Abstract
Lepidopteran baculovirus-specific protein FP25K performs many roles during the infection cycle, including functions in the production of occlusion bodies (OBs) and budded viruses (BVs), oral infection, and postmortem host degradation. To explore the common and specific functions of FP25K proteins among lepidopteran baculoviruses, we performed comparative analyses of FP25K proteins from group I and group II nucleopolyhedroviruses (NPVs) and granulovirus (GV). Using recombinant Bombyx mori NPVs (BmNPVs), we showed that the FP25Ks from NPVs were able to eliminate all the phenotypic defects observed in an infection with a BmNPV mutant lacking functional fp25K but that FP25K from GV did not show abilities to recover oral infectivity and postmortem host degradation. We also observed that introduction of Autographa californica multiple NPV (AcMNPV) fp25K into the BmNPV genome enhanced OB and BV production. According to these results, we generated a novel BmNPV-based expression vector with AcMNPV fp25K and examined its potential in BmN cells and B. mori larvae. Our results showed that the introduction of AcMNPV fp25K significantly increases the expression of foreign gene products in cultured cells and shortens the time for obtaining the secreted recombinant proteins from larval hemolymph.
Baculoviridae is a large family of pathogens that infect arthropods, particularly Lepidoptera. Nucleopolyhedroviruses (NPVs), members of a genus of Baculoviridae, have a large, circular genome of supercoiled, double-stranded DNA packaged into rod-shaped virions. NPVs produce two types of virions during their infection cycle to bring about efficient viral replication within infected host larvae and spread the virus from insect to insect. Budded viruses (BVs) spread the infection to neighboring cells (9, 30), whereas occlusion-derived viruses (ODVs), which are occluded in occlusion bodies (OBs), are transmitted from insect to insect via oral infection. The end of infection is marked by dramatic degradation of host larvae.
Serial passage of NPVs in established cultured cell lines often generates mutants called few-polyhedron (FP) mutants, which produce fewer OBs (polyhedra) in the nuclei of infected cells (42). FP mutants frequently acquire host genome fragments (often containing a transposon), lose a portion of the viral genome, or both (10). Mapping analysis has located a single gene, fp25K, encoding a 25-kDa protein involved in the FP phenotype of the Autographa californica multiple NPV (AcMNPV) (1). Sequencing of baculovirus genomes has revealed that the gene product FP25K is a lepidopteran baculovirus-specific protein (15), but the roles of FP25Ks from granuloviruses (GVs) are still unknown.
The common characteristics of the FP phenotype are a decrease in the number of OBs produced in each infected cell, an increase in the yield of BVs, and the occurrence of few or no ODVs within OBs in cells infected with FP mutants (10). Previous studies with Bombyx mori NPV (BmNPV) also showed that mutations in fp25K result in reduced postmortem host degradation due to a decrease in the expression of a viral cathepsin, V-CATH (22, 23, 28). Rescue experiments have confirmed that these phenotypes are related solely to the functional disruption of fp25K (10, 22, 28).
To identify the common and specific functions of FP25K proteins among lepidopteran NPVs and GVs, we generated recombinant BmNPVs in which fp25K was replaced with the corresponding gene from AcMNPV, Spodoptera litura NPV (SpltMNPV), or Xestia c-nigrum granulovirus (XecnGV) and compared the phenotypes of the recombinant viruses with those of wild-type virus T3, fp25K deletion mutant Bm25KD (10), and repair virus Bm25KD-Bm (28). We found that the FP25Ks from NPVs are able to eliminate all the phenotypic defects observed in infection with Bm25KD but that the FP25K from XecnGV does not eliminate the defects in oral infectivity and postmortem host degradation.
In the course of this study, we found that Bm25KD-Ac, a BmNPV possessing AcMNPV fp25K instead of BmNPV fp25K, produces more OBs in BmN cells than the wild-type virus through enhanced expression of the polyhedrin gene polh. In this study, we also generated a novel BmNPV-based expression vector with AcMNPV fp25K and tested whether this modified vector improves recombinant protein expression in BmN cells and B. mori larvae.
MATERIALS AND METHODS
Insects, cell lines, and viruses.
B. mori larvae were reared as described previously (25). The BmN (BmN-4) cells were cultured at 27°C in TC-100 medium supplemented with 10% fetal bovine serum. BmNPV T3 was used as the wild-type virus. Two recombinant BmNPVs, Bm25KD and Bm25KD-Bm, have been described previously (22, 28). Titers of T3 virus and recombinant BmNPVs were determined by plaque assays with BmN cells (25).
Generation of recombinant BmNPVs expressing His-tagged FP25K proteins.
To generate BmNPVs expressing His-tagged FP25K proteins from AcMNPV, SpltMNPV, and XecnGV, we amplified His sequence-tagged fp25K genes by using the primer sets listed in Table 1 and cloned these fragments into fp25Knull/pcDNA (28). BmN cells were cotransfected with the resultant plasmids, which express His-tagged FP25Ks under the control of the endogenous BmNPV fp25K promoter (see Fig. 1B), and Bsu36I-digested Bm25KD DNA by using Cellfectin (Invitrogen). Five days after transfection, the medium was collected and stored at 4°C until use. Three recombinant BmNPVs expressing His-tagged FP25K proteins from AcMNPV (Bm25KD-Ac), SpltMNPV (Bm25KD-Splt), and XecnGV (Bm25KD-Xc) were isolated by identification of plaques that did not express β-galactosidase (25). Insertion of His sequence-tagged fp25K genes at the fp25K locus of BmNPV was confirmed by PCR using the primers shown in Table 1.
TABLE 1.
List of primers
| Primer | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| Ac_fp_F | AAGAATTCATGGATCAATTTGAACAGTTG | Use in PCR for AcMNPV fp25K |
| Ac_fp_R | AAGGATCCTCAATGGTGATGGTGATGATGAATTAAATTTTGAAGCATTTTTTC | |
| Splt_fp_F | AAGAATTCATGGAAACCGATCTGATCAAC | Use in PCR for SpltMNPV fp25K |
| Splt_fp_R | AAGGATCCTCAATGGTGATGGTGATGATGAATTAATTTTATCACATC | |
| Xc_fp_F | AAGAATTCATGCAATATGACTTGGACAG | Use in PCR for XecnGV fp25K |
| Xc_fp_R | AAGGATCCTCAATGGTGATGGTGATGATGGGAGTTGTGTTGATCGATGCTATC | |
| FP_CF1 | TTACGCACCATATACGCATC | Genotyping of recombinant viruses |
| FP_CR1 | CATTACGAGATTCAACTTGATAC | |
| Bm31_F1 | ACTGTCGACAAGCTCTGTCC | |
| Bm31_R1 | ACAACGCACAGAATCTAACGC | |
| polh_F1 | CAAGAAGCACCTAGTCGAAC | Use in qRT-PCR for polh |
| polh_R1 | CGTGTTGAGCGAGGAACTTG | |
| ST-F | ACTGGTCGATTAAACTGTTG | Use in PCR for chimeric fp25K genes |
| ST-F1 | ATCTCACAGACAGCGTTGAG | |
| ST-F2 | CTGAATAATAAAAAAATTAGAAAC | |
| ST-F3 | AATCTTCGTTTGACAAAAAC | |
| ST-F4 | CTTAAAAAAACTCGTGACG | |
| ST-R | TTTGTAAACTACGATCGTAG | |
| ST-R1 | TGAATACCGTATATCTCAAC | |
| ST-R2 | GCAAATCTTTTTTAAATAATAGTTTC | |
| ST-R3 | GCCACAATGTGGTTTTTGTC | |
| ST-R4 | TAAAACGGCAACAGAGCGTC | |
| IL-3_F | AAAGATCTATGGTTCTTGCCAGCTCTACC | Cloning of mIL-3 gene |
| IL-3_R | AAACTAGATTACTTATCGTCGTCATCCTTGTAATCACATTCCACGGTTCCACGGTT |
FIG. 1.
