Abstract
Host cellular proteases induce influenza virus entry into cells by cleaving the viral surface envelope glycoprotein hemagglutinin (HA). However, details on the cellular proteases involved in this event are not fully available. We report here that ubiquitous type II transmembrane serine proteases, MSPL and its splice variant TMPRSS13, are novel candidates for proteases processing HA proteins of highly pathogenic avian influenza (HPAI) viruses, apart from the previously identified furin and proprotein convertases 5 and 6. HAs from all HPAI virus H5 and H7 strains have one of two cleavage site motifs, the R-X-K/R-R motif with R at position P4 and the K-K/R-K/T-R motif with K at position P4. In studies of synthetic 14-residue HPAI virus HA peptides with these cleavage site motifs, furin preferentially cleaved only HA peptides with the R-K-K-R motif in the presence of calcium and not peptides with the other motif, whereas MSPL and TMPRSS13 cleaved both types of HA peptides (those with the R/K-K-K-R motif) efficiently in the absence of calcium. Full-length recombinant HPAI virus HA with the K-K-K-R cleavage motif exhibited poor susceptibility to cleavage in the absence of MSPL or TMPRSS13 and the presence of furin in infected cells, but it was converted to mature HA subunits in transfected cells expressing MSPL or TMPRSS13, with membrane-fused giant-cell formation. This conversion and membrane fusion were suppressed by inhibitors of MSPL and TMPRSS13. Furthermore, infection with and multiplication of genetically modified live HPAI virus A/Crow/Kyoto/53/2004 (H5N1) with the K-K-K-R cleavage site motif were detected only in MSPL- and TMPRSS13-expressing cells.
The limited proteolysis of hemagglutinin (HA), a surface envelope glycoprotein, is an important step in the infection process of influenza A viruses. This cleavage induces maturation of HA protein and leads to fusion between virions and host cell membranes. However, influenza A virus cannot process HA by itself, because a HA-processing protease is not encoded in its genome. Therefore, viral entry into the cells is determined by the host cellular processing proteases (18, 19, 21, 22, 29). Proteolytic activation of human influenza A virus HA occurs extracellularly by airway proteases, such as tryptase Clara (19), miniplasmin (25), and ectopic pancreatic trypsin (22, 24, 39). In addition to these secretion-type serine proteases, transmembrane serine protease 2 (TMPRSS2) and human airway trypsin-like protease (HAT) are candidates for the processing proteases in the membrane of the human airway (5). In addition, it was reported recently that TMPRSS2 and TMPRSS4 proteolytically activate the 1918 influenza virus HA (6). These trypsin-type endoproteases recognize the carboxyl moiety of a single R residue within the sequence Q/E-X-R (where X is any amino acid except C and basic amino acids). Thus, the distributions of these proteases in cells and tissues and their proteolytic potentiation of various strains of the viruses (25) may determine the virus pathogenicity.
For all highly pathogenic avian influenza (HPAI) viruses of subtypes H5 and H7 known to date, the cleavage of HA occurs at the C-terminal R residue in the consensus multibasic motifs, such as R-X-K/R-R with R at position P4 and K-K/R-K/T-R with K at P4, and leads to systemic infection. Early studies demonstrated that the ubiquitously expressed furin and proprotein convertases (PCs) are activating proteases for HPAI viruses (2, 9, 13, 35, 40). Furin and PCs are calcium-dependent subtilisin-like serine proteases that cleave the consensus multibasic motif R-X-K/R/X-R with R at position P4 (12, 14, 33, 34, 38) and, as a result, convert proproteins into biologically active proteins. However, replacement of R by K and a nonbasic amino acid at P4 significantly suppresses the processing activities of furin and PCs (1, 33, 38). These findings suggest the possible involvement of a host cellular protease(s) other than furin and PCs in the processing of HA proteins of HPAI viruses with multibasic cleavage motifs including K at position P4.
The type II transmembrane serine proteases (TTSPs) have a common structure: a short cytoplasmic domain and a transmembrane domain at the N-terminal end and a serine protease domain at the extracellular C terminus. Although expression profiles and partial enzymatic characterization of TTSPs have been reported previously (26, 36, 37), there is little information on their physiological functions and natural substrates and inhibitors. Most of the TTSPs identified so far recognize a single R at position P1, but the newly isolated MSPL and its transcript variant TMPRSS13 have unique substrate specificities, with preferential recognition of paired basic residues at the cleavage site (17, 20). Thus, MSPL and TMPRSS13, which are ubiquitously expressed (17), can activate various bioactive polypeptides with multibasic residue motifs, such as growth factors, cytokines, prohormones, and toxins, as well as fusogenic viral envelope glycoproteins, in the membrane.
The present study was designed to characterize the proteolytic processing of the HPAI virus HA. For this reason, we searched for cells that express MSPL and TMPRSS13 at levels below detection and found human cell line ECV304. We then established ECV304 cells stably expressing MSPL and TMPRSS13. Both enzymes preferentially cleaved HA peptides of HPAI virus H5 and H7 strains at multibasic cleavage sites (N′-K/R-K-K-R↓-G-C′, where N′ and C′ indicate the N- and C-terminal ends and the arrow indicates cleavage). In addition, the results showed that cellular MSPL and TMPRSS13, but not furin, potentiate multiple infections with the HPAI virus carrying the cleavage site K-K-K-R motif in a live virus infection system, suggesting that these proteases facilitate the replication of a wide range of strains of HPAI virus subtypes in various organs.
MATERIALS AND METHODS
Proteins and chemicals.
4-Methylcoumaryl-7-amide (MCA) peptide substrates were purchased from the Peptide Institute (Osaka, Japan) and Japan Lamb Co. (Hiroshima, Japan). Furin (a recombinant form of the human protein) was obtained from Alexis Biochemicals (San Diego, CA). A furin inhibitor (decanoyl-R-V-K-R-chloromethylketone [dec-R-V-K-R-cmk]) was purchased from Calbiochem (San Diego, CA). Aprotinin was from Nacalai Tesque Inc. (Kyoto, Japan). All other protease inhibitors were obtained from Sigma (St. Louis, MO).
