Abstract
APOBEC3F (A3F) and APBOBEC3G (A3G) both are host restriction factors that can potently inhibit human immunodeficiency virus type 1 (HIV-1) replication. Their antiviral activities are at least partially mediated by cytidine deamination, which causes lethal mutations of the viral genome. We recently showed that A3G blocks viral plus-strand DNA transfer and inhibits provirus establishment in the host genome (J. L. Mbisa, R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak, J. Virol. 81:7099-7110, 2007). Here, we investigated whether A3F similarly interferes with HIV-1 provirus formation. We observed that both A3F and A3G inhibit viral DNA synthesis and integration, but A3F is more potent than A3G in preventing viral DNA integration. We further investigated the mechanisms by which A3F and A3G block viral DNA integration by analyzing their effects on viral cDNA processing using Southern blot analysis. A3G generates a 6-bp extension at the viral U5 end of the 3′ long terminal repeat (3′-LTR), which is a poor substrate for integration; in contrast, A3F inhibits viral DNA integration by reducing the 3′ processing of viral DNA at both the U5 and U3 ends. Furthermore, we demonstrated that a functional C-terminal catalytic domain is more critical for A3G than A3F function in blocking HIV-1 provirus formation. Finally, we showed that A3F has a greater binding affinity for a viral 3′-LTR double-stranded DNA (dsDNA) oligonucleotide template than A3G. Taking these results together, we demonstrated that mechanisms utilized by A3F to prevent HIV-1 viral DNA integration were different from those of A3G, and that their target specificities and/or their affinities for dsDNA may contribute to their distinct mechanisms.
APOBEC3F (A3F) and APOBEC3G (A3G) proteins belong to the APOBEC3 family of cytidine deaminases that exhibit broad antiviral activities (4, 12, 21, 33, 34, 37, 46-48, 56). Members of this family of proteins contain one or two zinc-binding catalytic domains with the consensus sequence (H/C)-X-E-X23-28-P-C-X2-4-C (15, 17), but A3F and A3G proteins are the two most potent inhibitors of HIV-1 replication. The incorporation of A3F or A3G protein into HIV-1Δvif virions causes extensive cytidine deamination on the viral minus-strand DNA during reverse transcription (RT), subsequently resulting in the massive G-to-A hypermutation of the viral genome (4, 10, 11, 18, 21, 23, 42, 48, 53, 56). The G-to-A hypermutation is partially responsible for the detrimental effects on viral replication, since they generate stop codons and nonfunctional viral proteins (18, 35, 53). In addition to the direct effects of G-to-A hypermutation, the presence of uridines in the nascent HIV-1 genome produced by A3 proteins might recruit cellular DNA repair machinery to degrade viral replication intermediates (51). We and others have shown that the cytidine deaminase activity of A3G is essential for its inhibition of HIV-1 replication (24, 26, 29, 38, 40); however, others also have reported that the antiviral activity of A3G is independent of its editing activity (3, 31). Our previous studies show that the A3G catalytic site mutant achieves antiviral activity similar to the physiological levels of wild-type A3G only when 50-fold or higher levels of the mutant A3G protein are incorporated into virions (26, 49). On the contrary, it was reported that the loss of cytidine deaminase activity did not significantly affect A3F's ability to block HIV-1 replication (14).
A3F and A3G proteins possess two catalytic domains: the N-terminal domain is involved in RNA binding and virion incorporation, whereas the C-terminal domain is associated primarily with cytidine deaminase activity and the substrate sequence specificity for deamination (4, 11, 21, 30, 48, 56). Despite the two proteins sharing approximately 49% identity in their amino acid sequences, they have distinct sequence specificities in editing substrates: A3F prefers 5′-TC dinucleotides, while A3G prefers 5′-CC dinucleotides (the deaminated nucleotide is underlined) (2, 4, 21, 48, 56). In addition to their substrate differences, A3G is a significantly more potent inhibitor of HIV-1 replication and causes a greater frequency of G-to-A mutations than A3F (4, 21, 48, 55, 56). Other distinctions between the two proteins include the following: A3F exhibits a more pronounced deamination-independent effect on HIV-1 replication than A3G (14, 26), and they differ in their dependency on HIV-1 genomic or nongenomic RNAs for virion incorporation (41). In spite of these differences, HIV-1 utilizes similar pathways to eliminate the restriction imposed by A3F and A3G on its replication capacity. This is accomplished by targeting the proteins for proteasomal degradation through interaction with the viral accessory protein Vif, thereby preventing the incorporation of the A3 proteins into nascent virions (7, 19, 22, 25, 27, 39, 42, 43, 54). Recently, we and others have shown that distinct regions of A3F and A3G are used to bind to different determinants of the Vif protein, suggesting independent evolutionary pathways for the two proteins (8, 13, 28, 36, 41).
The inhibition of HIV-1 replication by A3F and A3G proteins initially was attributed exclusively to extensive G-to-A hypermutations imposed during reverse transcription on the integrated proviral genome. However, A3F and A3G were observed also to suppress viral DNA synthesis. In addition, we and others recently described novel mechanisms by which the A3 proteins are capable of inhibiting viral DNA integration (23, 26). Our results indicated that A3G inhibits HIV-1 cDNA synthesis by reducing the efficiency of plus-strand DNA transfer. Other reports have described A3G-mediated antiviral effects at different stages of viral DNA synthesis, such as the inhibition of tRNA primer binding, the inhibition of the initiation of DNA synthesis, the inhibition of the elongation of reverse transcription, the inhibition of minus-strand DNA synthesis, and the inhibition of integration by binding to integrase (1, 5, 9, 10, 14, 20, 23). We also showed that A3G produces an aberrant 6-bp extension at the viral U5 end of the 3′-LTR that cannot be properly processed and subsequently results in the prevention of provirus formation. Likewise, A3F displays phenotypes similar to those of A3G in reducing the accumulation of HIV-1 reverse transcription products and preventing proviral DNA formation (3, 14, 23, 52); however, the mechanisms used by A3F to block HIV-1 replication have not been as extensively investigated as those for A3G.
In this study, we examined the effects of A3F on the metabolism of HIV-1 cDNA to further understand the mechanisms by which A3F inhibits HIV-1 replication. We report here that A3F inhibits viral DNA synthesis and integration by blocking plus-strand DNA transfer and reducing the efficiency of the 3′ processing of viral DNA ends, respectively. Furthermore, A3F exerts a greater inhibition of provirus formation than A3G. Our results show that A3F utilizes distinct mechanisms from A3G by interfering with the 3′-processing reactions at both U3 and U5 viral DNA ends.