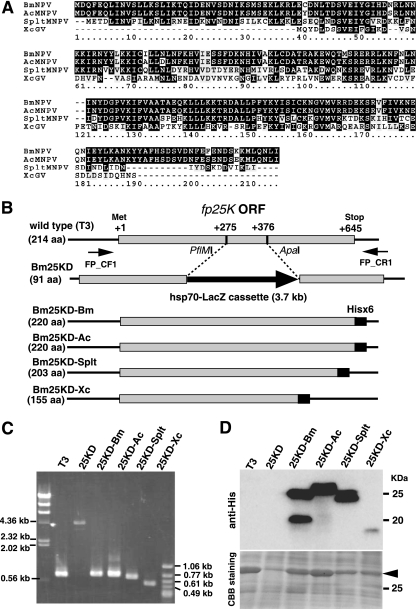
Generation of recombinant BmNPVs expressing fp25K genes from BmNPV, AcMNPV, SpltMNPV, and XecnGV. (A) Alignment of FP25K proteins from BmNPV, AcMNPV, SpltMNPV, and XecnGV (XcGV). In the sequences shown, black shading denotes identical residues and gray shading indicates similarities among the four FP25K proteins. (B) Schematic representation of recombinant BmNPVs. The set of PCR primers (FP_CF1 and FP_CR1) used in genotyping experiments is also shown. ORF, open reading frame. (C) PCR analysis of the genomes of T3, Bm25KD (25KD), Bm25KD-Bm (25KD-Bm), Bm25KD-Ac (25KD-Ac), Bm25KD-Splt (25KD-Splt), and Bm25KD-Xc (25KD-Xc). Each genotype was confirmed by PCR using the primers FP_CF1and FP_CR1. (D) Expression of His-tagged FP25K proteins in BmNPV-infected BmN cells. BmN cells were infected with T3 or recombinant BmNPVs, harvested at 3 dpi, and subjected to Western blot analysis with anti-His antibody. The gel stained with Coomassie brilliant blue (CBB) is shown in the lower panel. The molecular masses of protein standards are indicated to the right. The arrowhead indicates polyhedrin.
Western blot analysis of FP25K proteins.
BmN cells were infected with T3 or recombinant BmNPVs at a multiplicity of infection (MOI) of 5. At 3 days postinfection (dpi), the cells were collected and subjected to Western blot analysis with anti-His antibody (Qiagen) as described previously (28).
qRT-PCR.
Total RNA from BmN cells infected with Bm25KD-Bm or Bm25KD-Ac was prepared using Trizol reagent (Invitrogen). First-strand cDNA was synthesized from 0.2 μg of total RNA, and quantitative reverse transcription-PCR (qRT-PCR) was performed using Power SYBR green PCR master mix (Applied Biosystems) and the primers polh_F1 and polh_R1 (Table 1). Amplification was detected using the ABI PRISM 7000 sequence detection system (Applied Biosystems) as reported previously (25).
V-CATH assays.
V-CATH assays were performed as described previously (23). Hemolymph samples from virus-infected B. mori larvae were collected at 5 dpi and centrifuged at 20,000 × g for 10 min at 4°C, and the supernatants (30 μl/assay mixture) were used immediately for V-CATH assays. Protease activity was measured by an azocasein assay. V-CATH in the hemolymph was detected by Western blotting with an antibody against BmNPV V-CATH (27).
BV production and plaque size determination.
BmN cells were infected with T3 or mutant BmNPVs at an MOI of 5. After a 1-h incubation, virus-containing culture medium was removed and fresh medium was added after the cells were washed twice with serum-free TC-100 medium (this point was designated time zero). Small amounts of culture medium were harvested at 1 and 2 dpi. BV production was evaluated by a plaque assay. Plaques were photographed, and their sizes were determined using ImageJ software (National Institute of Health).
OB production in B. mori larvae and BmN cells.
Fifth-instar B. mori larvae were starved for several hours and then injected with 50 μl of a viral suspension containing 105 PFU and returned to an artificial diet at 27°C. At 4 dpi, hemolymph samples from infected larvae were collected and the OBs released were counted using a hemocytometer as described previously (27). BmN cells infected with T3 or recombinant BmNPVs were gently scraped with a rubber policeman at 5 dpi, and OB production was measured as described elsewhere (27).
Larval bioassays.
The 50% lethal concentration (LC50) of OBs for first-instar larvae was determined by feeding the larvae OBs at different concentrations. OBs were produced in BmN cells, purified by centrifugation, resuspended in distilled H2O, and quantified using a hemocytometer. First-instar larvae (within 12 h of molting) were orally inoculated with OBs by exposure to the artificial diet in a 960-mm2 area that was surface-contaminated with OBs (19). At least 20 larvae per dose were used in each of two independent experiments.
Generation of recombinant BmNPVs expressing chimeric FP25Ks.
To construct the plasmids possessing chimeric fp25K genes, the gene coding region was generated by overlapping PCR (26), checked by DNA sequencing, and cloned into fp25Knull/pcDNA. Primers used in the mutagenesis experiments are listed in Table 1. BmN cells were cotransfected with the resultant plasmids and Bsu36I-digested Bm25KD DNA, and recombinant BmNPVs were isolated as described above.
LC-MS/MS analysis of immunoprecipitated BmNPV FP25K.
BmN cells were infected with Bm25KD-Bm at an MOI of 5. At 2 dpi, the cells were collected and subjected to immunoprecipitation with anti-His antibody as described previously (24). The immunoprecipitated BmNPV FP25Ks were identified by SDS-PAGE analysis, and tryptic digests of the bands were analyzed by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) as described previously (49).
Generation of recombinant BmNPVs expressing mIL-3.
A mouse interleukin-3 (mIL-3) gene was amplified from mouse spleen cDNA. The FLAG sequence-tagged mIL-3 gene was cloned into the transfer vector pBm31 (31). BmN cells were cotransfected with the resultant plasmid and Bm25KD-Bm or Bm25KD-Ac genomic DNA by using Cellfectin (Invitrogen). Five days after transfection, the medium was collected and stored at 4°C until use. Recombinant BmNPVs expressing mIL-3 were isolated by identification of plaques that did not produce OBs (35). Insertion of the mIL-3 gene at the polh locus was confirmed by PCR using the primers shown in Table 1. Expression of mIL-3 in BmN cells and B. mori larvae was examined by Western blot analysis with anti-FLAG antibody (Sigma). Expression of actin in BmN cells was also examined as a loading control by Western blot analysis with anti-actin antibody (Santa Cruz Biotechnology).
