Abstract
The replication protein NS1 is essential for genome replication and protein production in parvoviral infection. Many of its functions, including recognition and site-specific nicking of the viral genome, helicase activity, and transactivation of the viral capsid promoter, are dependent on ATP. An ATP-binding pocket resides in the middle of the modular NS1 protein in a superfamily 3 helicase domain. Here we have identified key ATP-binding amino acid residues in canine parvovirus (CPV) NS1 protein and mutated amino acids from the conserved A motif (K406), B motif (E444 and E445), and positively charged region (R508 and R510). All mutations prevented the formation of infectious viruses. When provided in trans, all except the R508A mutation reduced infectivity in a dominant-negative manner, possibly by hindering genome replication. These results suggest that the conserved R510 residue, but not R508, is the arginine finger sensory element of CPV NS1. Moreover, fluorescence recovery after photobleaching (FRAP), complemented by computer simulations, was used to assess the binding properties of mutated fluorescent fusion proteins. These experiments identified ATP-dependent and -independent binding modes for NS1 in living cells. Only the K406M mutant had a single binding site, which was concluded to indicate ATP-independent binding. Furthermore, our data suggest that DNA binding of NS1 is dependent on its ability to both bind and hydrolyze ATP.
Canine parvovirus (CPV), an autonomous parvovirus, has two transcriptional units in its ∼5.3-kb single-stranded DNA genome (51). The right-side unit produces VP1 and VP2 proteins, from which the ∼26-nm icosahedral capsid is formed (70). The nonstructural proteins NS1 and NS2 are expressed from the left-side transcriptional unit. NS1 is a 76.7-kDa multifunctional nuclear phosphoprotein and is the only essential nonstructural protein in CPV (66). Many of its functions are necessary for parvoviral replication. NS1 initiates genomic replication by binding site specifically to the viral right-hand origin, together with endogenous high-mobility-group proteins (18), and to the left-hand origin, along with glucocorticoid modulatory element-binding proteins (11). Recognition at both origins leads to strand- and site-specific nicking of viral DNA (12, 45). These processes require ATP for tight binding and subsequent nicking (12, 13, 18). NS1 remains covalently linked to the 5′ end of nicked DNA, and the remaining 3′-hydroxyl group can be used for synthesis of the nascent strand (17, 45). Parvoviral replication is most likely mediated by polymerase δ, in a process that depends on the sliding clamp protein proliferating cell nuclear antigen, the single strand-binding protein replication protein A, and NS1 (15). In this process, NS1 is needed as an ATP-powered helicase to resolve terminal hairpin structures of the viral genome (68). Transcription from a viral capsid promoter, P38, is also enhanced by the NS1 protein, in an ATP-dependent manner (10, 41, 52). In addition to these functions, NS1 is responsible for the cytopathic effect of parvoviruses (2, 14) and has been proposed to assist in packing DNA into newly formed capsids (8, 19).
Parvovirus NS1 proteins belong to the superfamily III (SF3) helicases, as do many other viral helicases (26, 27). All characterized members of this family travel along DNA in a 3′-to-5′ direction (58). There are four conserved sequence motifs in SF3 helicases (A, B, B′, and C), from which the nucleoside triphosphate (NTP)-binding pocket, the metal ion coordination site, the DNA-binding site, and the sensory element are formed. These motifs reside in a stretch of approximately 100 amino acid residues in the middle of NS1 (27, 69). SF3 helicases surround DNA as a ring of six or eight subunits, and the ATP-binding pocket lies between adjacent subunits (30, 42). The first subunit provides the A and B motifs, and the arginine residue of the second subunit functions as a trans-acting “arginine finger” sensor for ATP binding and hydrolysis status (56, 58). Structures of representative SF3 helicases from simian virus 40 (39), bovine papillomavirus (23), and adeno-associated virus (34) support this model. At the sequence level, the arginine finger lies after the C motif, and at the structural level, it is often embedded in a cluster of positively charged amino acids (25, 29, 58). In a ring configuration, this area interacts with the ATP-binding pocket of the neighboring subunit.