MATERIALS AND METHODS
Plasmids.
pFLAG-A3F(E251Q) was constructed using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and confirmed by DNA sequencing. The HIV-1-based vector pHDV-eGFP and plasmid constructs of pHCMV-G, pFLAG-A3G, pcDNA3.1-A3G(E259Q), and pFLAG-A3F have been described previously (26, 36, 50). pHDV-eGFP expresses HIV-1 Gag-Pol proteins and enhanced green fluorescent protein (eGFP); virus produced by this plasmid is referred to as HIV-1Δvif virus.
Cells, transfection, and virus production.
The human 293T cell line was obtained from the American Type Culture Collection (ATCC) and maintained as previously described (32). Virus stocks were prepared by the transfection of 293T cells, seeded at a density of 5 × 106 cells per 100-mm-diameter dish, with pHDV-eGFP, pHCMV-G, pFLAG-A3F, or pcDNA-APO3G (or salmon sperm DNA) using a CalPhos Mammalian Transfection kit (Clontech). For real-time PCR experiments, the transfected cell monolayers were rinsed with phosphate-buffered saline (PBS) containing 1% fetal calf serum three times each at 6 and 24 h posttransfection and once at 30 h posttransfection to reduce or eliminate contamination with transfected pHDV-eGFP DNA. Virus supernatants were harvested at 48 h posttransfection, clarified by centrifugation, filtered through 0.45-μm filters, and quantified by p24 enzyme-linked immunosorbent assay (ELISA) (Perkin-Elmer).
Infection and viral DNA quantification by real-time PCR.
The 293T cells were seeded at a density of 1 × 105 cells per 60-mm-diameter dish, and 24 h later they were infected with HIV-1 preparations containing equal amounts of p24 capsid for 3 h as previously described (32). A duplicate of each virus sample was heat inactivated at 65°C for 1 h and used to infect cells in parallel as a background control to determine the level of contamination with transfected DNA. In general, the values for the heat-inactivated viruses were <1%, indicating a low background. Cells were harvested at 6, 24, and 120 h postinfection, and total cellular DNA was extracted using a QIAamp DNA Blood Mini kit (Qiagen). The DNA then was digested with DpnI, which digests methylated plasmid DNA but not viral or cellular DNA, to further reduce plasmid DNA contamination. There are two DpnI sites in the amplicon (nucleotide [nt] 557 to 701), and DpnI digestion is expected to effectively suppress the amplification of the transfected plasmid DNA. The amounts of unintegrated viral DNA and proviruses were quantified using ABI Prism 7700 sequence detection as described previously (26) or using the Roche LightCycler 480 Real-Time PCR system and LightCycler 480 Probe Master reaction mix according to the manufacturer's instructions. Briefly, the copy numbers in each sample were adjusted for DNA input by the quantification of cellular chemokine (C-C motif) receptor 5 (CCR5) gene copy numbers and the subtraction of background signal as determined by infection with heat-inactivated virus. The primer-probe sets were purchased from Integrated DNA Technologies and were described previously (26). Cells also were harvested 48 h postinfection and analyzed by flow cytometry (FACScan; Becton Dickinson) for eGFP expression to determine the efficiency of infection.
Analysis of G-to-A hypermutation.
Total cellular DNA was extracted from infected cells, digested with DpnI to remove plasmid DNA, and used as a template to amplify the eGFP-encoding sequences with a forward primer (5′-CCACAAGTTCAGCGTGTCC-3′) and a reverse primer (5′-GGCACAAGCAGCATTGTTAG-3′). The resulting PCR product was resolved on a 1% agarose gel and purified by a QIAquick gel extraction kit (Qiagen). The purified PCR product was cloned into pGEM-T Easy vector (Promega). The plasmid DNA extracted from white colonies using the QIAprep Turbo kit (Qiagen) was sequenced. The G-to-A hypermutation was analyzed by Hypermut 2.0 software from Los Alamos National Laboratory.
Southern blot analysis of HIV-1 viral DNA ends in PICs.
To isolate preintegration complexes (PICs) for Southern blot analysis, 293T cells were seeded at a density of 3 × 106 cells per 100-mm-diameter dish and 24 h later were infected with HIV-1Δvif virus produced in the presence or absence of A3F or A3G. Cytoplasmic DNA was extracted 6 h postinfection by washing the cells twice in buffer K−/− (20 mM HEPES, pH 7.6, 150 mM KCl, 5 mM MgCl2) and lysed in buffer K+/+ (buffer K−/− plus 0.025% [wt/vol] digitonin, 1 mM dithiothreitol, and 20 μg/ml aprotinin) for 10 min at room temperature. Supernatant was collected by sequential centrifugation at 1,500 × g for 4 min and 19,000 × g for 1 min at 4°C and treated with RNase A (20 μg/ml). The DNA samples were digested with either HindIII or EarI for the viral U5 or U3 end, respectively, resolved by electrophoresis through a denaturing polyacrylamide gel, transferred to a Duralon-UV membrane (Stratagene), and analyzed by Southern blotting as described previously (44).
Analysis of A3F and A3G binding to HIV-1 DNA.
To determine the binding affinities of A3F and A3G to the HIV-1 U5 DNA end, we reconstituted the viral U5 DNA end by annealing a forward biotinylated oligonucleotide (5′-biotin-TCCCTCAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGT-3′) and a reverse oligonucleotide (5′-ACTGCTAGAGATTTTCCACACTGACTAAAAGGGTCTGAGGGA-3′). 293T cells were transfected with control empty vector or pFLAG-A3F, pFLAG-A3G, or pFLAG-A3F-E251Q expression plasmid, and the transfected cells were lysed 48 h posttransfection in cell extraction buffer (50). The cell extracts were adjusted to equivalent protein concentrations by using Bradford reagent (Bio-Rad), and equal aliquots were incubated with either forward single-stranded DNA (ssDNA) oligonucleotides or annealed double-stranded DNA (dsDNA) oligonucleotides at 4°C overnight with rotation. Equal volumes of Dynabead M-280 Streptavidin (Invitrogen) were added, and the samples were incubated at room temperature for 15 min with rotation. Dynabeads were washed five times with 0.5 ml cell extraction buffer, and bound proteins were eluted by being heated in 1× lithium dodecyl sulfate sample buffer (Invitrogen). The bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting using rabbit anti-FLAG antibody (Sigma).