Cell lines and culture conditions.
ECV304 cells (a human vascular endothelial cell line), A549 cells (a human lung adenocarcinoma cell line), and LoVo cells (a human colorectal adenocarcinoma cell line) were purchased from the American Type Culture Collection (ATCC). ECV304 cells were cultured at 37°C in 5% CO2 in minimum essential medium (MEM; Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS; Roche Molecular Biochemicals, Indianapolis, IN) and 50 μg/ml gentamicin (Invitrogen). A549 cells were maintained at 37°C in 5% CO2 in MEM containing 10% FBS, nonessential amino acids, and 50 μg/ml gentamicin. LoVo cells were maintained at 37°C in 5% CO2 in Ham's F-12 medium containing 10% FBS and 50 μg/ml gentamicin. Human umbilical vein endothelial cells (HUVEC) were purchased from Cambrex Bio Science Walkersville Inc. (Walkersville, MD) and maintained at 37°C in 5% CO2 in supplemented medium (endothelial growth medium 2 [EGM2]).
Virus and cells.
HPAI virus A/Crow/Kyoto/53/2004 (H5N1) was isolated from embryonated eggs inoculated with tracheal homogenates from dead crows (41). Chicken embryonic fibroblasts (CEF) were prepared from 10-day-old embryonated eggs and maintained in MEM containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. HEK293T cells (a human embryonic kidney cell line) were purchased from GenHunter Corporation (Nashville, TN) and cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Generation of recombinant virus.
Viral RNA was isolated using Trizol reagent (Invitrogen), and cDNA was synthesized using random hexamers. The full-length HA sequence of HPAI virus A/Crow/Kyoto/53/2004 (H5N1) was constructed by PCR. Then the mutant HA sequence was constructed by changing the R residue to a K residue (N′-R-K-K-R-C′ was changed to N′-K-K-K-R-C′) at the HA cleavage site by site-directed mutagenic PCR. The constructed HA genes were cloned into the pPOLI plasmid (10), yielding pPOLI-HA plasmids. Recombinant virus was generated by using a previously described reverse-genetics system (3, 7, 10) with slight modification. Briefly, HEK293T cells that had been cocultured with CEF (at a 7:3 ratio) were transfected with each constructed pPOLI-HA plasmid, together with seven other viral RNA segments (corresponding to PB2, PB1, PA, NP, neuraminidase [NA], M, and NS proteins) from the A/Crow/Kyoto/53/2004 (H5N1) genome and pCAGGS expression plasmids (3) for influenza virus A/WSN/33 (H1N1) PA, PB1, PB2, and NP. At 96 h postinfection, recombinant viruses in the supernatant were collected and inoculated into 10-day-old embryonated eggs. The HA gene of recombinant virus was checked by sequencing.
Analysis of MSPL and TMPRSS13 expression in cultured cells.
To analyze the MSPL and TMPRSS13 expression levels in cultured human cells, reverse transcription-PCR (RT-PCR) was performed as described previously (32). The following gene-specific primers were used for amplification by PCR (with 35 cycles): 5′-CGGACGGGTCAGATTAGGCCAG-3′ and 5′-GGAGCCAATGAGACAGATGTGGA-3′ for human MSPL (hMSPL); 5′-GGCAGGCCCCACTTCCCAGGT-3′ and 5′-GGGCAATCATTCCTGGATGTTCTG-3′ for human TMPRSS13 (hTMPRSS13); 5′-GACATCCTCACCGAGCCCAAAGA-3′ and 5′-GCCTTCCTCGCACACCACACA-3′ for human furin; and 5′-CAAATCAAGACAATCCGCCCTAA-3′ and 5′-GACCATTCTTTCAATTTACCTGGA-3′ for human PC5/PC6. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific primers (5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and 5′-CATGTGGGCCATGAGGTCCACCAC-3′) were used for the positive control reaction (PCR with 22 cycles). The PCR products were applied to 2% agarose-Tris-acetate-EDTA gels for electrophoresis and stained with ethidium bromide solution. The specificities of the amplified PCR products were confirmed by sequencing.
Construction and expression of recombinant MSPL/TMPRSS13.
Expression clones were constructed from hMSPL/hTMPRSS13 full-length cDNA by standard PCR amplification methods. For protein expression, the sequences encoding the extracellular regions of MSPL and TMPRSS13 (i.e., the scavenger receptor cysteine-rich [SRCR] domain and the serine protease domain) were amplified by PCR and subcloned into p3xFLAG-CMV9 at a site immediately after the region encoding the enteropeptidase recognition sequence (D-D-D-D-K). The resulting constructs were verified by sequence analysis. Transient transfection of HEK293T cells was performed with FuGENE-HD reagent according to the instructions supplied by the manufacturer (Roche Molecular Biochemicals, Indianapolis, IN), and then the recombinant proteins were induced in the serum-free culture medium (SFCM). Under the conditions employed, purified recombinant soluble hMSPL and hTMPRSS13 from SFCM exhibited proteolytic activities.
To establish a stable cell line expressing MSPL/TMPRSS13, full-length MSPL/TMPRSS13 cDNA with a FLAG tag sequence was subcloned into the pIRES-puro vector (BD Bioscience Clontech) at the NotI site and ECV304 cells were transfected with the vector and screened for resistance to puromycin.
Enzyme and inhibitor assay.