In this study, we mutated conserved amino acids from the A and B motifs to clarify the role of ATP binding and hydrolysis in the intranuclear dynamics of NS1. In addition, two conserved arginine residues, one of which is most likely the arginine finger, were mutated. These mutants were expressed as fluorescent fusion proteins, and their dynamics were studied in living cells by use of fluorescent recovery after photobleaching (FRAP). The resulting data were reconstructed in a virtual cell modeling environment to determine the binding properties of individual mutants. Moreover, the effects of these mutations on replication efficacy, P38 promoter transactivation, ATP binding, and progression of CPV infection were measured.
MATERIALS AND METHODS
Comparative modeling.
Structures for comparative modeling were retrieved from the Protein Data Bank (PDB) (7) database, using a protein BLAST (3) search with the CPV NS1 sequence (51). Alignment of NS1 and adeno-associated virus 2 (AAV2) REP40 (34) sequences and visual inspection of models were done in Bodil (38). A comparative model of CPV NS1 was built with the Modeler 9v1 program (55), using an alignment of amino acid residues 277 to 490 of REP40 (PDB accession no. 1S9H and 1U0J) (33, 34) and residues 338 to 556 of NS1 proteins. The simian virus 40 (SV40) LTag structure (PDB accession no. 1N25) (39) and the protease-chaperone complex from Haemophilus influenzae (chains A to F; PDB accession no. 1G3I [62]) were used to model the NS1 hexameric ring by superimposing structures in Bodil. The ATP molecule from the bacterial ABC transporter protein (PDB accession no. 1L2T [60]) was added to hexameric models to clarify ATP positioning at the proposed binding pocket. Images of protein structures were produced with the PyMOL program (21), and images of protein alignment were produced with the Alscript program (5).
Mutagenesis, fusion proteins, and plasmids.
First, the CPV NS1 gene was PCR amplified from the infectious plasmid clone CPV-d (47) and cloned into the pEYFP-N3 plasmid (Clontech) between BglII and BamHI restriction enzyme sites. This construct was named NS1-EYFP and has the enhanced yellow fluorescent protein (EYFP) gene fused to the 3′ terminus of the NS1 gene. Next, conserved mutations of the TATA box area of the NS1 P38 promoter (1718-tataaat-1724 to GaTcAAc) and methionine-to-threonine mutation of the starting codon of EYFP (atg to aCg) were introduced. This modified construct, NS1-deYFP, was characterized earlier (31). All ATP-binding mutations were made to the NS1 region of both NS1-EYFP and NS1-deYFP constructs and were named according to the amino acid change (e.g., K406M-deYFP). Mutations were made by a PCR-based site-directed mutagenesis method or a two-step PCR method. Primer sequences are available upon request. Mutated regions of NS1-EYFP constructs were cut with the BseRI restriction enzyme and ligated to an infectious plasmid clone. These constructs were named with the prefix “pIC-” followed by the name of the mutation (e.g., pIC-K406M). Correctness of all clones was confirmed by sequencing. H2B-ECFP and H2B-EYFP plasmids were generous gifts from J. Langowski (German Cancer Research Center, Heidelberg, Germany) (67).
Cells, viruses, and Western blotting.
Norden Laboratory feline kidney (NLFK) cells were grown in Dulbecco's modified Eagle medium containing 10% fetal bovine serum, 1% penicillin-streptomycin, 1% l-glutamine, and 1% nonessential amino acids. Viruses were purified from NLFK cells transfected with the CPV-d clone as previously described (63). Anti-green fluorescent protein (anti-GFP) rabbit antibody (Ab) (1:2,000; Molecular Probes) and anti-NS1 mouse monoclonal antibody (MAb) (1:500; a gift from Caroline Astell, University of British Columbia, Vancouver, Canada [71]) were used for Western blotting, together with goat anti-rabbit IgG-alkaline phosphatase and goat anti-mouse IgG-alkaline phosphatase secondary Abs (1:2,000; Bio-Rad), respectively. Development of signals was done with nitroblue tetrazolium (NBT; Sigma) and BCIP (5-bromo-4-chloro-3-indolylphosphate; Sigma). Alternatively, Western blots were done as described for P38 transactivation experiments.