Statistical analysis.
We used the two-sample Student's t test to determine if viral infectivity, DNA synthesis, and integration efficiency were significantly different between two viruses. The null hypothesis was rejected in favor of the alternative hypothesis if the P value was above the significance threshold level of 0.05.
RESULTS
Effects of A3F and A3G on HIV-1Δvif virus infectivity and DNA synthesis.
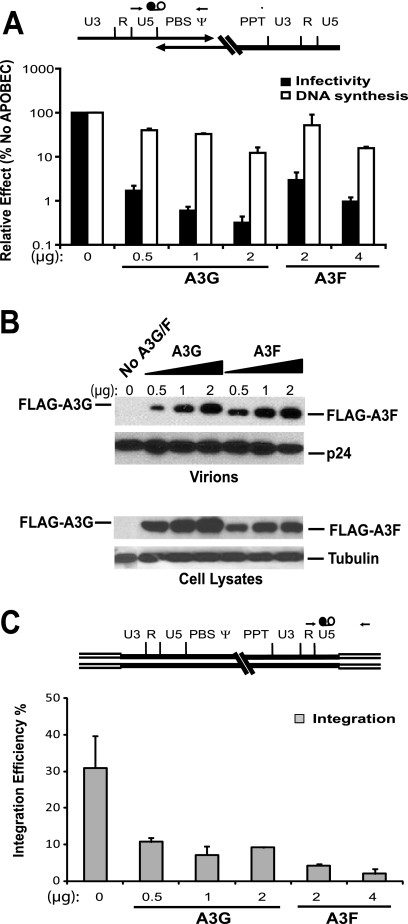
We first analyzed the effect of A3F and A3G on the accumulation of late-stage viral DNA and provirus formation using real-time PCR assays (Fig. 1). Since it has been reported that A3G is a more potent inhibitor of HIV-1Δvif virus than A3F, we compared their effects on viral DNA synthesis using equivalent amounts of transfected plasmid DNA, as well as amounts that caused similar reductions in viral infectivity. The infection of 293T target cells with the transfection-derived HIV-1Δvif viruses resulted in a single round of replication, and the infection rate was determined by analyzing the level of GFP expression using flow cytometry. The multiplicity of infection (MOI) of HIV-1Δvif viruses in the absence of A3F or A3G ranged from 1.37 to 1.51. The effect of A3F and A3G on HIV-1Δvif viral replication then was compared to that of HIV-1Δvif virus in the absence of the A3 proteins (Fig. 1A). This experiment showed that the cotransfection of 2 μg of pFLAG-A3F produced a reduction in infectivity comparable to that of the cotransfection of 0.5 μg pFLAG-A3G (30-fold versus 33-fold), while 4 μg of pFLAG-A3F produced a reduction comparable to that of 1 μg of pFLAG-A3G (102-fold and 138-fold, respectively). However, when equivalent amounts (2 μg) of pFLAG-A3F and pFLAG-A3G were used, A3G was approximately 10-fold more potent than A3F, with a 297-fold reduction in infectivity, whereas that for A3F was 30-fold. These results are in agreement with previously published data using equivalent amounts of A3F and A3G that showed pronounced differences in their effects on both infectivity and mutational load (3, 21, 55). This difference in potency is not due to a difference in the packaging efficiency of the two proteins, since Western blot analysis showed that equivalent amounts of the A3F and A3G proteins are incorporated into the HIV-1Δvif virions (Fig. 1B, upper). This is in spite of cellular A3F proteins being expressed at slightly lower levels than those of A3G proteins when equivalent amounts of expression plasmids were used (Fig. 1B, lower), suggesting that A3F proteins are packaged more efficiently than A3G proteins.
FIG. 1.
Comparison of the effects of A3F and A3G on viral infectivity, DNA synthesis, and integration. (A) Viral infectivity was determined by the proportion of GFP+ cells to total cells (black bar). Human 293T cells were transfected with pHDV-eGFP in the absence or presence of A3 expression plasmids or different amounts of A3 expression plasmids as indicated below the bar graphs. Viral infectivity without A3 proteins was set as 100%. DNA synthesis was determined at 6 h postinfection (empty bar) by the quantification of viral late reverse transcription products probing for the U5-ψ region, which is indicated at the top. Thin arrows indicate primers, and black and white circles connected with a line indicate the probe for quantitative real-time PCR. (B) Virion incorporation and cellular expression of A3F and A3G proteins were determined by Western blot probing with anti-FLAG antibody. Viral p24 and cellular tubulin proteins were used as the loading control for virions and cells, respectively. (C) The integration efficiency was determined by the proportion of integrated proviruses at 5 days postinfection to the viral late RT products at 6 h postinfection (Alu-LTR-5d/U5-ψ-6 h). The location of Alu-LTR primers and probe are indicated at the top; double lines represent host sequences flanking proviral DNA. Error bars in all graphs represent the standard deviations of at least two independent experiments.
Viral DNA was isolated 6 h postinfection, at which point maximal viral DNA is detected (45), and quantified by real-time PCR using a primer-probe set spanning the U5-Ψ region to detect late reverse transcription products (Fig. 1A, top schematic). The effect on HIV-1Δvif viral DNA levels in the presence of either A3F or A3G was compared to that in the absence of the A3 proteins (set at 100%) (Fig. 1A). The amounts of A3F or A3G that caused similar reductions in infectivity, 2 μg versus 0.5 μg and 4 μg versus 1 μg, respectively, also resulted in comparable reductions in the accumulation of viral late RT products (2.3-fold versus 2.8-fold and 6.4-fold versus 3.9-fold, respectively). In contrast, when equivalent amounts of A3F or A3G (2 μg) were used, A3G had a greater effect on reducing the accumulation of viral DNA than A3F (10.6-fold and 2.3-fold, respectively).
A3G induces more G-to-A hypermutation than A3F.