We analyzed the amidolytic activities of the enzyme preparations toward various synthetic peptides listed in Table 1. The amount of 7-amino-4-methylcoumarin liberated from the substrate was determined as described previously (32). One unit of enzyme activity was defined as the amount that degraded 1 μmol of the substrate per min. To determine the effects of inhibitors, enzyme preparations were preincubated for 5 min with various inhibitors at 37°C and the residual enzyme activity was measured. The kinetic parameter (Km) was determined from double-reciprocal plots by using different concentrations of fluorogenic peptides (10 to 100 μM).
TABLE 1.
Km values of typical synthetic peptide substrates and Ki values of typical inhibitors for recombinant soluble hMSPL and hTMPRSS13
| Substrate or inhibitor |
Km (μM) or Ki (nM) for: |
|
|---|---|---|
| hMSPL | hTMPRSS13 | |
| Substrates | ||
| Boc-Q-A-R-MCA | 982.0 | 1789.0 |
| Boc-Q-R-R-MCA | 25.0 | 13.7 |
| Boc-L-K-R-MCA | 130.0 | 230.0 |
| Boc-E-K-K-MCA | 890.0 | 910.0 |
| Boc-R-V-R-R-MCA | 10.0 | 8.6 |
| Pyr-R-T-K-R-MCA | 8.7 | 7.4 |
| Pyr-K-K-K-R-MCA | 7.5 | 6.0 |
| Pyr-R-K-K-R-MCA | 12.0 | 7.1 |
| Inhibitors | ||
| Aprotinin | 10.5 | 16.3 |
| BBI | 19.2 | 31.7 |
| Dec-R-V-K-R-cmk | 2.9 | 0.9 |
| α1-PDX | NI | NI |
Km values of typical synthetic substrates derived from monobasic and multibasic substrates for hMSPL and hTMPRSS13 were measured as described previously (31). Ki values for Boc-Q-R-R-MCA, one of the efficient substrates, were measured. Pyr, pyroglutamyl; NI, no inhibition.
Processing of peptide substrates by MSPL/TMPRSS13.
To determine the cleavage specificities of MSPL/TMPRSS13 and furin, peptides (20 μg each) were incubated with 0.25 mU of MSPL/TMPRSS13 for 1 h and furin for 8 h at 37°C. After incubation, the samples were separated by reverse-phase (RP)-high-performance liquid chromatography (HPLC) with the use of a C18 column (YMC-Pack; column size, 250 by 4.6 mm [internal diameter]; particle size, 10 μm [YMC Co., Kyoto, Japan]). The buffer system comprised an aqueous 0.1% (vol/vol) trifluoroacetic acid (TFA) solution and an organic phase of acetonitrile containing 0.1% (vol/vol) TFA. The peptides were eluted with a 0.33%/min linear gradient (15 to 25%) of 0.1% (vol/vol) TFA-acetonitrile at a flow rate adjusted to 0.8 ml/min. The elution samples were then identified both by amino acid sequence analysis with an Applied Biosystems model 492 gas-phase sequencer/140C system and by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) with an Applied Biosystems model 4700 system according to the instructions provided by the manufacturer.
SDS-PAGE and immunoblotting.
SDS-PAGE was performed with gradient gels (10 to 20% acrylamide) under reducing conditions according to the method of Laemmli (23). Immunoblot analyses were performed as described previously (32). After blocking of the membranes with 3.5% skim milk in a solution of 50 mM Tris-HCl and 150 mM NaCl, pH 7.4 (TBS), the membranes were probed with a 1:1,000 dilution of anti-H5N2 polyclonal antibody or anti-H5N1 monoclonal antibody (4C12) (8) in 3.5% skim milk in TBS by incubation for 2 h at room temperature. Then the membranes were incubated for 1 h at room temperature with a 1:2,000 dilution of peroxidase-labeled anti-rabbit IgG or a 1:4,000 dilution of peroxidase-labeled anti-mouse IgG in 3.5% skim milk in TBS. Immunoreactive proteins were visualized using the ECL detection system (Amersham Pharmacia Biotech, Uppsala, Sweden).
Processing of recombinant HA in MSPL/TMPRSS13-expressing stable transfectant cells.
Simian virus 40 (SV40) vectors encoding HPAI virus HA proteins (A/chicken/Penn/1370/83 H5 HA [16] and A/FPV/Rostock/34 H7 HA), prepared as described previously (28), were used to infect wild-type (WT) ECV304 cells or MSPL/TMPRSS13-expressing stable transfectant cells. After 48 h of infection, the cells were rinsed with phosphate-buffered saline (PBS), placed into SFCM with or without inhibitors of MSPL/TMPRSS13, and cultured for another 24 h. Cells were lysed and then applied to gels for SDS-PAGE. The HA-processing activities were analyzed by immunoblotting.
Analysis of giant-cell formation.
A fusion assay was performed as described previously (30). Briefly, the SV40 vectors encoding HA proteins (A/chicken/Penn/1370/83 H5 HA and A/FPV/Rostock/34 H7 HA) were used to infect ECV304 cells or MSPL/TMPRSS13-expressing stable transfectant cells. After 24 h of infection, the medium was replaced with SFCM and the cells were further incubated for 24 h at 37°C. Then the cells were rinsed with PBS and incubated with the reaction buffer (DMEM, pH 5.0) for 5 min at 37°C. After low-pH treatment, the cells were further incubated with the growth medium for 8 h at 37°C. The cells were then fixed and stained with Giemsa solution. Giant cells that contained more than three nuclei were regarded as fused cells. Fusion activities were monitored by counting the fused cells in an entire field.
Viral infection.
