. A., and J. H. LaVail. 1999. Differential anterograde transport of HSV type 1 viral strains in the murine optic pathway. J. Neurovirol. 5:140-150. [DOI] [PubMed] [Google Scholar]
- 25.He, B., J. Chou, D. A. Liebermann, B. Hoffman, and B. Roizman. 1996. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the γ134.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J. Virol. 70:84-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hillier, L. W., G. T. Marth, A. R. Quinlan, D. Dooling, G. Fewell, D. Barnett, P. Fox, J. I. Glasscock, M. Hickenbotham, W. Huang, V. J. Magrini, R. J. Richt, S. N. Sander, D. A. Stewart, M. Stromberg, E. F. Tsung, T. Wylie, T. Schedl, R. K. Wilson, and E. R. Mardis. 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5:183-188. [DOI] [PubMed] [Google Scholar]
- 27.Holt, K. E., J. Parkhill, C. J. Mazzoni, P. Roumagnac, F. X. Weill, I. Goodhead, R. Rance, S. Baker, D. J. Maskell, J. Wain, C. Dolecek, M. Achtman, and G. Dougan. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella typhi. Nat. Genet. 40:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarosinski, K. W., N. G. Margulis, J. P. Kamil, S. J. Spatz, V. K. Nair, and N. Osterrieder. 2007. Horizontal transmission of Marek's disease virus requires US2, the UL13 protein kinase, and gC. J. Virol. 81:10575-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jovasevic, V., L. Liang, and B. Roizman. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kienzle, T. E., J. S. Henkel, J. Y. Ling, M. C. Banks, D. R. Beers, B. Jones, and W. G. Stroop. 1995. Cloning and restriction endonuclease mapping of herpes simplex virus type-1 strains H129 and +GC. Arch. Virol. 140:1663-1675. [DOI] [PubMed] [Google Scholar]
- 31.Leege, T., W. Fuchs, H. Granzow, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1. J. Virol. 83:896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, H., J. Ruan, and R. Durbin. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling, J. Y., T. E. Kienzle, T. M. Chen, J. S. Henkel, G. C. Wright, and W. G. Stroop. 1997. Comparative analyses of the latency-associated transcript promoters from herpes simplex virus type 1 strains H129, +GC and KOS-63. Virus Res. 50:95-106. [DOI] [PubMed] [Google Scholar]
- 34.Loh, J., G. Zhao, R. M. Presti, L. R. Holtz, S. R. Finkbeiner, L. Droit, Z. Villasana, C. Todd, J. M. Pipas, B. Calgua, R. Girones, D. Wang, and H. W. Virgin. 2009. Detection of novel sequences related to african Swine Fever virus in human serum and sewage. J. Virol. 83:13019-13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2006. Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 80:10534-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacLean, D., J. D. Jones, and D. J. Studholme. 2009. Application of ‘next-generation’ sequencing technologies to microbial genetics. Nat. Rev. Microbiol. 7:287-296. [DOI] [PubMed] [Google Scholar]
- 37.Maertzdorf, J., L. Remeijer, A. Van Der Lelij, J. Buitenwerf, H. G. Niesters, A. D. Osterhaus, and G. M. Verjans. 1999. Amplification of reiterated sequences of herpes simplex virus type 1 (HSV-1) genome to discriminate between clinical HSV-1 isolates. J. Clin. Microbiol. 37:3518-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao, H., and K. S. Rosenthal. 2002. An N-terminal arginine-rich cluster and a proline-alanine-threonine repeat region determine the cellular localization of the herpes simplex virus type 1 ICP34.5 protein and its ligand, protein phosphatase 1. J. Biol. Chem. 277:11423-11431. [DOI] [PubMed] [Google Scholar]
- 39.Mardis, E., J. McPherson, R. Martienssen, R. K. Wilson, and W. R. McCombie. 2002. What is finished, and why does it matter. Genome Res. 12:669-671. [DOI] [PubMed] [Google Scholar]
- 40.McGeoch, D. J. 2009. Lineages of varicella-zoster virus. J. Gen. Virol. 90:963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 42.McGeoch, D. J., A. Dolan, S. Donald, and D. H. Brauer. 1986. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 14:1727-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGraw, H. M., S. Awasthi, J. A. Wojcechowskyj, and H. M. Friedman. 2009. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not Us9. J. Virol. 83:8315-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.National Center for Biotechnology Information. 2002. Chapter 18, The Reference Sequence (RefSeq) Project. The NCBI handbook. National Library of Medicine, Bethesda, MD. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books.