We analyzed the G-to-A hypermutation frequency in virions produced in the presence of levels of A3G and A3F that induced similar levels of viral inhibition (Table 1). As expected, the majority of A3G-induced G-to-A mutations were in the context of GG dinucleotides (66.3 to 72.9%), whereas the majority of A3F-induced G-to-A mutations were in the GA dinucleotide context (72.0 to 75.5%). The G-to-A mutation frequency was 2.54 and 0.90% when 0.5 or 2.0 μg of A3G and A3F were cotransfected, respectively; similarly, the mutation frequency was 3.63 and 0.94% when 1.0 and 4.0 μg of A3G and A3F were cotransfected, respectively. These results indicated that A3F induces approximately 3- to 4-fold fewer G-to-A mutations than A3G to achieve a similar level of viral inhibition and suggest that A3F uses mechanisms other than hypermutation to inhibit viral replication.
TABLE 1.
Dinucleotide context of G-to-A mutations and their frequency
| Restriction factora (concn, in μg) | Dinucleotide context (%) of G-to-A mutations |
G-to-A mutations/total nucleotides (frequency, %) | |||
|---|---|---|---|---|---|
| GGb | GA | GC | GT | ||
| No A3G or A3F | 0 | 0 | 0 | 0 | 0/11,920 (<0.008) |
| A3G (0.5) | 199 (72.9) | 68 (24.9) | 5 (1.8) | 1 (0.4) | 273/10,728 (2.54)c |
| A3G (1.0) | 287 (66.3) | 126 (29.1) | 12 (2.8) | 8 (1.8) | 433/11,920 (3.63)c |
| A3F (2.0) | 6 (5.6) | 77 (72.0) | 24 (22.4) | 0 | 107/11,920 (0.90)d |
| A3F (4.0) | 4 (3.8) | 80 (75.5) | 22 (20.8) | 0 | 106/11,324 (0.94)d |
HIV-1Δvif was cotransfected with different amounts of A3G- or A3F-expressing plasmid, and the virus produced was used to infect 293T cells. The viral DNAs produced were analyzed by sequencing to determine the G-to-A hypermutation frequency and dinucleotide context of mutations.
The first G nucleotide in the GG dinucleotide is the target of G-to-A mutation.
The G-to-A hypermutation frequency is higher when the amount of A3G is increased from 0.5 to 1.0 μg (P = 0.000002).
The G-to-A hypermutation frequency is not significantly higher when the amount of A3F is increased from 2.0 to 4.0 μg (P = 0.78).
A3F decreases the integration of HIV-1Δvif virus more potently than A3G.
We compared the effects of A3F and A3G on the integration efficiency of HIV-1Δvif virus by quantifying total cellular DNA and integrated viral DNA from infected cells (Fig. 1C) (6). The integration efficiency was determined by calculating the amount of integrated viral DNA copies as a proportion of the viral late RT DNA products at 6 h postinfection. In contrast to the effect on viral DNA accumulation, the amounts of A3F that resulted in reductions in infectivity comparable to those of A3G (2 μg versus 0.5 μg and 4 μg versus 1 μg, respectively) showed a greater impact on HIV-1Δvif viral DNA integration efficiency (7.2-fold versus 2.9-fold and 15.5-fold versus 4.4-fold, respectively). Accordingly, we conclude that A3F is a more potent inhibitor of viral DNA integration than A3G, because when equivalent amounts of A3F or A3G (2 μg) were used, A3F caused a 7.2-fold reduction while A3G resulted in a 3.4-fold decrease (P = 0.004). The greater antiviral activity of A3G compared to that of A3F could be attributed to its more potent ability to induce G-to-A hypermutation.
A3F decreases the efficiency of 3′ processing but does not produce a 6-bp extension at the U5 3′ end of viral DNA.
We investigated whether A3F uses mechanisms distinct from those of A3G to more potently inhibit viral DNA integration. The integration of HIV-1 cDNA into the host genome requires the removal of GT dinucleotides at the 3′ end of both viral DNA strands by HIV-1 integrase (IN), a process that occurs in the cytoplasm and is called 3′ processing (Fig. 2A, vi). The 3′-processing step is followed by the transfer of the viral DNA into the nucleus, where it is ligated to the host DNA in a process called DNA strand transfer. We previously showed that A3G inhibits viral DNA integration prior to nuclear import by producing a 6-bp extension at the U5 3′ end of viral DNA, thereby producing a poor substrate for 3′ processing by IN (Fig. 2A, vii to ix). We performed Southern blot analysis of the U5 3′ end of viral DNA from cells infected with HIV-1Δvif virus in the absence of A3 proteins or in the presence of A3F or A3G. Consistently with our previous studies, we observed a 111-nt-long fragment in the viral PICs produced from A3G-incorporated virus, which represents a 6-bp extension at the U5 3′ end that is produced by the aberrant removal of primer tRNA by RNase H (Fig. 2A and B). It is hypothesized that this 6-bp extension blocks proper 3′ processing and subsequently inhibits viral DNA integration. The absence of this fragment from the viral PICs produced from A3F-expressing cells suggests that A3F does not interfere with the removal of the primer tRNA and uses a different mechanism from A3G to suppress viral DNA integration.
FIG. 2.