Briefly, WT ECV304 cells or MSPL-expressing stable transfectant cells were infected with HPAI virus A/Crow/Kyoto/53/2004 (H5N1) (HA motif, N′-R-K-K-R-C′) and the corresponding genetically modified recombinant virus (HA motif, N′-K-K-K-R-C′) at a multiplicity of infection (MOI) of 1. After 1 h of infection, the cells were rinsed with PBS, placed into SFCM with or without inhibitors of MSPL, and cultured for another 24 h. Newly prepared ECV304 WT cells and MSPL-expressing stable transfectant cells were infected with the conditioned SFCM. After 24 h of infection, both cell types were analyzed by an immunofluorescence assay. The cells were fixed with 4% paraformaldehyde in PBS containing 0.1% Triton X-100. To detect viral antigen, a polyclonal antibody obtained from a rabbit immunized with A/Duck/Hong Kong/342/78 (H5N2) (7) was used as the primary antibody (diluted 1:2,000 in PBS containing 1% bovine serum albumin). Polyclonal antibodies binding to the viral protein were detected with an Alexa Fluor 488-conjugated secondary antibody (diluted 1:500 in PBS containing 1% bovine serum albumin). Cell lysates prepared from primary infected cells were analyzed by SDS-PAGE. The HA molecule-processing activities were analyzed by immunoblotting.
Statistical analysis.
Values were expressed as means ± standard deviations (SD). Differences between groups were examined for statistical significance using Student's t test, while differences among several groups were examined by one-way analysis of variance (ANOVA). A P value of less than 0.05 was considered statistically significant.
RESULTS
Characterization of recombinant MSPL/TMPRSS13.
To clarify the enzymatic features of hMSPL and hTMPRSS13 with respect to the multibasic substrates, we analyzed the kinetics of purified soluble recombinant proteases free of the transmembrane domain and the N-terminal cytoplasmic region. These enzymes had a pH optimum of 7.0 to 8.8 and no calcium requirement. Among the synthetic substrates tested (Table 1), those with paired basic residues at the cleavage sites, particularly those with R at position P1, were hydrolyzed favorably by hMSPL, with Km values significantly lower than the value for a monobasic substrate, t-butyloxycarbonyl (Boc)-Q-A-R-MCA (Km = 982.0 μM), although furin does not cleave dibasic peptides (2, 12). In addition, hMSPL preferentially cleaved substrates with cleavage site sequences mimicking the amino acid sequences of HPAI virus H5 and H7 HAs, even peptides containing the K-K-K-R motif, which have poor susceptibility to furin. It is noteworthy that the basic residue at position P4 in the multibasic residue motifs greatly enhanced the efficiency of processing, corresponding to the lowest Km value range. A substrate with K at positions P1 and P2 was hydrolyzed to a lesser extent by hMSPL, with the Km value being almost equivalent to the value for a monobasic substrate. Similar results were obtained with hTMPRSS13. Among the inhibitors tested, dec-R-V-K-R-cmk, also known as a furin inhibitor (35), markedly inhibited the enzymatic activities, with inhibitor constant (Ki) values of 2.9 nM for hMSPL and 0.9 nM for hTMPRSS13. However, another potent inhibitor of furin, α1-antitrypsin Portland (α1-PDX) (38), did not inhibit these enzymes. Trypsin inhibitors, such as aprotinin and Bowman-Birk trypsin inhibitor (BBI), inhibited the activities of these enzymes with low Ki values.
Cleavage of HA peptides by MSPL/TMPRSS13.
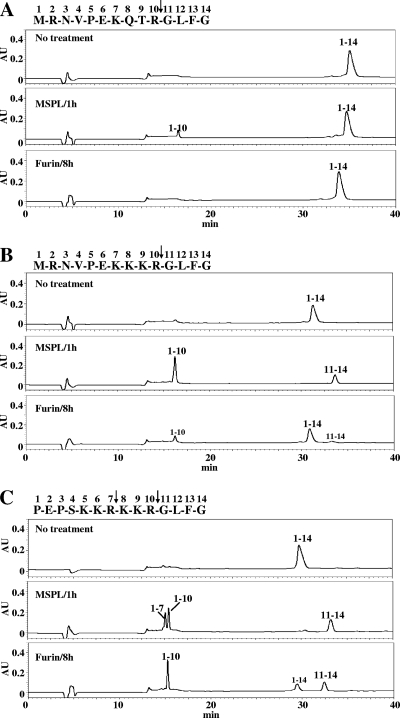
We next analyzed the cleavability of 14-residue synthetic peptides derived from HA cleavage sites of HPAI virus strains, such as A/chicken/Penn/1370/83 (H5N2) (16) and A/FPV/Rostock/34 (H7N1) (9), and weakly pathogenic influenza virus strain A/Aich/2/58 (H3N2). After incubation with hMSPL or human furin, the digested samples were separated by RP-HPLC and peptide fragments were characterized by mass spectrometry and protein sequencing. In contrast to the H3 HA peptide with a single R at the cleavage site, which corresponded to low cleavage efficiencies (Fig. 1A), both the H5 HA peptide with the K-K-K-R motif (Fig. 1B) and the H7 HA peptide with the R-K-K-R motif (Fig. 1C) were fully processed at the correct positions by MSPL within 1 h. In the case of H7 HA peptides with multiple basic residues, MSPL cleaved two R residues on the carboxyl-terminal side in the cleavage site sequence N′-K-K-R↓-K-K-R↓-G-C′ while furin cleaved R only at a single site in a sequence with R at position P4, N′-K-K-R-K-K-R↓-G-C′, in the presence of 1 mM CaCl2. These cleavage site specificities of furin were consistent with that reported previously for the H5 HA peptide of HPAI virus A/Hong Kong/156/97 (H5N1) with the R-K-K-R motif (2). However, the H5 HA peptide with K at position P4 (Fig. 1B) was hardly cleaved by furin under the same experimental conditions. TMPRSS13 showed similar results (data not shown). These findings suggest that MSPL and TMPRSS13 cover diverse cleavage specificities, including specificities for cleavage sites not susceptible to furin.
FIG. 1.