Generation of recombinant BmNPVs expressing firefly luciferase.
A firefly luciferase gene was excised from the pGL3-Basic vector (Promega) and cloned into the transfer vector pBm31. BmN cells were cotransfected with the resultant plasmid and Bm25KD-Bm or Bm25KD-Ac genomic DNA by using Cellfectin. Five days after transfection, the medium was collected and stored at 4°C until use. Recombinant BmNPVs expressing luciferase were isolated as described above. A luciferase reporter assay was carried out using a luciferase assay system (Promega) as described previously (29). In brief, BmN cells were grown in 60-mm dishes and infected with recombinant BmNPVs at an MOI of 5. At 1, 2, and 3 dpi, luciferase activity was measured.
Statistical analysis.
In most experiments, we performed one-way analyses of variance (ANOVA) with post hoc Dunnett's tests comparing each of the treatment group means with the mean for the control group. In qRT-PCR and luciferase assays, comparisons were performed with Student's t test.
RESULTS
Generation of recombinant BmNPVs expressing the fp25K gene from BmNPV, AcMNPV, SpltMNPV, or XecnGV.
To identify the common and specific functions of FP25K proteins among lepidopteran baculoviruses, we performed comparative analyses of four FP25Ks, those from BmNPV (group I NPV), AcMNPV (group I NPV), SpltMNPV (group II NPV), and XecnGV (GV), by using recombinant BmNPVs. The amino acid sequence of BmNPV FP25K (214 amino acids [aa]) was aligned with the sequences of FP25Ks from AcMNPV (214 aa), SpltMNPV (197 aa), and XecnGV (147 aa). As seen in Fig. 1A, BmNPV FP25K has 96, 55, and 34% amino acid sequence identity to FP25Ks from AcMNPV, SpltMNPV, and XecnGV, respectively.
By cotransfection of BmN cells with Bm25KD DNA and plasmids containing His sequence-tagged fp25K genes from BmNPV, AcMNPV, SpltMNPV, and XecnGV, we generated recombinant BmNPVs expressing His-tagged FP25K proteins (Fig. 1B). Insertion and expression of His sequence-tagged fp25K genes were verified by PCR and Western blot analysis, respectively (Fig. 1C and D). Two bands were detected in samples from BmN cells infected with Bm25KD-Bm (24- and 19-kDa bands) and Bm25KD-Splt (23- and 24-kDa bands), whereas a single band was observed in samples from cells infected with Bm25KD-Ac (a 25-kDa band) and Bm25KD-Xc (a 17-kDa band) (Fig. 1D). We also noticed that the expression level of XecnGV FP25K is low compared to that of other FP25Ks (Fig. 1D).
OB production in and release from BmN cells.
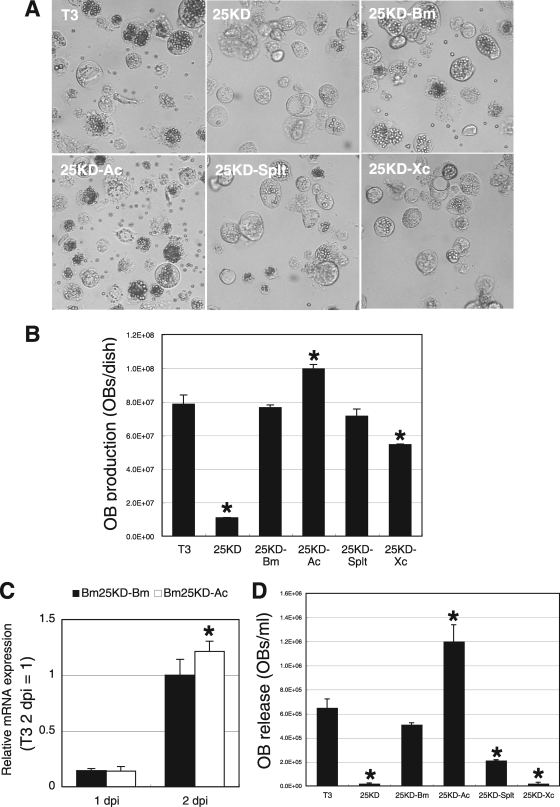
To examine the effect of fp25K substitution on OB production, we first examined the numbers of OBs in BmN cells infected with recombinant BmNPVs at 3 dpi. As reported previously, an fp25K deletion BmNPV, Bm25KD, produces fewer OBs than wild-type T3 in BmN cells (22) (Fig. 2A and B). Bm25KD-Bm and Bm25KD-Splt produced numbers of OBs similar to those observed in T3-infected cells (Fig. 2A and B). In Bm25KD-Xc-infected BmN cells, OB production was markedly enhanced compared with that in Bm25KD-infected cells but lower than that in T3-infected cells (Fig. 2A and B). In addition, we noticed that Bm25KD-Ac produced more OBs in BmN cells than T3 and Bm25KD-Bm. qRT-PCR analysis showed that polh expression in Bm25KD-Ac-infected cells at 2 dpi was significantly enhanced compared with that in Bm25KD-Bm-infected cells (Fig. 2C), suggesting that the increase in OB production by the introduction of AcMNPV fp25K is due to the transcriptional upregulation of polh. Collectively, these data demonstrate that FP25Ks from group I and II NPVs as well as GV can eliminate the defect in OB production in Bm25KD-infected BmN cells.
FIG. 2.
Comparison of OB production levels in BmN cells. (A) Light microscopic observations of BmNPV-infected BmN cells at 5 dpi. (B) OB production. BmNPV-infected BmN cells at 3 dpi were gently scraped using a rubber policeman, and total OB production was measured. Data shown are means ± standard deviations (SD; n = 3). *, P < 0.05 by one-way ANOVA and Dunnett's posttests using T3 as a control. (C) polh expression in Bm25KD-Bm- and Bm25KD-Ac-infected BmN cells. BmN cells were infected with Bm25KD-Bm and Bm25KD-Ac at an MOI of 5. Total RNA was reverse transcribed, and qRT-PCR analysis of polh was performed. Data shown are means ± SD (n = 3). *, P < 0.05 by Student's t test. (D) OB release from BmNPV-infected BmN cells at 5 dpi. Data shown are means ± SD (n = 3). *, P < 0.05 by one-way ANOVA and Dunnett's posttests using T3 as a control.
As shown in Fig. 2A and D, OB release from Bm25KD-infected cells was heavily reduced compared with that from T3-infected cells. Thus, we next examined the effect of substitution of fp25K on OB release. Bm25KD-Bm-infected cells released numbers of OBs similar to those released from T3-infected cells (Fig. 2A and D). OB release from Bm25KD-Splt-infected BmN cells was markedly enhanced compared with that from Bm25KD-infected cells but lower than that from T3-infected cells (Fig. 2A and D). Bm25KD-Xc-infected cells did not release any OBs, as observed for Bm25KD-infected cells, whereas Bm25KD-Ac-infected cells released more OBs into the medium than T3- and Bm25KD-Bm-infected cells (Fig. 2A and D). These results suggest that FP25Ks from group I and II NPVs but not GV can eliminate the defect in OB release from Bm25KD-infected BmN cells.