Comparison of effects of A3F and A3G on U5 viral DNA ends of the 3′-LTR. (A) Schematic representation of HIV-1 reverse transcription and U5 viral DNA end fragments detected after restriction enzyme digestion with HindIII. (i) Virion-incorporated tRNA binds to the primer binding site (PBS) of viral genomic RNA and initiates minus-strand DNA synthesis. (ii) Minus-strand strong stop (−SSS) DNA is transferred to the 3′ end of genomic RNA and continues the synthesis of viral minus-strand DNA. (iii) Viral plus-strand DNA synthesis initiates from the polypurine tract (PPT) and is elongated until the end of the PBS region of minus-strand DNA. (iv) In the absence of A3G or in the presence of A3F, RNase H cleaves the tRNA primer between the 3′ adenosine and cytosine nucleotides. (v) The released plus-strand strong-stop (+SSS) DNA is transferred and annealed to the PBS of minus-strand DNA. (vi) Both strands are extended to complete full-length cDNA synthesis, and integrase (IN) removes the 3′ GT dinucleotide from the proviral DNA. After digestion with HindIII, the processed U5 plus-strand DNA generates a 103-nt fragment, the unprocessed end generates a 105-nt fragment, and the 122-nt fragment corresponds to viral +SSS DNA; A3F inhibits the 3′-processing reaction. (vii) In the presence of A3G, an aberrant cleavage of the tRNA primer by RNase H occurs 6 nt upstream. (viii) The partially released +SSS DNA is transferred and annealed to the PBS of minus-strand DNA. (ix) Both strands are extended to the complete full-length cDNA synthesis, and reverse transcription of the fragment of the tRNA primer that remains associated with the minus-strand DNA generates a 6-nt extension at the 3′ end of the plus-strand DNA. This generates an aberrant 111-nt fragment following digestion with HindIII. The thin line represents RNA; the thick line, DNA; dashed thin lines, RNA template degraded by RNase H; arrows, the direction of DNA synthesis; the black box, the riboprobe that hybridizes to the plus strand of U5 at the 3′-LTR. (B) Southern blot analysis of U5 viral DNA plus strand at the 3′-LTR. Viral preintegration complexes (PICs) were harvested at 6 h postinfection, and viral late RT products were quantified by real-time PCR probing the U5-ψ region. The virions were produced in the absence of A3G/A3F or in the presence of 0.5 or 2 μg of A3G or 2 μg of A3F. Viral late RT products (3 × 107 copies) from each sample were digested with HindIII and analyzed by Southern blotting. The +SSS DNA fragment (122 nt), aberrant DNA fragment (111 nt), unprocessed 3′-U5 plus-strand DNA fragment (105 nt), and the processed 3′-U5 plus-strand DNA fragment (103 nt) are shown. The 3′-processing efficiency was calculated by the proportion of processed band (103 nt) to the processed plus unprocessed bands (103 nt and 105 plus 106 nt). The 106-nt band is not labeled. The values are averages from two independent experiments ± standard errors.
We next determined the efficiency of 3′ processing by quantifying the intensities of the 103-nt and the 105- and 106-nt bands representing the processed and unprocessed U5 viral DNA ends, respectively. As described previously (26, 44), the 106-nt band results from the presence of an adenine ribonucleotide that remains attached to the 3′ end of some viral DNAs. The comparison of A3F (2 μg) to A3G (0.5 μg), which results in comparable reductions in infectivity (Fig. 1A), showed no effect in 3′-processing efficiency for A3G-containing virus (51% ± 5%) but a significant reduction for A3F-containing virus (23% ± 2%) compared to that of the no-A3G/A3F control (53% ± 3%) (Fig. 2B). These results suggested that A3F (P = 0.004), but not A3G (P = 0.48), significantly interferes with viral U5 3′ processing. The expression of equivalent amounts of the A3 proteins by transfecting 2 μg of A3F- or A3G-expressing plasmid showed that A3F is more potent at reducing viral U5 3′ processing than A3G (23% ± 2% and 37% ± 2%, respectively). In addition to the 103-nt and 105/106-nt bands, we also observed a 122-nt fragment produced by HindIII digestion that corresponds to the plus-strand strong-stop DNA (+SSS DNA) (Fig. 2A, iii). The intensities of the 122-nt band were significantly increased in the presence of the A3 proteins compared to that of the no-A3G/A3F control (Fig. 2B). The increase was 4-fold for 0.5 and 2 μg of A3G and A3F, indicating that both A3G and A3F display comparable inhibitory effects on +SSS DNA transfer. The observation suggests that when A3G and A3F inhibit replication to the same extent, a similar mechanism is involved in the inhibition of this reverse transcription step.
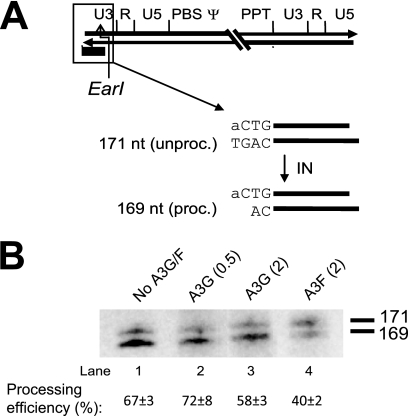
A3F, but not A3G, interferes with U3 3′ processing.
We next analyzed the 3′-processing efficiency at the U3 end of HIV-1 DNA in the presence or absence of A3F or A3G. A 171-nt fragment representing the unprocessed U3 viral DNA end and a 169-nt fragment representing a 3′-processed U3 viral DNA end were observed (Fig. 3A and B). The 3′-processing efficiency at the U3 end in the absence of the A3 proteins was 67% ± 3%. The expression of 0.5 or 2 μg of A3G had no obvious effects on processing reactions, since their processing efficiencies were comparable to those of virus without A3 proteins (72% ± 8% and 58% ± 3%, respectively). However, the expression of 2 μg of A3F significantly reduced the 3′-processing efficiency to 40% ± 2% (P = 0.002). These experiments demonstrated that A3F interferes with viral U3 3′ processing while A3G does not. Taken together, these data showed that A3F blocks viral DNA integration by significantly interfering with the 3′ processing at both viral DNA ends.
FIG. 3.
Comparison of effects of A3F and A3G proteins on U3 viral DNA ends of the 5′-LTR. (A) Reverse transcription products of U3 viral DNA of the 5′-LTR were detected after digestion with the restriction enzyme EarI and by using an RNA probe (black box) that hybridizes to the U3 minus strand of the viral 5′-LTR. The unprocessed fragment of the 3′ end of the minus strand after EarI treatment is 171 nt in length; the processed fragment is 169 nt in length. (B) Southern blot analysis of the U3 minus strand at 5′-LTR. Viral PICs were harvested at 6 h postinfection and quantified by real-time PCR to determine the viral late RT products by probing the U5-ψ region. A total of 3 × 107 copies of viral late RT products from each sample listed at the top and as described for panel A were digested with EarI and analyzed by Southern blotting. The processing efficiency was calculated by the proportion of processed band (169 nt) to both processed and unprocessed bands (169 and 171 nt). The values are averages from three independent experiments ± standard deviations.
The role of cytidine deamination activity in the A3G- and A3F-mediated inhibition of HIV-1 DNA integration.