Processing of synthetic HA peptides with a single basic R in the cleavage motif (A) and with multibasic cleavage motifs (B and C) by MSPL and furin. Synthetic 14-residue peptides (20 μg each) with the sequences indicated at the tops of the panels were incubated with or without MSPL for 1 h at 37°C or furin for 8 h at 37°C. After incubation, the samples were separated by RP-HPLC with the use of a C18 column. The intensities of the separated peaks monitored by absorbance at 215 nm (expressed in arbitrary units [AU]) were recorded, and each peak was characterized by MALDI-TOF MS and protein sequencer analysis. Peaks in panel C with retention times of 15.2 and 15.5 min were found to correspond to N-terminal residues 1 to 7 and N-terminal residues 1 to 10, respectively. The peptides were derived from A/Aichi/2/68 (H3N2) (A), A/chicken/Penn/1370/83 (H5N2) (B), and A/FPV/Rostock/34 (H7N1) (C).
Proteolytic activation of HPAI virus recombinant HAs in MSPL/TMPRSS13-expressing cells.
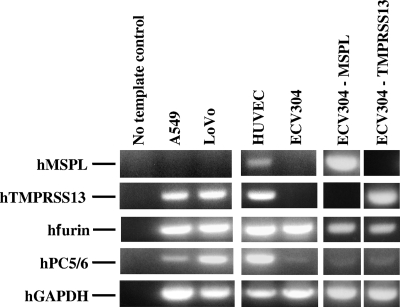
Before the start of the ex vivo experiments, RT-PCR was carried out to analyze the MSPL and TMPRSS13 mRNA expression levels in various human cells in culture (Fig. 2). Among the cell lines tested, ECV304 cells had MSPL and TMPRSS13 mRNA expression levels that were below detection levels, despite the ubiquitous expression of furin in these cells. Therefore, we established stable transfectant ECV304 cells expressing full-length hMSPL and hTMPRSS13 (ECV304-MSPL and ECV304-TMPRSS13 cells, respectively). To confirm the role of MSPL/TMPRSS13 in the activation of HPAI virus infectivity, we analyzed the cleavage of full-length HPAI virus HA with the K-K-K-R motif, which has poor susceptibility to furin, and subsequent cell fusion activity. ECV304-MSPL, ECV304-TMPRSS13, and ECV304 WT cells were infected with the A/chicken/Penn/1370/83 (H5N2) HA-expressing recombinant SV40 vector for 48 h, and the HA cleavage efficiencies and cell fusion activities, monitored by evaluating giant-cell formation, were analyzed. Although the recombinant HA precursor (HA0) with the K-K-K-R cleavage motif was poorly converted to the HA1 subunit in ECV304 WT cells, conversion of HA0 to the HA1 subunit was observed in ECV304-MSPL cells (Fig. 3). Similar results were obtained with ECV304-TMPRSS13 cells. In contrast, significant cleavage of furin-susceptible HPAI virus H7 HA0 with the R-K-K-R motif was found in both WT cells and MSPL/TMPRSS13-expressing transfectant cells, with almost similar efficiencies, after infection with the A/FPV/Rostock/34 H7 HA-expressing recombinant SV40 vector (27). After proteolytic activation by MSPL, the HPAI virus A/chicken/Penn/1370/83 (H5N2) recombinant HA protein with the K-K-K-R motif facilitated cell fusion (Fig. 3B). Cell fusion after infection with the A/FPV/Rostock/34 H7 HA-expressing recombinant SV40 vector was also detected among ECV304 WT and ECV304-MSPL cells with almost similar efficiencies. To further analyze the relevance of the proteases, inhibitors of MSPL/TMPRSS13 were applied. As shown in Fig. 4, MSPL/TMPRSS13 inhibitors, such as aprotinin, BBI, and substrate-mimicking inhibitor dec-R-V-K-R-cmk with R at position P4 (11), strongly suppressed HA cleavage and membrane-fused giant-cell formation. These results suggest that MSPL and TMPRSS13 are novel proteolytic enzymes for HAs of diverse HPAI viruses, not only those with the K-K-K-R motif but also those with the R-K-K-R motif.
FIG. 2.
The RT-PCR approach was used to evaluate the expression of hMSPL, hTMPRSS13, human furin (hfurin), and human PC5/PC6 (hPC5/6) mRNAs in the indicated human cell lines. Human cDNAs synthesized from total RNA (1 μg each) were used as templates. The level of expression of human GAPDH (hGAPDH) mRNA in each sample was used as an internal control. The results shown are representative of findings from three experiments with similar results.
FIG. 3.
Cleavage of HPAI virus A/chicken/Penn/1370/83 (H5N2) recombinant HA with the K-K-K-R motif, which is weakly susceptible to furin, by MSPL and subsequent fusion activity. (A) ECV304-MSPL and ECV304 WT cells were infected with the A/chicken/Penn/1370/83 (H5N2) HA-expressing recombinant SV40 vector for 48 h, and cell lysates from two separate experiments were subjected to SDS-PAGE and analyzed by Western immunoblotting with anti-H5N2 antibodies, the antibody epitopes being in the HA1 subunit. Molecular mass markers are on the right. Western blots were scanned, and the HA0/HA1 ratio for each lane was calculated and is shown at the bottom. (B) A fusion test was performed as described previously (30), and the results for H5 HA (top) and H7 HA (bottom) are shown. Giant cells containing more than three nuclei were regarded as fused cells. Arrows indicate the fused cells. Bars, 50 μm. Fusion activities presented in each graph were determined by counting the fused cells in an entire field. Values are means ± SD. **, P < 0.05.
FIG. 4.
Effects of inhibitors of MSPL on the cleavage and fusion activities of HPAI virus A/chicken/Penn/1370/83 (H5N2) recombinant HA. (A) ECV304-MSPL and ECV304 WT cells were infected with the A/chicken/Penn/1370/83 (H5N2) HA-expressing recombinant SV40 vector for 48 h with or without protease inhibitors (aprotinin, BBI, and dec-R-V-K-R-cmk), and cell lysates from two separate experiments were subjected to SDS-PAGE and analyzed by Western immunoblotting with anti-H5N2 antibodies, the antibody epitopes being in the HA1 subunit. Molecular mass markers are on the right. (B) A fusion test was performed as described previously (30). Fusion activities were estimated by counting the fused cells in an entire field. Values are means ± SD. *, P < 0.01; **, P < 0.05.