BV production in BmN cells.
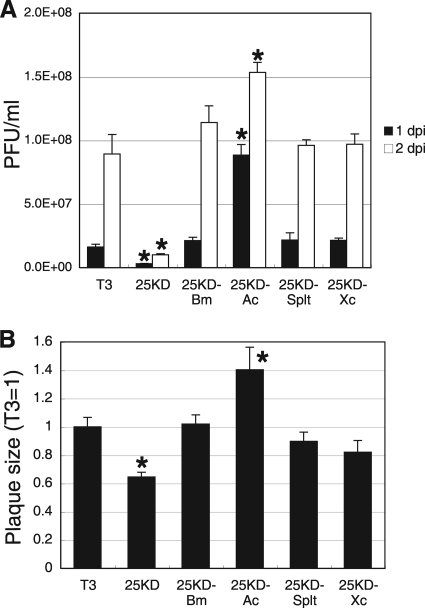
One of the common characteristics of the FP phenotype is an increase in the yield of BVs (10). We next examined BV production in BmN cells infected with recombinant BmNPVs. Unlike other NPVs, Bm25KD produces fewer infectious BVs in BmN cells than T3 (Fig. 3A). Bm25KD-Bm, Bm25KD-Splt, and Bm25KD-Xc produced numbers of BVs similar to those produced in T3-infected cells. We also observed that Bm25KD-Ac produced more BVs in BmN cells than T3 (Fig. 3A). Furthermore, we determined the sizes of plaques formed by recombinant BmNPVs. As shown in Fig. 3B, the data corresponded well to the levels of BV production. These results suggest that FP25Ks from group I and II NPVs as well as GV can eliminate the defect in BV production in Bm25KD-infected BmN cells.
FIG. 3.
Comparison of BV production levels and plaque sizes in BmN cells. (A) BV production. BmN cells were infected with T3 or recombinant BmNPVs at an MOI of 5. BV titers at 1 and 2 dpi were determined by plaque assays. Data shown are means ± SD (n = 3). *, P < 0.05 by one-way ANOVA and Dunnett's posttests using T3 as a control. (B) Plaque size. Plaques were photographed, and their sizes were quantified using ImageJ software. Data shown are means ± SD (n = 10). *, P < 0.05 by one-way ANOVA and Dunnett's posttests using T3 as a control.
Infection of B. mori larvae with recombinant BmNPVs.
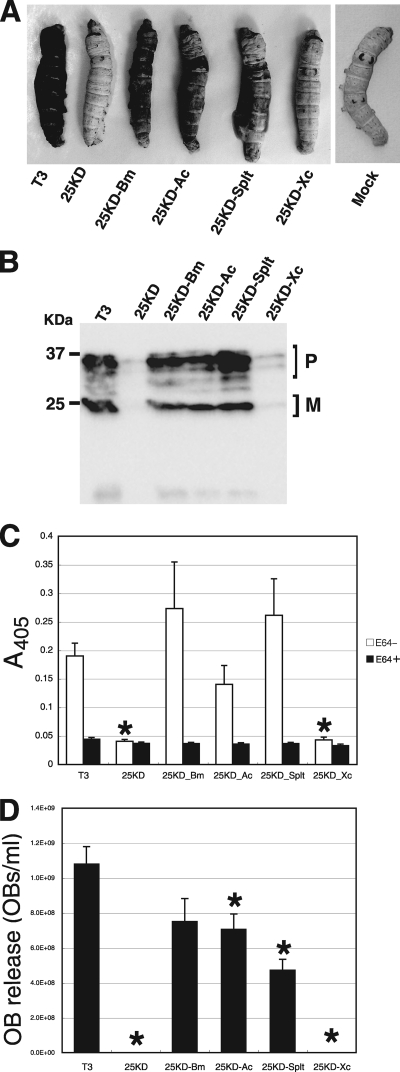
BmNPV infection results in host degradation after death by activation of V-CATH (cathepsin) and V-CHIA (chitinase) (23, 41). Previous studies have shown that mutations in fp25K of BmNPV result in the inhibition of larval degradation after death due to a decrease in V-CATH expression (22, 23, 28). Thus, we investigated the phenotypes of recombinant BmNPVs infecting B. mori larvae. As illustrated in Fig. 4A, typical postmortem degradation of T3-, Bm25KD-Bm-, Bm25KD-Ac-, and Bm25KD-Splt-infected larvae was observed. In contrast, Bm25KD or Bm25KD-Xc infection resulted in significant inhibition of postmortem degradation. Next, we examined hemolymph samples from infected larvae for the existence of V-CATH. Previous studies using anti-V-CATH antiserum showed that BmNPV expresses three forms of pro-V-CATH and two forms of putative mature V-CATH (27, 28). Western blot analyses of hemolymph proteins from BmNPV-infected larvae detected V-CATH in the hemolymph samples from larvae infected with each virus at 4 dpi, but the levels of V-CATH in samples from Bm25KD- and Bm25KD-Xc-infected larvae were markedly reduced compared to those in samples from other groups (Fig. 4B). This finding was consistent with the levels of V-CATH activity (Fig. 4C) and OB release (Fig. 4D) observed in the hemolymph samples from infected larvae. Taken together, these results suggest that FP25Ks from group I and II NPVs but not GV can eliminate the defect in postmortem degradation of Bm25KD-infected larvae.
FIG. 4.
Effects of substitution of fp25K genes on in vivo infection phenotypes. (A) Postmortem host degradation of BmNPV-infected larvae. B. mori larvae were subcutaneously injected with BVs from T3 or recombinant BmNPVs. Host degradation was assessed visually, and infected larvae were photographed at 7 dpi. (B) V-CATH secretion into the hemolymph of BmNPV-infected larvae. B. mori larvae were injected with BVs from T3 or recombinant BmNPVs, and hemolymph samples were collected at 4 dpi. Western blot analysis of the hemolymph samples was performed with anti-V-CATH antibody. The molecular masses of protein standards are indicated to the left. P, pro-V-CATH; M, mature form of V-CATH. (C) V-CATH activities of hemolymph samples from virus-infected larvae. Hemolymph samples from T3- or recombinant BmNPV-infected larvae at 4 dpi were assayed for V-CATH activity in the presence (black bars) or absence (white bars) of E64, a cysteine protease inhibitor. Data shown are means ± SD (n = 10). *, P < 0.05 by one-way ANOVA and Dunnett's posttests using T3 as a control. (D) Quantification of OB release into the hemolymph of BmNPV-infected larvae at 4 dpi. Data shown are means ± SD (n = 10). *, P < 0.05 by one-way ANOVA and Dunnett's posttests using T3 as a control.