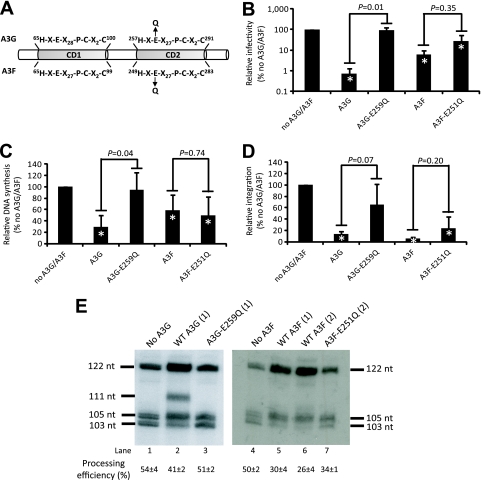
To determine if cytidine deamination activity plays a role in blocking HIV-1 DNA integration, we investigated the effects of A3F and A3G cytidine deamination mutants on the processing of viral DNA ends. As shown in Fig. 4A, the glutamic acid residues of the H-X-E-X27-P-C-X2-C motifs in the C-terminal catalytic domain of the A3 proteins, which act as the proton donors in the deamination reaction, were mutated to glutamine to abolish their cytidine deaminase activities and generate mutants A3G-E259Q and A3F-E251Q (14, 24, 26, 40). First, we observed that unlike wild-type A3G, the A3G-E259Q mutant did not inhibit HIV-1 replication, suggesting that the cytidine deaminase activity is indispensable for A3G antiviral activity (Fig. 4B). Furthermore, the infectivity of HIV-1Δvif virus in the presence of wild-type A3G was significantly lower than the infectivity in the presence of the A3G-E259Q mutant (P = 0.01). In contrast to A3G, both wild-type A3F and the A3F-E251Q mutant inhibited HIV-1Δvif replication, whereas the no-A3F control did not (Fig. 4B). In addition, the HIV-1Δvif infectivity in the presence of wild-type A3F was not significantly different from the infectivity in the presence of the A3F-E251Q mutant (P = 0.35).
FIG. 4.
Effects of the catalytic-site mutations of A3F and A3G on viral infectivity, DNA synthesis, integration, and viral DNA processing. (A) Schematic representation of the E-to-Q mutations of A3G and A3F proteins. (B) Relative viral infectivity was determined by the proportion of GFP+ cells compared to the proportion of GFP+ cells in the absence of A3F or A3G (set to 100%). (C) Relative DNA synthesis efficiency was determined by the quantification of viral late RT products at 6 h postinfection compared to the viral late RT products in the absence of A3F or A3G (set to 100%). (D) Relative integration efficiency was determined by quantifying the proportion of integrated proviruses at 5 days postinfection to the viral late RT products at 6 h postinfection (U5-ψ-120 h/U5-ψ-6 h). The integration efficiency in the absence of A3F or A3G was set to 100%. Error bars in all graphs represent the standard deviations from at least three independent experiments. All P values were determined using the two-sample t test. The asterisks indicate significant P values (<0.05) compared to results for the no-A3G/A3F control. (E) Southern blot analysis of the effects of CD2 mutations on viral DNA ends. Viral preintegration complexes were harvested and analyzed as shown in Fig. 2. The numbers in parentheses above each lane indicate the micrograms of the indicated plasmid used for transfection. The processing efficiencies were calculated as shown in Fig. 2 by the analysis of this blot. The values are averages from two independent experiments ± standard errors.
The results shown in Fig. 4C reveal that the A3G cytidine deamination mutant does not significantly affect viral DNA synthesis compared to that of the no-A3G/A3F control. Additionally, the amount of viral DNA detected in the presence of wild-type A3G is significantly lower than the amount of DNA detected in the presence of the A3G-E259Q mutant (P = 0.04). In contrast to A3G, both the wild-type A3F and the A3F-E251Q mutant substantially reduced the amount of detectable viral DNA (P < 0.05), and there was no difference in the amount of viral DNA in the presence of the wild-type or mutant A3F proteins (P = 0.74).
To determine the effects of the catalytic site mutants of A3F and A3G on viral DNA integration, the numbers of integrated proviruses were quantified (Fig. 4D). Unlike the wild-type A3G, the A3G-E259Q mutant did not significantly affect the integration efficiency of HIV-1Δvif virus. In contrast, both wild-type A3F and the A3F-E251Q mutant significantly inhibited viral DNA integration. These data indicate that the cytidine deamination activity is crucial for A3G's but not A3F's ability to inhibit HIV-1 infectivity, viral DNA synthesis, and viral DNA integration.
Southern blot analysis was performed to further examine the requirement of the cytidine deamination activity of the A3 proteins in preventing HIV-1 DNA 3′ processing (Fig. 4E). First, we observed that the 111-nt aberrant band that is produced in the presence of wild-type A3G was absent in A3G-E259Q-containing viruses (Fig. 4E, lanes 2 and 3). This observation indicated that the A3G catalytic domain is required for the generation of the 6-bp extension at the U5 3′ end of viral DNA. Second, we noted no change in the amount of +SSS DNA produced in the presence of mutant A3G but a 3-fold increase in the presence of wild-type A3G, indicating that the inhibition of +SSS DNA transfer also required a functional A3G catalytic domain (Fig. 4E, lanes 2 and 3). Finally, we observed that viruses containing the mutant A3F displayed an intermediate phenotype regarding the accumulation of +SSS DNA and U5 3′-processing efficiency. Thus, the accumulation of +SSS DNA in the presence of 2 μg of mutant A3F increased by 2-fold, and it was 6-fold in the presence of 2 μg of wild-type A3F (Fig. 4E, lanes 6 and 7). Similarly, viral U5 3′-processing efficiency in the presence of 2 μg of mutant A3F was 34% ± 1%, and it was 50% ± 2% and 26% ± 4% in the absence of A3F and presence of 2 μg of wild-type A3F, respectively (Fig. 4E, lanes 4, 6, and 7). Overall, these data indicate that the catalytic site mutant of A3F has an intermediate phenotype with respect to the inhibition of viral DNA synthesis and 3′ processing; thus, both cytidine deaminase-dependent and -independent activities of A3F contribute to the inhibition of HIV-1 replication.
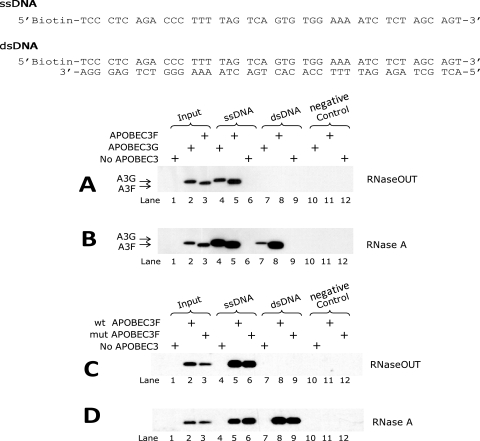
A3F and A3G display different binding affinities to dsDNA.