Role of MSPL/TMPRSS13 in live HPAI virus infection and multiplication.
To confirm the involvement of MSPL/TMPRSS13 in HPAI virus HA cleavage and virus infection, ECV304 WT and ECV304-MSPL cells were infected with the HPAI virus A/Crow/Kyoto/53/2004 (H5N1) (HA cleavage motif, N′-R-R-K-K-R↓G-C′) (41) and a genetically modified mutant recombinant virus (HA cleavage motif, N′-R-K-K-K-R↓G-C′) (Fig. 5). Although processing of HA from the mutant recombinant virus (with the motif N′-K-K-K-R-C′) in ECV304 WT cells was hardly detected, conversion of HA0 to the mature form was detected in ECV-MSPL cells. In addition, this conversion was suppressed by BBI, a high-molecular-mass inhibitor of MSPL/TMPRSS13 that cannot permeate cell membranes. To test for the generation of infective virus, the conditioned media from 1-day cultures of ECV304 WT and ECV304-MSPL cells infected with WT and mutant HPAI H5N1 viruses were inoculated into newly prepared cells and the cells were cultured for 24 h. Although infection with WT virus carrying the HA cleavage motif R-K-K-R spread from the conditioned media of both ECV304 WT and ECV304-MSPL cells, infection with mutant virus carrying the HA cleavage motif K-K-K-R spread only from the conditioned medium of ECV304-MSPL cells. These results strongly suggest that the expression of MSPL, but not furin, potentiates multicycle replication of HPAI virus with the K-K-K-R HA cleavage motif.
FIG. 5.
Cleavage of HA proteins from WT HPAI virus A/Crow/Kyoto/53/2004 (H5N1) and the corresponding mutant virus by MSPL and multiple infections with the viruses. (A) ECV304-MSPL and ECV304 WT cells were infected with HPAI virus A/Crow/Kyoto/53/2004 (H5N1) (H5N1-WT; carrying the HA motif R-K-K-R) and genetically modified recombinant virus (H5N1-Mut; carrying the HA motif K-K-K-R) for 24 h, and cell lysates were subjected to SDS-PAGE and analyzed by Western immunoblotting with anti-H5N1 monoclonal antibody (4C12). Molecular mass markers are on the right. (B) An immunofluorescence assay was performed as described previously (8). The cells were fixed with 4% paraformaldehyde in PBS containing 0.1% Triton X-100. Polyclonal antibodies binding to the viral protein were detected with an Alexa Fluor 488-conjugated secondary antibody (diluted 1:500). Bars, 50 μm.
DISCUSSION
Seasonal human influenza A virus HAs have a consensus monobasic cleavage site sequence, N′-Q/E-X-R↓-G-C′, and all HPAI virus HAs have one of two types of cleavage site sequences with multiple basic amino acids, N′-R-K/R-K/R/X-R↓-G-C′ with R at position P4, present in a large number of HPAI viruses, and N′-K-K/R-K/T-R↓-G-C′ with K at position P4, present in a small number of HPAI viruses. HAs of HPAI viruses have been postulated to be cleaved by ubiquitously expressed subtilisin-like serine proteases, such as furin and PC5/PC6, with infectious viruses generated by the production of fusogenic HAs. However, the cleavage preferences of furin and PC5/PC6 are R at P1 and P4 positions in the cleavage site motif R-X-(R/K/X)-R, and replacement of P4 R by K significantly suppresses their processing activities. Figure 1 shows that furin efficiently cleaved a synthetic HPAI virus A/Hong Kong/156/97 (H5N1) HA cleavage site peptide with the R-K-K-R motif but hardly cleaved the HPAI virus A/chicken/Penn/1370/83 HA cleavage site peptide with the K-K-K-R motif. Furthermore, cleavage of the full-length HA of HPAI virus with the R-K-K-R motif was detected but cleavage of HPAI virus HA with the K-K-K-R motif was hardly detected in ECV304 WT cells (Fig. 3 and 5). These substrate specificities of furin for HA cleavage add support to the previously reported results (1, 33, 38) and suggest that proteases other than furin and PC5/PC6 play a role in the processing of HAs of HPAI virus with the K-K/R-K/T-R cleavage motif.
In the present study, we proposed TTSPs MSPL and TMPRSS13 as novel candidate HA-processing proteases for diverse HPAI subtype viruses with both K-K-K-R and R-K-K-R cleavage motifs. In contrast to furin, which is localized in the trans-Golgi network, MSPL and TMPRSS13 are localized in the plasma membrane, with optimum pHs in the neutral and weakly alkaline range and no calcium requirement (17). These enzymes show unique cleavage site specificities for double basic residues at the cleavage site, and the presence of R or K at position P4 greatly enhanced the efficiency of cleavage (Table 1); none of the other TTSPs have shown similar substrate specificities so far. The diverse cleavage specificities of MSPL for the HA peptides from HPAI virus A/chicken/Penn/1370/83 (H5N2), with the K-K-K-R motif, and HPAI virus A/FPV/Rostock/34 (H7N1), with the R-K-K-R motif, support the results (Fig. 1). Furthermore, processing of the full-length recombinant HA of HPAI virus A/chicken/Penn/1370/83 (H5N2) and generation of fusogenic HA products by MSPL and TMPRSS13 were confirmed in ECV304-MSPL cells but not in ECV304 WT cells. Multicycle replication of live virus, along with HA processing, was also noted for the genetically modified mutant recombinant live HPAI virus A/Crow/Kyoto/53/2004 (H5N1) with the K-K-K-R cleavage motif in ECV304-MSPL cells (Fig. 5). In analyses of HA processing, two processed HA1 bands were observed in ECV304-MSPL cell samples whereas only one HA1 band, from the WT virus with the R-K-K-R motif, was detected in ECV304 WT cell samples (Fig. 5); the WT band may reflect processing by furin or PC5/PC6. These results were supported by the data for two peptides cleaved by MSPL, depicted in Fig. 1C. These findings suggest that MSPL has diverse cleavage specificities and may cleave HA at at least two sites, although multiplicity of the mutant HPAI virus was observed under the experimental conditions. These results also suggest that MSPL and TMPRSS13 in the membrane may potently activate the membrane fusion activity of HPAI virus HAs and promote virus spread.