Our previous study showed that few or no ODVs are occluded within OBs produced by Bm25KD (22). Thus, we next examined the oral infectivity of OBs from T3 and recombinant BmNPVs by using first-instar larvae. As shown in Table 2, no significant differences in the LC50s of T3, Bm25KD-Bm, Bm25KD-Ac, and Bm25KD-Splt were observed whereas OBs from Bm25KD and Bm25KD-Xc showed no oral infectivity after inoculation at the highest tested concentration (105 OBs/ml), suggesting that FP25Ks from group I and II NPVs but not GV can eliminate the defect in the oral infectivity of Bm25KD for B. mori larvae.
TABLE 2.
Dose-mortality relationships for T3, Bm25KD, Bm25KD-Bm, Bm25KD-Ac, Bm25KD-Splt, and Bm25KD-Xc in B. mori larvae
| Virus | LC50 (OBs/ml) | 95% fiducial limit |
|
|---|---|---|---|
| Lower | Upper | ||
| T3 | 2.0 × 104 | 1.2 × 104 | 3.0 × 104 |
| Bm25KD | >1.0 × 105 | ||
| Bm25KD-Bm | 2.3 × 104 | 1.5 × 104 | 3.2 × 104 |
| Bm25KD-Ac | 2.5 × 104 | 1.7 × 104 | 3.6 × 104 |
| Bm25KD-Splt | 2.0 × 104 | 8.8 × 103 | 3.3 × 104 |
| Bm25KD-Xc | >1.0 × 105 | ||
Generation of recombinant BmNPVs expressing chimeric fp25K genes.
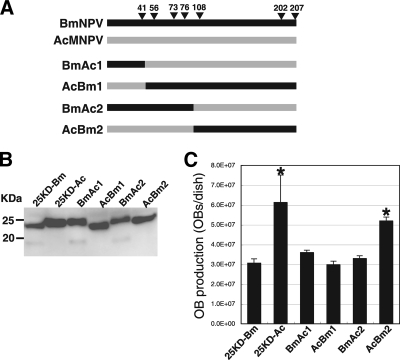
Comparative studies of baculovirus fp25K genes using recombinant BmNPVs showed that OB production in BmNPV-infected BmN cells is enhanced when AcMNPV fp25K is introduced into the BmNPV genome (Fig. 2 and 3). To determine the region required for AcMNPV-type OB production, we generated four recombinant BmNPVs, BmAc1, BmAc2, AcBm1, and AcBm2, expressing chimeric FP25Ks (Fig. 5A). As shown in Fig. 5B, we detected all of these chimeric proteins by Western blot analysis with anti-His antibody. Like samples from Bm25KD-Bm-infected cells (Fig. 1D), samples from BmAc1- and BmAc2-infected cells exhibited two bands (25 and 19 kDa) (Fig. 5B). In contrast, AcBm1 and AcBm2 each produced a single form of FP25K in BmN cells (Fig. 5B). These results clearly suggest that the N-terminal region of BmNPV FP25K is required for production of the smaller form (19 kDa) and that the cysteine residue at position 41 is essential for this process. LC-MS/MS analysis of the immunoprecipitates from Bm25KD-Bm-infected cells with anti-His antibody confirmed that the smaller form is FP25K lacking its N terminus (data not shown).
FIG. 5.
Generation of recombinant BmNPVs expressing chimeric fp25K genes. (A) Diagrammatic representation of chimeric FP25Ks. Four viruses (BmAc1, BmAc2, AcBm1, and AcBm2) expressing chimeras generated from BmNPV and AcMNPV FP25Ks were constructed. Arrowheads indicate the amino acid residues in BmNPV and AcMNPV FP25Ks that are not identical. (B) Expression of His-tagged chimeric FP25K proteins in BmNPV-infected BmN cells. BmN cells infected with recombinant BmNPVs were collected and subjected to Western blot analysis with anti-His antibody. The molecular masses of protein standards are indicated to the left. (C) OB production. BmNPV-infected BmN cells at 3 dpi were gently scraped using a rubber policeman, and total OB production was measured. Data shown are means ± SD (n = 3). *, P < 0.05 by one-way ANOVA and Dunnett's posttests using Bm25KD-Bm as a control.
We next assessed OB production in BmN cells infected with recombinant BmNPVs. As shown in Fig. 5C, AcBm2 produced more OBs than Bm25KD-Bm and numbers of OBs similar to those produced by Bm25KD-Ac. The other three chimeras produced numbers of OBs similar to those observed in Bm25KD-Bm-infected cells (Fig. 5C). These results suggest that a single amino acid change, from tyrosine to cysteine at position 41, is sufficient for a decrease in OB production. This is due presumably to production of the truncated form of FP25K. However, AcBm1, which expresses BmNPV FP25K with tyrosine at position 41, did not show AcMNPV-type OB production, suggesting that this residue is not sufficient for enhanced OB production.
Development of a novel BmNPV expression vector with AcMNPV fp25K.
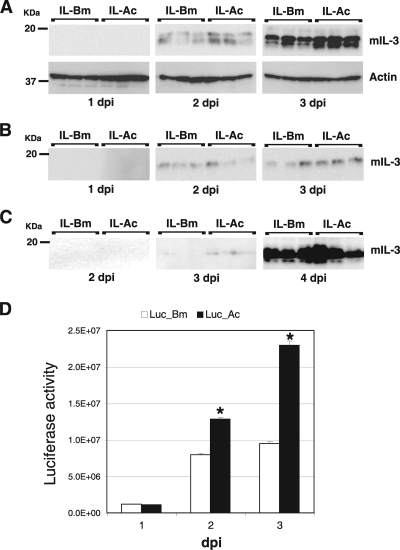
The above-described results suggest that a novel BmNPV-based expression vector could be generated by inserting AcMNPV fp25K into the fp25K locus of BmNPV. To validate this possibility, we used the Bm25KD-Bm and Bm25KD-Ac genomes to generate two recombinant BmNPVs expressing mIL-3 (IL-Bm and IL-Ac), because mIL-3 is abundantly expressed in a BmNPV expression system (38). We first examined mIL-3 expression in BmN cells. As shown in Fig. 6A, mIL-3 expression was observed from 2 dpi and the level of expression in IL-Ac-infected cells was higher than that in IL-Bm-infected cells. In contrast, there was no difference in mIL-3 secretion into the medium between these two viruses (Fig. 6B). We also performed Western blot analyses of hemolymph proteins from larvae injected with BVs from IL-Bm and IL-Ac. mIL-3 secretion was first detected in hemolymph samples from IL-Ac-infected larvae at 3 dpi, but secretion into the hemolymph of IL-Bm-infected larvae at 3 dpi was not significantly detectable (Fig. 6C). At 4 dpi, the levels of mIL-3 secretion by the two viruses were indistinguishable.
FIG. 6.