We investigated the possible mechanisms that contribute to A3F's inhibition of 3′ processing and integration by analyzing the affinity of A3F and A3G to single-stranded DNA (ssDNA) and double-stranded (dsDNA). We examined the association of A3F and A3G with a 42-bp biotinylated DNA fragment from the unprocessed 3′ end of HIV-1Δvif using a magnetic bead capture assay (Fig. 5A). Equal amounts of total protein from 293T cells transfected with a control plasmid, pFLAG-A3F, or pFLAG-A3G were incubated with biotinylated ssDNA or dsDNA and precipitated using streptavidin-conjugated beads. The input cell lysate and precipitated samples were analyzed by Western blotting using an anti-FLAG antibody. Since A3F and A3G form ribonucleoprotein complexes in vivo, the cell lysates were treated with either RNaseOUT (an inhibitor of RNase A) to retain the complexes (Fig. 5A and C) or RNase A to dissociate the RNA-protein complexes (Fig. 5B and D). In the presence of RNaseOUT or RNase A, we observed that the amounts of A3F and A3G proteins bound to ssDNA correlated with the input cell lysates, suggesting that their binding affinities for ssDNA are not significantly different (Fig. 5A and B, lanes 1 to 6). In contrast, neither A3F nor A3G bound to dsDNA in the presence of RNaseOUT (Fig. 5A, lanes 7 to 9). Following RNase A treatment, however, we observed that significantly more A3F was bound to dsDNA than A3G (Fig. 5B, lanes 7 to 9). This observation suggests that RNA-dissociated A3F has a higher affinity for dsDNA than does RNA-dissociated A3G. These results are consistent with the hypothesis that A3F has a higher affinity than A3G for dsDNA, and as a result, A3F binds to the viral DNA ends and inhibits the 3′-processing reaction catalyzed by IN more efficiently than A3G.
FIG. 5.
Binding affinities of A3G, A3F, and mutant A3F proteins to ssDNA and dsDNA of U5 of the 3′-LTR. The sequences of ssDNA and annealed dsDNA of the viral U5 end of the 3′LTR are indicated. Equal amounts of total cell lysates treated with RNase (A and C) or RNaseOUT (B and D) were incubated with ssDNA, dsDNA, and no oligonucleotides (as a negative control) and precipitated by streptavidin-conjugated beads. An approximately 1/20 volume of each sample was loaded on the gel to compare the input amounts of each protein. Input and immunoprecipitated samples were analyzed by Western blotting and probed with anti-FLAG antibody.
We used this assay to analyze the potential effect of the E251Q mutation in A3F on its dsDNA binding affinity (Fig. 5C and D). The results showed that the wild-type A3F and the A3F-E251Q mutant both bound to ssDNA and dsDNA with a similar efficiency. This result was consistent with the observation that the E251Q mutation had little effect on the ability of A3F to inhibit the efficiency of 3′ processing and viral DNA integration.
DISCUSSION
In these studies, we compared the mechanisms by which A3F and A3G inhibit HIV-1 replication and found significant differences. Surprisingly, we found that even though these two proteins share 49% amino acid identity, they inhibit HIV-1 integration and provirus formation through different mechanisms. A3F primarily inhibits the 3′-processing reaction catalyzed by HIV-1 IN and displays a partial dependence on cytidine deaminase activity. In contrast, A3G has a minimal effect on 3′ processing and is associated with an aberrant 6-bp extension at the viral DNA end; furthermore, A3G's cytidine deaminase activity plays a critical role in its ability to generate the 6-bp extension at the viral DNA end and to inhibit provirus formation. These studies show for the first time that despite their structural similarities, A3F and A3G inhibit HIV-1 integration using different mechanisms.
In addition to their different effects on provirus formation, A3F and A3G display other differences in their antiviral activities. First, consistently with previous studies, we observed that A3F is less potent at suppressing HIV-1 replication than A3G, given that 4-fold more A3F plasmid DNA is needed to achieve similar antiviral activity (4, 21, 48, 55, 56). We show that this is not due to differences in the packaging efficiencies of the two A3 proteins. Second, A3F displays a reduced cytidine deaminase activity but a greater cytidine deaminase-independent antiviral activity than A3G. In agreement with previous findings (14, 24, 26), A3F and A3G both reduced the accumulation of HIV-1 reverse transcription products; furthermore, A3F and A3G amounts that caused similar reductions in viral infectivity also caused similar reductions in viral DNA accumulation as determined by quantitative real-time PCR. We previously observed that the effect of A3G on viral DNA accumulation was dependent on its cytidine deaminase activity (26). Correspondingly, Southern blot analysis indicated that the inhibition of plus-strand DNA transfer and an increase in the amount of +SSS DNA by both A3F and A3G also was dependent on cytidine deaminase activity.
Although previous studies have shown that both A3F and A3G proteins partly inhibit HIV-1 replication by reducing provirus formation (23, 26), the mechanisms used by A3F and how much this effect contributes to their overall antiviral activities has not been extensively investigated. Contrary to their effects on viral DNA synthesis, our data clearly show that A3F is a more potent inhibitor of viral DNA integration, despite the overall magnitude of antiviral activity of A3F being 10-fold less than that of A3G. An A3G cytidine deamination mutant was defective in inhibiting integration and did not generate the 6-bp extension at the U5 end. In contrast, an A3F cytidine deamination mutant, A3F-E251Q, still was able to inhibit 3′ processing and integration, albeit to a lesser extent than wild-type A3F, indicating that the A3F cytidine deamination activity plays a minor role in the inhibition of viral DNA integration. In summary, the block to viral DNA integration by A3G but not A3F is almost entirely dependent on cytidine deamination activity.
We explored the cytidine deaminase-independent mechanism by which A3F inhibits 3′ processing by determining the relative affinities of A3F and A3G to single- and double-stranded nucleic acids. Our results showed that cell-derived A3F and A3G bound to single-stranded DNA with similar efficiency before and after RNase A treatment; in contrast, A3F bound to dsDNA with a higher affinity than A3G upon the RNase A treatment of the cell lysates. These results are consistent with the hypothesis that A3F binds to viral DNA ends more tightly than A3G, and as a result it blocks 3′ processing by IN more efficiently than A3G. These results also are consistent with previous reports that A3G has a similar affinity for single-stranded RNA and DNA and a much lower affinity for double-stranded DNA (16, 53). A recent study also showed that both A3F and A3G interact with IN in an RNA-dependent manner, suggesting that the interactions with IN are indirect (23).