HPAI viruses replicate in various organs in birds, and the HA-processing proteases may be widely distributed in these organs. Indeed, TMPRSS13 and MSPL are ubiquitously expressed in almost all bird and human organs tested and are highly expressed in leukocytes and lung, pancreas, spleen, and placenta tissues (17). In addition, MSPL and TMPRSS13 are strictly localized in the plasma membranes, suggesting that proteolytic activation of HPAI virus HA occurs not only in the trans-Golgi network by furin and PC5/PC6 but also on the cell surface by MSPL and TMPRSS13.
Specific inhibitors of host cellular processing proteases would be useful therapeutic tools that should prevent the frequent appearance of drug-resistant viruses. Stieneke-Gröber et al. (35) reported that the peptide substrate analogue furin inhibitor dec-R-V-K-R-cmk blocked the replication of H7 HPAI virus in cell cultures. In addition, the use of the inhibitor has been considered for the treatment of HIV and human cytomegalovirus infections (11, 15). However, dec-R-V-K-R-cmk is not specific for furin and PCs and is also known to inhibit the enzyme activities of MSPL/TMPRSS13 and other trypsin-type proteases such as plasmin (32), and thus, it may have broad effects by inhibiting the production of bioactive polypeptides. In contrast to the low-molecular-mass peptide-based inhibitors, nonpermeating protein-based inhibitors, such as aprotinin and human mucosal protease inhibitor, suppressed influenza A virus replication in human airway epithelial cells (42) and in an animal model (4) due to the inhibition of HA cleavage. Although aprotinin and BBI are antigenic by themselves, a natural inhibitor(s) of MSPL and TMPRSS13 from humans may be useful for the inhibition of HPAI virus infection. Screening studies for natural inhibitors of MSPL and TMPRSS13 in human organs, as well as analytical studies of the crystal structures of these enzymes, are currently in progress.
Acknowledgments
This study was supported in part by a grant-in-aid from the Ministry of Education, Science and Culture of Japan (21249061) and the Special Coordination Funds for Promoting Science and Technology of MEXT of Japan.
Footnotes
Published ahead of print on 10 March 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Barr, P. J. 1991. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell 66:1-3. [DOI] [PubMed] [Google Scholar]
- 2.Basak, A., M. Zhong, J. S. Munzer, M. Chrétien, and N. G. Seidah. 2001. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 353:537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. García-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. U. S. A. 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beppu, Y., Y. Imamura, M. Tashiro, T. Towatari, H. Ariga, and H. Kido. 1997. Human mucus protease inhibitor in airway fluids is a potential defensive compound against infection with influenza A and Sendai viruses. J. Biochem. 121:309-316. [DOI] [PubMed] [Google Scholar]
- 5.Böttcher, E., T. Matrosovich, M. Beyerle, H. D. Klenk, W. Garten, and M. Matrosovich. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaipan, C., D. Kobasa, S. Bertram, I. Glowacka, I. Steffen, T. S. Tsegaye, M. Takeda, T. H. Bugge, S. Kim, Y. Park, A. Marzi, and S. Pöhlmann. 2009. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 83:3200-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daidoji, T., T. Koma, A. Du, C. S. Yang, M. Ueda, K. Ikuta, and T. Nakaya. 2008. H5N1 avian influenza virus induces apoptotic cell death in mammalian airway epithelial cells. J. Virol. 82:11294-11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du, A., T. Daidoji, T. Koma, M. S. Ibrahim, S. Nakamura, U. C. de Silva, M. Ueda, C. S. Yang, T. Yasunaga, K. Ikuta, and T. Nakaya. 2009. Detection of circulating Asian H5N1 viruses by a newly established monoclonal antibody. Biochem. Biophys. Res. Commun. 378:197-202. [DOI] [PubMed] [Google Scholar]
- 9.Feldmann, A., M. K. Schäfer, W. Garten, and H. D. Klenk. 2000. Targeted infection of endothelial cells by avian influenza virus A/FPV/Rostock/34 (H7N1) in chicken embryos. J. Virol. 74:8018-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallenberger, S., V. Bosch, H. Angliker, E. Shaw, H. D. Klenk, and W. Garten. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358-361. [DOI] [PubMed] [Google Scholar]
- 12.Hatsuzawa, K., M. Nagahama, S. Takahashi, K. Takada, K. Murakami, and K. Nakayama. 1992. Purification and characterization of furin, a Kex2-like processing endoprotease, produced in Chinese hamster ovary cells. J. Biol. Chem. 267:16094-16099. [PubMed] [Google Scholar]
- 13.Horimoto, T., K. Nakayama, S. P. Smeekens, and Y. Kawaoka. 1994. Proprotein-processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J. Virol. 68:6074-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosaka, M., M. Nagahama, W. S. Kim, T. Watanabe, K. Hatsuzawa, J. Ikemizu, K. Murakami, and K. Nakayama. 1991. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J. Biol. Chem. 266:12127-12130. [PubMed] [Google Scholar]
- 15.Jean, F., L. Thomas, S. S. Molloy, G. Liu, M. A. Jarvis, J. A. Nelson, and G. Thomas. 2000. A protein-based therapeutic for human cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A. 97:2864-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaoka, Y., C. W. Naeve, and R. G. Webster. 1984. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology 139:303-316. [DOI] [PubMed] [Google Scholar]