Expression of mIL-3 and luciferase using a novel BmNPV vector with AcMNPV fp25K. (A) Expression of mIL-3 in BmN cells. BmN cells were infected with IL-Bm and IL-Ac and harvested at 1 to 3 dpi in cell lysis buffer. The lysates were then analyzed by Western blotting with anti-FLAG antibody. Western blotting using anti-actin was also performed. The molecular masses of protein standards are indicated to the left. (B) Secretion of mIL-3 in BmN cells. BmN cells were infected with IL-Bm and IL-Ac, and the culture medium was harvested at 1 to 3 dpi. Western blotting of the medium samples was performed with anti-FLAG antibody. The molecular masses of protein standards are indicated to the left. (C) Secretion of mIL-3 into the larval hemolymph. B. mori larvae were injected with BVs from IL-Bm and IL-Ac, and hemolymph samples were collected at 2 to 4 dpi. Western blot analysis of the hemolymph samples was performed with anti-FLAG antibody. (D) Expression of luciferase in BmN cells. BmN cells were infected with Luc-Bm and Luc-Ac and harvested at 1 to 3 dpi in cell lysis buffer. The lysates were then subjected to a luciferase assay. Data shown are means ± SD (n = 3). *, P < 0.05 by Student's t test.
To accurately quantify the expression of recombinant proteins, we next examined luciferase activities in BmN cells infected with recombinant BmNPVs expressing firefly luciferase (designated Luc-Bm and Luc-Ac). As shown in Fig. 6D, luciferase expression was observed from 1 dpi and the level of expression in Luc-Ac-infected cells was significantly higher than that in Luc-Bm-infected cells at 2 and 3 dpi. Taken together, these results indicate that the introduction of AcMNPV fp25K into the BmNPV genome significantly increases the expression of foreign gene products in BmN cells and accelerates secretion of these products into B. mori larval hemolymph.
DISCUSSION
FP mutants of group I and II NPVs, including AcMNPV (13), Trichoplusia ni MNPV (42), Galleria mellonella MNPV (8), Lymantria dispar MNPV (44), BmNPV (22), and Helicoverpa armigera NPV (HearNPV) (4, 47), have been reported previously. In AcMNPV, mutations in fp25K result in a decreased polh mRNA level and altered transport of polyhedrin protein in the nucleus (11, 16), decreased amounts of ODV-E66 and impaired transportation of ODV-E66 in the nucleus (2, 3, 43), and a marked increase in the synthesis of some structural viral proteins of BVs such as GP64, BV/ODV-E26, and VP39 (2). Biochemical experiments also revealed that FP25K forms a protein complex containing ODV-E66, ODV-E25, and p39 (2), demonstrating that FP25K may work in a complex that regulates the expression and transport of viral proteins in infected cells. Involving such diverse pathways, deletion of fp25K causes various phenotypic defects during viral infection.
The FP phenotype is characterized by a decrease in the number of OBs, an increase in the yield of BVs, and the occurrence of few or no ODVs within OBs. Previous and present studies revealed that some of these phenotypic defects in AcMNPV FP mutants are not observed in other NPVs: Bm25KD, a BmNPV fp25K mutant, produces significantly fewer BVs in BmN cells (Fig. 3A) and B. mori larvae (data not shown) than T3, and deletion of fp25K in HearNPV does not result in an increase in GP64 expression (47). In this study, to compare the functions of FP25Ks among lepidopteran baculoviruses, we constructed recombinant BmNPVs harboring fp25K genes from the other baculoviruses AcMNPV, SpltMNPV, and XecnGV and examined the phenotypes of these viruses in BmN cells and B. mori larvae. From the results obtained, we conclude that FP25Ks from group I and group II NPVs can compensate for all of the phenotypic defects in Bm25KD to various degrees but that the FP25K of GV can eliminate only the defects in OB and BV production in a BmNPV expression system.
In contrast to NPVs, GVs have been subjected to very few expression or biochemical characterization studies of genes or proteins, due mainly to the lack of cell lines permissive toward GV (46). At present, functional studies of GV genes are often performed using a heterologous NPV system (7, 20, 31, 34, 48). Cydia pomonella GV (CpGV) V-CATH and V-CHIA are able to eliminate defects in BmNPV v-cath and v-chiA deletion mutants, respectively (7, 20). The F protein of Agrotis segetum GV can readily rescue the infectivity of gp64 null AcMNPV (48), whereas that of Plutella xylostella GV cannot compensate for the absence of GP64 (34). In our present study, we investigated the functions of XecnGV FP25K in a BmNPV system. Bm25KD-Xc produced numbers of OBs and BVs similar to those produced by T3 (Fig. 2B and 3A), suggesting that XecnGV FP25K is functionally expressed in this system. Taken together with findings from other experiments, these results indicate that XecnGV FP25K lacks the ability to occlude ODVs into OBs and does not activate V-CATH in a BmNPV system (Fig. 4C and Table 2). However, it is difficult to conclude that the inability of Bm25KD-Xc to substitute for these functions is due to the functional difference between NPV and GV FP25Ks since the expression level of XecnGV FP25K is relatively low (Fig. 1D). To completely eliminate the possibility that the expression level of XecnGV FP25K results in the phenotypic defects observed in Bm25KD-Xc, additional experiments are required to examine the phenotype of an XecnGV fp25K disruption mutant in host insects by using a GV bacmid system recently developed with CpGV (12).
Although AcMNPV FP25K has a markedly high degree of homology (96% identity) to BmNPV FP25K, Bm25KD-Ac produces more OBs and BVs than T3 and Bm25KD-Bm (Fig. 2B and 3A). Combined with the qRT-PCR results for polh (Fig. 2C), these findings suggest that AcMNPV FP25K has greater potency in the activation of late and very late gene expression than its BmNPV homolog. Also, using recombinant BmNPVs expressing chimeric FP25Ks, we showed that the N-terminal region of FP25K plays an important role in OB production (Fig. 5). To date, there have been several studies of the functional differences of BmNPV and AcMNPV genes. P35 is a well-known baculoviral antiapoptosis protein that prevents apoptosis by inhibiting caspase activity (5). A p35 mutant of AcMNPV does not produce OBs in Spodoptera frugiperda cells, whereas the BmNPV p35 mutant can grow normally and produce OBs in BmN cells (18). Biochemical studies revealed that AcMNPV P35 efficiently blocks apoptosis induced by caspase overexpression but that BmNPV P35 does so very poorly, suggesting that BmNPV P35 is weaker than AcMNPV P35 in terms of the suppression of caspase-induced apoptosis (40). Also, a BmNPV possessing AcMNPV gp64 was shown to produce more BVs in BmN cells than repaired BmNPV (21). Furthermore, homologous recombination studies using AcMNPV and BmNPV showed that expansion of the AcMNPV host range to include B. mori cells is established by the recombination of a BmNPV-derived 572-bp-long fragment within a DNA helicase gene (6, 37). AcMNPV has the widest host range (comprising more than 30 host species) among baculoviruses, while BmNPV has very narrow (1-species) host specificity. Phylogenetic studies have suggested that BmNPV has evolved within the last 1,000 years from ancestors that include AcMNPV. Amino acid substitutions found in AcMNPV and BmNPV proteins may represent important evolutionary steps in the adaptation of BmNPV to its host, B. mori.