The role of the cytidine deamination of A3G in its antiviral activity is controversial, since conflicting conclusions have been reached by different groups. Several reports show that the antiviral function of A3G is not associated with a functional cytidine deaminase activity (3, 31), but we and others have shown that functional editing activity is indispensable for A3G to inhibit viral infection (24, 26, 38, 40). Here, we again show that editing activity is absolutely required for A3G's antiviral activity; furthermore, in agreement with Holmes et al. (14), we observed that A3F antiviral activity is not dependent on its catalytic activity.
These observations suggest that the A3 proteins have developed into a versatile and multifaceted arm of the host innate immune response that defends against retroviruses as well as other viruses. Our studies support a novel concept by which two of these A3 proteins have evolved to share common antiviral pathways but use distinct cytidine deaminase-dependent and -independent mechanisms. We speculate that, through the result of virus-host interactions over evolutionary time, A3G and A3F have developed distinct but overlapping mechanisms to provide broad and robust protection against a variety of viral pathogens.
Acknowledgments
We especially thank Wei-Shau Hu for intellectual input throughout the project. We also thank Jianbo Chen and Jessica Smith for valuable discussions and critical comments during manuscript preparation.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (V.K.P. and J.L.M.) and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E (W.B.).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Anderson, J. L., and T. J. Hope. 2008. APOBEC3G restricts early HIV-1 replication in the cytoplasm of target cells. Virology 375:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beale, R. C., S. K. Petersen-Mahrt, I. N. Watt, R. S. Harris, C. Rada, and M. S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585-596. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, K. N., M. Verma, E. Y. Kim, S. M. Wolinsky, and M. H. Malim. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 7.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 8.Dang, Y., X. Wang, T. Zhou, I. A. York, and Y. H. Zheng. 2009. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J. Virol. 83:8544-8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, F., S. Cen, M. Niu, Y. Yang, R. J. Gorelick, and L. Kleiman. 2007. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J. Virol. 81:11322-11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haché, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280:10920-10924. [DOI] [PubMed] [Google Scholar]
- 12.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 13.He, Z., W. Zhang, G. Chen, R. Xu, and X. F. Yu. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381:1000-1011. [DOI] [PubMed] [Google Scholar]
- 14.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282:2587-2595. [DOI] [PubMed] [Google Scholar]
- 15.Huthoff, H., and M. H. Malim. 2005. Cytidine deamination and resistance to retroviral infection: towards a structural understanding of the APOBEC proteins. Virology 334:147-153. [DOI] [PubMed] [Google Scholar]
- 16.Iwatani, Y., H. Takeuchi, K. Strebel, and J. G. Levin. 2006. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 80:5992-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 18.Jern, P., R. A. Russell, V. K. Pathak, and J. M. Coffin. 2009. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 5:e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 282:32065-32074. [DOI] [PubMed] [Google Scholar]
- 21.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 22.Liu, B., P. T. Sarkis, K. Luo, Y. Yu, and X. F. Yu. 2005. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 79:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 81:7238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 25.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 26.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 28.Mehle, A., H. Wilson, C. Zhang, A. J. Brazier, M. McPike, E. Pery, and D. Gabuzda. 2007. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J. Virol. 81:13235-13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyagi, E., S. Opi, H. Takeuchi, M. Khan, R. Goila-Gaur, S. Kao, and K. Strebel. 2007. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 81:13346-13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navarro, F., B. Bollman, H. Chen, R. Konig, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 31.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 32.Nikolenko, G. N., E. S. Svarovskaia, K. A. Delviks, and V. K. Pathak. 2004. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J. Virol. 78:8761-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pion, M., A. Granelli-Piperno, B. Mangeat, R. Stalder, R. Correa, R. M. Steinman, and V. Piguet. 2006. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J. Exp. Med. 203:2887-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rösler, C., J. Kock, M. Kann, M. H. Malim, H. E. Blum, T. F. Baumert, and F. von Weizsacker. 2005. APOBEC-mediated interference with hepadnavirus production. Hepatology 42:301-309. [DOI] [PubMed] [Google Scholar]
- 35.Russell, R. A., M. D. Moore, W. S. Hu, and V. K. Pathak. 2009. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell, R. A., and V. K. Pathak. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasada, A., A. Takaori-Kondo, K. Shirakawa, M. Kobayashi, A. Abudu, M. Hishizawa, K. Imada, Y. Tanaka, and T. Uchiyama. 2005. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology 2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher, A. J., G. Hache, D. A. Macduff, W. L. Brown, and R. S. Harris. 2008. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 82:2652-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 40.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412-44416. [DOI] [PubMed] [Google Scholar]
- 41.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, J. L., W. Bu, R. C. Burdick, and V. K. Pathak. 2009. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol. Sci. 30:638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 44.Svarovskaia, E. S., R. Barr, X. Zhang, G. C. Pais, C. Marchand, Y. Pommier, T. R. Burke, Jr., and V. K. Pathak. 2004. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat-circle junctions. J. Virol. 78:3210-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, D. C., Y. A. Voronin, G. N. Nikolenko, J. Chen, W. S. Hu, and V. K. Pathak. 2007. Determination of the ex vivo rates of human immunodeficiency virus type 1 reverse transcription by using novel strand-specific amplification analysis. J. Virol. 81:4798-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 47.Wiegand, H. L., and B. R. Cullen. 2007. Inhibition of alpharetrovirus replication by a range of human APOBEC3 proteins. J. Virol. 81:13694-13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, H., E. Chertova, J. Chen, D. E. Ott, J. D. Roser, W. S. Hu, and V. K. Pathak. 2007. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology 360:247-256. [DOI] [PubMed] [Google Scholar]
- 50.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, B., K. Chen, C. Zhang, S. Huang, and H. Zhang. 2007. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 282:11667-11675. [DOI] [PubMed] [Google Scholar]
- 52.Yang, Y., F. Guo, S. Cen, and L. Kleiman. 2007. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology 365:92- 100. [DOI] [PubMed] [Google Scholar]
- 53.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 54.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 55.Zennou, V., and P. D. Bieniasz. 2006. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology 349:31-40. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]