- 17.Kido, H., and Y. Okumura. 2008. MSPL/TMPRSS13. Front. Biosci. 13:754-758. [DOI] [PubMed] [Google Scholar]
- 18.Kido, H., M. Murakami, K. Oba, Y. Chen, and T. Towatari. 1999. Cellular proteinases trigger the infectivity of influenza A and Sendai viruses. Mol. Cells 9:235-244. [PubMed] [Google Scholar]
- 19.Kido, H., Y. Yokogoshi, K. Sakai, M. Tashiro, Y. Kishino, A. Fukutomi, and N. Katunuma. 1992. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J. Biol. Chem. 267:13573-13579. [PubMed] [Google Scholar]
- 20.Kim, D. R., S. Sharmin, M. Inoue, and H. Kido. 2001. Cloning and expression of novel mosaic serine proteases with and without a transmembrane domain from human lung. Biochim. Biophys. Acta 1518:204-209. [DOI] [PubMed] [Google Scholar]
- 21.Klenk, H. D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 22.Klenk, H. D., R. Rott, M. Orlich, and J. Blödorn. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426-439. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Le, T. Q., M. Kawachi, H. Yamada, M. Shiota, Y. Okumura, and H. Kido. 2006. Identification of trypsin I as a candidate for influenza A virus and Sendai virus envelope glycoprotein processing protease in rat brain. Biol. Chem. 387:467-475. [DOI] [PubMed] [Google Scholar]
- 25.Murakami, M., T. Towatari, M. Ohuchi, M. Shiota, M. Akao, Y. Okumura, M. A. Parry, and H. Kido. 2001. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur. J. Biochem. 268:2847-2855. [DOI] [PubMed] [Google Scholar]
- 26.Netzel-Arnett, S., J. D. Hooper, R. Szabo, E. L. Madison, J. P. Quigley, T. H. Bugge, and T. M. Antalis. 2003. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 22:237-258. [DOI] [PubMed] [Google Scholar]
- 27.Ohuchi, M., A. Cramer, M. Vey, R. Ohuchi, W. Garten, and H. D. Klenk. 1994. Rescue of vector-expressed fowl plague virus hemagglutinin in biologically active form by acidotropic agents and coexpressed M2 protein. J. Virol. 68:920-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohuchi, M., M. Orlich, R. Ohuchi, B. E. Simpson, W. Garten, H. D. Klenk, and R. Rott. 1989. Mutations at the cleavage site of the hemagglutinin alter the pathogenicity of influenza virus A/chick/Penn/83 (H5N2). Virology 168:274-280. [DOI] [PubMed] [Google Scholar]
- 29.Ohuchi, R., M. Ohuchi, W. Garten, and H. D. Klenk. 1991. Human influenza virus hemagglutinin with high sensitivity to proteolytic activation. J. Virol. 65:3530-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohuchi, R., M. Ohuchi, W. Garten, and H. D. Klenk. 1997. Oligosaccharides in the stem region maintain the influenza virus hemagglutinin in the metastable form required for fusion activity. J. Virol. 71:3719-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okumura, Y., M. Hayama, E. Takahashi, M. Fujiuchi, A. Shimabukuro, M. Yano, and H. Kido. 2006. Serase-1B, a new splice variant of polyserase-1/TMPRSS9, activates urokinase-type plasminogen activator and the proteolytic activation is negatively regulated by glycosaminoglycans. Biochem. J. 400:551-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okumura, Y., H. Sato, M. Seiki, and H. Kido. 1997. Proteolytic activation of the precursor of membrane type 1 matrix metalloproteinase by human plasmin. A possible cell surface activator. FEBS Lett. 402:181-184. [DOI] [PubMed] [Google Scholar]
- 33.Remacle, A. G., S. A. Shiryaev, E. S. Oh, P. Cieplak, A. Srinivasan, G. Wei, R. C. Liddington, B. I. Ratnikov, A. Parent, R. Desjardins, R. Day, J. W. Smith, M. Lebl, and A. Y. Strongin. 2008. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J. Biol. Chem. 283:20897-20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiner, D. F. 1998. The proprotein convertases. Curr. Opin. Chem. Biol. 2:31-39. [DOI] [PubMed] [Google Scholar]
- 35.Stieneke-Gröber, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H. D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szabo, R., and T. H. Bugge. 2008. Type II transmembrane serine proteases in development and disease. Int. J. Biochem. Cell Biol. 40:1297-1316. [DOI] [PubMed] [Google Scholar]
- 37.Szabo, R., Q. Wu, R. B. Dickson, S. Netzel-Arnett, T. M. Antalis, and T. H. Bugge. 2003. Type II transmembrane serine proteases. Thromb. Haemost. 90:185-193. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3:753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towatari, T., M. Ide, K. Ohba, Y. Chiba, M. Murakami, M. Shiota, M. Kawachi, H. Yamada, and H. Kido. 2002. Identification of ectopic anionic trypsin I in rat lungs potentiating pneumotropic virus infectivity and increased enzyme level after virus infection. Eur. J. Biochem. 269:2613-2621. [DOI] [PubMed] [Google Scholar]
- 40.Walker, J. A., S. S. Molloy, G. Thomas, T. Sakaguchi, T. Yoshida, T. M. Chambers, and Y. Kawaoka. 1994. Sequence specificity of furin, a proprotein-processing endoprotease, for the hemagglutinin of a virulent avian influenza virus. J. Virol. 68:1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.WHO/OIE/FAO H5N1 Evolution Working Group. 2008. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhirnov, O. P., M. R. Ikizler, and P. F. Wright. 2002. Cleavage of influenza A virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases. J. Virol. 76:8682-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]