The baculovirus expression vector system (BEVS) has been greatly improved in terms of host range expansion (39), development of a bacmid system (33), and stabilization of the products by deletion of v-cath and v-chiA from the viral genome (17, 32, 45). However, little genetic modification has been performed to increase the amount of foreign products or enhance the activity of the polh promoter in the BEVS. A recent study demonstrated that the deletion of five nonessential genes, v-cath, v-chiA, p26, p10, and p74, from the AcMNPV genome significantly increases the expression levels of recombinant proteins (14). Unlike most AcMNPV-based BEVSs, the BmNPV-based BEVS is particularly suitable for the mass production of foreign proteins in larvae and pupae of B. mori (36). To improve this system, our group has been attempting to identify the genes modifying the promoter activity of polh. We recently found that Bm34, a homolog of AcMNPV Ac43, enhances transcription of late and very late genes, especially that of polh (29). In this study, we showed that a BmNPV-based vector with AcMNPV fp25K has great potential for increasing the production of recombinant proteins in BmN cells and shortening the time for obtaining the secreted recombinant proteins from larval hemolymph. Multiple genetic modifications in the BmNPV genome will give us a novel, high-potential BEVS to produce recombinant proteins in B. mori.
Acknowledgments
This work was supported by grants from MEXT (no. 17018007 to T.S. and no. 19688004 to S.K.), the Professional Program for Agricultural Bioinformatics), and MAFF-NIAS (Agrigenome Research Program), Japan.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Beames, B., and M. D. Summers. 1989. Location and nucleotide sequence of the 25K protein missing from baculovirus few polyhedra (FP) mutants. Virology 168:344-353. [DOI] [PubMed] [Google Scholar]
- 2.Braunagel, S. C., J. K. Burks, G. Rosas-Acosta, R. L. Harrison, H. Ma, and M. D. Summers. 1999. Mutations within the Autographa californica nucleopolyhedrovirus FP25K gene decrease the accumulation of ODV-E66 and alter its intranuclear transport. J. Virol. 73:8559-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braunagel, S. C., S. T. Williamson, S. Saksena, Z. Zhong, W. K. Russell, D. H. Russell, and M. D. Summers. 2004. Trafficking of ODV-E66 is mediated via a sorting motif and other viral proteins: facilitated trafficking to the inner nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 101:8372-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty, S., and S. Reid. 1999. Serial passage of a Helicoverpa armigera nucleopolyhedrovirus in Helicoverpa zea cell cultures. J. Invertebr. Pathol. 73:303-308. [DOI] [PubMed] [Google Scholar]
- 5.Clem, R. J., M. Fechheimer, and L. K. Miller. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388-1390. [DOI] [PubMed] [Google Scholar]
- 6.Croizier, G., L. Croizier, O. Argaud, and D. Poudevigne. 1994. Extension of Autographa californica nuclear polyhedrosis virus host range by interspecific replacement of a short DNA sequence in the p143 helicase gene. Proc. Natl. Acad. Sci. U. S. A. 91:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daimon, T., S. Katsuma, W. K. Kang, and T. Shimada. 2007. Functional characterization of chitinase from Cydia pomonella granulovirus. Arch. Virol. 152:1655-1664. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, M. J., and W. F. Hink. 1982. The isolation and characterization of the MP and FP plaque variants of Galleria mellonella nuclear polyhedrosis virus. Virology 117:366-378. [DOI] [PubMed] [Google Scholar]
- 9.Granados, R. R., and K. A. Lawler. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108:297-308. [DOI] [PubMed] [Google Scholar]
- 10.Harrison, R. L., and M. D. Summers. 1995. Mutations in the Autographa californica multinucleocapsid nuclear polyhedrosis virus 25 kDa protein gene result in reduced virion occlusion, altered intranuclear envelopment and enhanced virus production. J. Gen. Virol. 76:1451-1459. [DOI] [PubMed] [Google Scholar]
- 11.Harrison, R. L., D. L. Jarvis, and M. D. Summers. 1996. The role of the AcMNPV 25K gene, “FP25,” in baculovirus polh and p10 expression. Virology 226:34-46. [DOI] [PubMed] [Google Scholar]
- 12.Hilton, S., E. Kemp, G. Keane, and D. Winstanley. 2008. A bacmid approach to the genetic manipulation of granuloviruses. J. Virol. Methods 152:56-62. [DOI] [PubMed] [Google Scholar]
- 13.Hink, W. F., and P. V. Vail. 1973. A plaque assay for titration of alfalfa looper nuclear polyhedrosis virus in a cabbage looper (TN-368) cell line. J. Invertebr. Pathol. 22:168-174. [Google Scholar]
- 14.Hitchman, R. B., R. D. Possee, A. T. Crombie, A. Chambers, K. Ho, E. Siaterli, O. Lissina, H. Sternard, R. Novy, K. Loomis, L. E. Bird, R. J. Owens, and L. A. King. 5 August 2009. Genetic modification of a baculovirus vector for increased expression in insect cells. Cell Biol. Toxicol. 26:57-68. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 15.Ikeda, M., M. Shikata, N. Shirata, S. Chaeychomsri, and M. Kobayashi. 2006. Gene organization and complete sequence of the Hyphantria cunea nucleopolyhedrovirus genome. J. Gen. Virol. 87:2549-2562. [DOI] [PubMed] [Google Scholar]
- 16.Jarvis, D. L., D. A. Bohlmeyer, and A. Garcia, Jr. 1992. Enhancement of polyhedrin nuclear localization during baculovirus infection. J. Virol. 66:6903-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaba, S. A., A. M. Salcedo, P. O. Wafula, J. M. Vlak, and M. M. van Oers. 2004. Development of a chitinase and v-cathepsin negative bacmid for improved integrity of secreted recombinant proteins. J. Virol. Methods 122:113-118. [DOI] [PubMed] [Google Scholar]
- 18.Kamita, S. G., K. Majima, and S. Maeda. 1993. Identification and characterization of the p35 gene of Bombyx mori nuclear polyhedrosis virus that prevents virus-induced apoptosis. J. Virol. 67:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamita, S. G., K. Nagasaka, J. W. Chua, T. Shimada, K. Mita, M. Kobayashi, S. Maeda, and B. D. Hammock. 2005. A baculovirus-encoded protein tyrosine phosphatase gene induces enhanced locomotory activity in a lepidopteran host. Proc. Natl. Acad. Sci. U. S. A. 102:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, W., M. Tristem, S. Maeda, N. E. Crook, and D. R. O'Reilly. 1998. Identification and characterization of the Cydia pomonella granulovirus cathepsin and chitinase genes. J. Gen. Virol. 79:2283-2292. [DOI] [PubMed] [Google Scholar]
- 21.Katou, Y., M. Ikeda, and M. Kobayashi. 2006. Abortive replication of Bombyx mori nucleopolyhedrovirus in Sf9 and High Five cells: defective nuclear transport of the virions. Virology 347:455-465. [DOI] [PubMed] [Google Scholar]