- 45.Nishiyama, Y., H. Kimura, and T. Daikoku. 1991. Complementary lethal invasion of the central nervous system by nonneuroinvasive herpes simplex virus types 1 and 2. J. Virol. 65:4520-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norberg, P. 2010. Divergence and genotyping of human alpha-herpesviruses: an overview. Infect. Genet. Evol. 10:14-25. [DOI] [PubMed] [Google Scholar]
- 47.Norberg, P., T. Bergstrom, and J. A. Liljeqvist. 2006. Genotyping of clinical herpes simplex virus type 1 isolates by use of restriction enzymes. J. Clin. Microbiol. 44:4511-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norberg, P., T. Bergstrom, E. Rekabdar, M. Lindh, and J. A. Liljeqvist. 2004. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J. Virol. 78:10755-10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norberg, P., S. Olofsson, M. A. Tarp, H. Clausen, T. Bergstrom, and J. A. Liljeqvist. 2007. Glycoprotein I of herpes simplex virus type 1 contains a unique polymorphic tandem-repeated mucin region. J. Gen. Virol. 88:1683-1688. [DOI] [PubMed] [Google Scholar]
- 50.Orvedahl, A., D. Alexander, Z. Talloczy, Q. Sun, Y. Wei, W. Zhang, D. Burns, D. A. Leib, and B. Levine. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23-35. [DOI] [PubMed] [Google Scholar]
- 51.Overton, H. A., D. J. McMillan, L. S. Klavinskis, L. Hope, A. J. Ritchie, and P. Wong-kai-in. 1992. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology 190:184-192. [DOI] [PubMed] [Google Scholar]
- 52.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 53.Peters, G. A., S. D. Tyler, C. Grose, A. Severini, M. J. Gray, C. Upton, and G. A. Tipples. 2006. A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J. Virol. 80:9850-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Presti, R. M., G. Zhao, W. L. Beatty, K. A. Mihindukulasuriya, A. P. da Rosa, V. L. Popov, R. B. Tesh, H. W. Virgin, and D. Wang. 2009. Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J. Virol. 83:11599-11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pyles, R. B., and R. L. Thompson. 1994. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J. Virol. 68:4963-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quail, M. A., I. Kozarewa, F. Smith, A. Scally, P. J. Stephens, R. Durbin, H. Swerdlow, and D. J. Turner. 2008. A large genome center's improvements to the Illumina sequencing system. Nat. Methods 5:1005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rekabdar, E., P. Tunback, J. A. Liljeqvist, and T. Bergstrom. 1999. Variability of the glycoprotein G gene in clinical isolates of herpes simplex virus type 1. Clin. Diagn. Lab Immunol. 6:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riley, M., and M. Buckley. 2009. Large-scale sequencing: the future of genomic sciences? American Academy of Microbiology, Washington, DC. [PubMed]
- 59.Rinaman, L., and G. Schwartz. 2004. Anterograde transneuronal viral tracing of central viscerosensory pathways in rats. J. Neurosci. 24:2782-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 61.Schacherer, J., D. M. Ruderfer, D. Gresham, K. Dolinski, D. Botstein, and L. Kruglyak. 2007. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS One 2:e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shanda, S. K., and D. W. Wilson. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J. Virol. 82:7388-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snyder, A., K. Polcicova, and D. C. Johnson. 2008. Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons. J. Virol. 82:10613-10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sommer, D. D., A. L. Delcher, S. L. Salzberg, and M. Pop. 2007. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spatz, S. J., C. Rue, D. Schumacher, and N. Osterrieder. 2008. Clustering of mutations within the inverted repeat regions of a serially passaged attenuated gallid herpesvirus type 2 strain. Virus Genes 37:69-80. [DOI] [PubMed] [Google Scholar]
- 67.Spatz, S. J., and C. A. Rue. 2008. Sequence determination of a mildly virulent strain (CU-2) of gallid herpesvirus type 2 using 454 pyrosequencing. Virus Genes 36:479-489. [DOI] [PubMed] [Google Scholar]
- 68.Spatz, S. J., Y. Zhao, L. Petherbridge, L. P. Smith, S. J. Baigent, and V. Nair. 2007. Comparative sequence analysis of a highly oncogenic but horizontal spread-defective clone of Marek's disease virus. Virus Genes 35:753-766. [DOI] [PubMed] [Google Scholar]