- 22.Katsuma, S., Y. Noguchi, C. L. Zhou, M. Kobayashi, and S. Maeda. 1999. Characterization of the 25K FP gene of the baculovirus Bombyx mori nucleopolyhedrovirus: implications for post-mortem host degradation. J. Gen. Virol. 80:783-791. [DOI] [PubMed] [Google Scholar]
- 23.Katsuma, S., S. Tanaka, T. Shimada, and M. Kobayashi. 2004. Reduced cysteine protease activity of the hemolymph of Bombyx mori larvae infected with fp25K-inactivated Bombyx mori nucleopolyhedrovirus results in the reduced postmortem host degradation. Arch. Virol. 149:1773-1782. [DOI] [PubMed] [Google Scholar]
- 24.Katsuma, S., T. Daimon, K. Mita, and T. Shimada. 2006. Lepidopteran ortholog of Drosophila Breathless is a receptor for the baculoviral fibroblast growth factor. J. Virol. 80:5474-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katsuma, S., S. Horie, T. Daimon, M. Iwanaga, and T. Shimada. 2006. In vivo and in vitro analyses of a Bombyx mori nucleopolyhedrovirus mutant lacking functional vfgf. Virology 355:62-70. [DOI] [PubMed] [Google Scholar]
- 26.Katsuma, S., T. Daimon, S. Horie, M. Kobayashi, and T. Shimada. 2006. N-linked glycans of Bombyx mori nucleopolyhedrovirus fibroblast growth factor are crucial for its secretion. Biochem. Biophys. Res. Commun. 350:1069-1075. [DOI] [PubMed] [Google Scholar]
- 27.Katsuma, S., T. Nakanishi, T. Daimon, and T. Shimada. 2009. N-linked glycans located in the pro-region of Bombyx mori nucleopolyhedrovirus V-CATH are essential for the proper folding of V-CATH and V-CHIA. J. Gen. Virol. 90:170-176. [DOI] [PubMed] [Google Scholar]
- 28.Katsuma, S., T. Nakanishi, and T. Shimada. 2009. Bombyx mori nucleopolyhedrovirus FP25K is essential for maintaining a steady-state level of v-cath expression throughout the infection. Virus Res. 140:155-160. [DOI] [PubMed] [Google Scholar]
- 29.Katsuma, S., and T. Shimada. 2009. Bombyx mori nucleopolyhedrovirus ORF34 is required for efficient transcription of late and very late genes. Virology 392:230-237. [DOI] [PubMed] [Google Scholar]
- 30.Keddie, B. A., G. W. Aponte, and L. E. Volkman. 1989. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science 243:1728-1730. [DOI] [PubMed] [Google Scholar]
- 31.Ko, R., K. Okano, and S. Maeda. 2000. Structural and functional analysis of the Xestia c-nigrum granulovirus matrix metalloproteinase. J. Virol. 74:11240-11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, K. S., Y. H. Je, S. D. Woo, H. D. Sohn, and B. R. Jin. 2006. Production of a cellulase in silkworm larvae using a recombinant Bombyx mori nucleopolyhedrovirus lacking the virus-encoded chitinase and cathepsin genes. Biotechnol. Lett. 28:645-650. [DOI] [PubMed] [Google Scholar]
- 33.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lung, O., M. Westenberg, J. M. Vlak, D. Zuidema, and G. W. Blissard. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 76:5729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda, S., T. Kawai, M. Obinata, H. Fujiwara, T. Horiuchi, Y. Saeki, Y. Sato, and M. Furusawa. 1985. Production of human alpha-interferon in silkworm using a baculovirus vector. Nature 315:592-594. [DOI] [PubMed] [Google Scholar]
- 36.Maeda, S. 1989. Expression of foreign genes in insects using baculovirus vectors. Annu. Rev. Entomol. 34:351-372. [DOI] [PubMed] [Google Scholar]
- 37.Maeda, S., S. G. Kamita, and A. Kondo. 1993. Host range expansion of Autographa californica nuclear polyhedrosis virus (NPV) following recombination of a 0.6-kilobase-pair DNA fragment originating from Bombyx mori NPV. J. Virol. 67:6234-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyajima, A., J. Schreurs, K. Otsu, A. Kondo, K. Arai, and S. Maeda. 1987. Use of the silkworm, Bombyx mori, and an insect baculovirus vector for high-level expression and secretion of biologically active mouse interleukin-3. Gene 58:273-281. [DOI] [PubMed] [Google Scholar]
- 39.Mori, H., H. Nakazawa, N. Shirai, N. Shibata, M. Sumida, and F. Matsubara. 1992. Foreign gene expression by a baculovirus vector with an expanded host range. J. Gen. Virol. 73:1877-1880. [DOI] [PubMed] [Google Scholar]
- 40.Morishima, N., K. Okano, T. Shibata, and S. Maeda. 1998. Homologous p35 proteins of baculoviruses show distinctive anti-apoptotic activities which correlate with the apoptosis-inducing activity of each virus. FEBS Lett. 427:144-148. [DOI] [PubMed] [Google Scholar]
- 41.Ohkawa, T., K. Majima, and S. Maeda. 1994. A cysteine protease encoded by the baculovirus Bombyx mori nuclear polyhedrosis virus. J. Virol. 68:6619-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potter, J. N., P. Faulkner, and E. A. MacKinnon. 1976. Strain selection during serial passage of Trichoplusia ni nuclear polyhedrosis virus. J. Virol. 18:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosas-Acosta, G., S. C. Braunagel, and M. D. Summers. 2001. Effects of deletion and overexpression of the Autographa californica nuclear polyhedrosis virus FP25K gene on synthesis of two occlusion-derived virus envelope proteins and their transport into virus-induced intranuclear membranes. J. Virol. 75:10829-10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slavicek, J. M., J. Podgwaite, and C. Lanner-Herrera. 1992. Properties of two Lymantria dispar nuclear polyhedrosis virus isolates obtained from microbial pesticide Gypchek. J. Invertebr. Pathol. 59:142-148. [Google Scholar]
- 45.Suzuki, T., T. Kanaya, H. Okazaki, K. Ogawa, A. Usami, H. Watanabe, K. Kadono-Okuda, M. Yamakawa, H. Sato, H. Mori, S. Takahashi, and K. Oda. 1997. Efficient protein production using a Bombyx mori nuclear polyhedrosis virus lacking the cysteine proteinase gene. J. Gen. Virol. 78:3073-3080. [DOI] [PubMed] [Google Scholar]
- 46.Winstanley, D., and N. E. Crook. 1993. Replication of Cydia pomonella granulosis virus in cell cultures. J. Gen. Virol. 74:1599-1609. [DOI] [PubMed] [Google Scholar]
- 47.Wu, D., F. Deng, X. Sun, H. Wang, L. Yuan, J. M. Vlak, and Z. Hu. 2005. Functional analysis of FP25K of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. J. Gen. Virol. 86:2439-2444. [DOI] [PubMed] [Google Scholar]
- 48.Yin, F., M. Wang, Y. Tan, F. Deng, J. M. Vlak, Z. Hu, and H. Wang. 2008. A functional F analogue of Autographa californica nucleopolyhedrovirus GP64 from the Agrotis segetum granulovirus. J. Virol. 82:8922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida, M., K. Higashi, L. Jin, Y. Machi, T. Suzuki, A. Masuda, N. Dohmae, A. Suganami, Y. Tamura, K. Nishimura, T. Toida, H. Tomitori, K. Kashiwagi, and K. Igarashi. 2010. Identification of acrolein-conjugated protein in plasma of patients with brain infarction. Biochem. Biophys. Res. Commun. 391:1234-1239. [DOI] [PubMed] [Google Scholar]