- 69.Sun, N., M. D. Cassell, and S. Perlman. 1996. Anterograde, transneuronal transport of herpes simplex virus type 1 strain H129 in the murine visual system. J. Virol. 70:5405-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takakuwa, H., F. Goshima, N. Nozawa, T. Yoshikawa, H. Kimata, A. Nakao, A. Nawa, T. Kurata, T. Sata, and Y. Nishiyama. 2003. Oncolytic viral therapy using a spontaneously generated herpes simplex virus type 1 variant for disseminated peritoneal tumor in immunocompetent mice. Arch. Virol. 148:813-825. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka, M., Y. Nishiyama, T. Sata, and Y. Kawaguchi. 2005. The role of protein kinase activity expressed by the UL13 gene of herpes simplex virus 1: the activity is not essential for optimal expression of UL41 and ICP0. Virology 341:301-312. [DOI] [PubMed] [Google Scholar]
- 72.Tyler, S. D., G. A. Peters, C. Grose, A. Severini, M. J. Gray, C. Upton, and G. A. Tipples. 2007. Genomic cartography of varicella-zoster virus: a complete genome-based analysis of strain variability with implications for attenuation and phenotypic differences. Virology 359:447-458. [DOI] [PubMed] [Google Scholar]
- 73.Ugolini, G. 2008. Use of rabies virus as a transneuronal tracer of neuronal connections: implications for the understanding of rabies pathogenesis. Dev. Biol. (Basel) 131:493-506. [PubMed] [Google Scholar]
- 74.Umene, K., and T. Kawana. 2003. Divergence of reiterated sequences in a series of genital isolates of herpes simplex virus type 1 from individual patients. J. Gen. Virol. 84:917-923. [DOI] [PubMed] [Google Scholar]
- 75.Umene, K., S. Oohashi, M. Yoshida, and Y. Fukumaki. 2008. Diversity of the a sequence of herpes simplex virus type 1 developed during evolution. J. Gen. Virol. 89:841-852. [DOI] [PubMed] [Google Scholar]
- 76.Umene, K., R. J. Watson, and L. W. Enquist. 1984. Tandem repeated DNA in an intergenic region of herpes simplex virus type 1 (Patton). Gene 30:33-39. [DOI] [PubMed] [Google Scholar]
- 77.Umene, K., and M. Yoshida. 1989. Reiterated sequences of herpes simplex virus type 1 (HSV-1) genome can serve as physical markers for the differentiation of HSV-1 strains. Arch. Virol. 106:281-299. [DOI] [PubMed] [Google Scholar]
- 78.Ushijima, Y., C. Luo, F. Goshima, Y. Yamauchi, H. Kimura, and Y. Nishiyama. 2007. Determination and analysis of the DNA sequence of highly attenuated herpes simplex virus type 1 mutant HF10, a potential oncolytic virus. Microbes Infect. 9:142-149. [DOI] [PubMed] [Google Scholar]
- 79.Velicer, G. J., G. Raddatz, H. Keller, S. Deiss, C. Lanz, I. Dinkelacker, and S. C. Schuster. 2006. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc. Natl. Acad. Sci. U. S. A. 103:8107-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Warren, R. L., G. G. Sutton, S. J. Jones, and R. A. Holt. 2007. Assembling millions of short DNA sequences using SSAKE. Bioinformatics 23:500-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wen, K. W., D. P. Dittmer, and B. Damania. 2009. Disruption of LANA in rhesus rhadinovirus generates a highly lytic recombinant virus. J. Virol. 83:9786-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Whitley, R. J., E. R. Kern, S. Chatterjee, J. Chou, and B. Roizman. 1993. Replication, establishment of latency, and induced reactivation of herpes simplex virus gamma 1 34.5 deletion mutants in rodent models. J. Clin. Invest. 91:2837-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu, F., M. R. Sternberg, B. J. Kottiri, G. M. McQuillan, F. K. Lee, A. J. Nahmias, S. M. Berman, and L. E. Markowitz. 2006. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA 296:964-973. [DOI] [PubMed] [Google Scholar]
- 84.Yeh, P. C., D. G. Meckes, Jr., and J. W. Wills. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J. Virol. 82:10693-10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zelnik, V. 2003. Marek's disease virus research in the post-sequencing era: new tools for the study of gene functions and virus-host interactions. Avian Pathol. 32:323-333. [DOI] [PubMed] [Google Scholar]
- 86.Zemanick, M. C., P. L. Strick, and R. D. Dix. 1991. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc. Natl. Acad. Sci. U. S. A. 88:8048-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.