Fixed-cell imaging.
Infection percentages were calculated from NLFK cells stably expressing H2B-ECFP (67). Cells were grown on coverslips to ∼80% confluence and transfected with NS1-deYFP constructs. Transfection was done with TransIT-LT1 transfection reagent according to the manufacturer's instructions (Mirus Bio, Madison, WI). In infectivity studies, CPV was added to culture medium after transfection. After 24 h or 48 h of transfection/infection, cells were fixed with 4% paraformaldehyde (PFA) (20 min, room temperature), immunolabeled, and mounted with Mowiol-Dabco (Sigma). CPV capsids were detected with a MAb (a gift from Collin Parrish, Cornell University, Ithaca, NY). Goat anti-mouse Alexa 633-conjugated secondary Ab was used (Molecular Probes, Eugene, OR).
In infectivity experiments, a total of 60 cells transfected with a construct from the NS1-deYFP series in two different experiments were used for calculations. Fixed samples were imaged using an Olympus FV-1000 confocal microscope with a UPLSAPO 60× oil immersion objective (numerical aperture [NA] = 1.35). The image size was 512 by 512 pixels, with a resolution of 69 nm/pixel. H2B-ECFP, NS1-EYFP, and Alexa 633 fluorophores were excited with 405-nm, 515-nm, and 633-nm laser lines, respectively. Fluorescence was detected with 460- to 500-nm-band-pass (enhanced cyan fluorescent protein [ECFP]), 530- to 570-nm-band-pass (EYFP and deYFP), and 650-nm-long-pass (Alexa 633) filters. For publication figures, linear adjustments of the image brightness and contrast and Gaussian blurring with a 3-by-3 kernel were performed using ImageJ software (1). Distribution of the capsids in CPV-infected cells was studied in NLFK cells stably expressing H2B-ECFP. Cells were fixed at 48 h postinfection (p.i.) with 4% PFA (20 min, room temperature). CPV capsids were detected with a MAb (a gift from Collin Parrish, Cornell University, Ithaca, NY) followed by Alexa 488-conjugated anti-mouse antibody (Molecular Probes). ECFP was imaged with a 405-nm laser line and a 425- to 475-nm-band-pass filter, and Alexa 488 was imaged with a 488-nm laser line and a 500- to 600-nm-band-pass filter.
Replicative efficacy.
The effect of ATP-binding-pocket mutations on replicative efficacy of the virus was examined by secondary infection studies. NLFK cells were plated on 3-cm culture dishes and transfected on the following day with pIC constructs. Medium from transfected cells (2 ml) was collected at 2, 3, or 4 days posttransfection (p.t.) and concentrated to 50 μl in 100-kDa filter tubes (Amicon Ultra-4; Millipore). Secondary infection was studied as follows. Twenty-five microliters of concentrated medium was applied to cells on a coverslip and incubated at 37°C for 15 min, 2 ml of fresh culture medium was added, and cells were moved back to the incubator. At 48 h postinoculation, cells were fixed with 4% PFA, labeled with anti-NS1 (Alexa 488-conjugated anti-mouse secondary antibody), and embedded with ProLong antifade reagent containing DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes). Cells were imaged with an Olympus FV-1000 microscope using a 20× objective (NA, 0.75). Ten randomly selected fields of cells were imaged from two individual experiments with each pIC construct and each medium concentrate. The infectivity percentage was calculated by dividing the number of NS1-positive nuclei by the total number of DAPI-stained nuclei (over 5,100 cells in each sample). NS1- and DAPI-positive nuclei were segmented using the k-means clustering algorithm in ImageJ.
Live-cell imaging and fluorescence recovery after photobleaching.
In live-cell imaging experiments, cells were cultivated on glass-bottom dishes (MatTek, Ashland, MA) and transfected ∼20 h before experiments. Confocal laser scanning microscopic images were acquired with a Zeiss LSM 510 inverted laser scanning microscope (Carl Zeiss AG, Jena, Germany) using a Plan-Neofluar 63× (NA = 1.25) oil immersion objective. The sample holder and the objective were heated to 37°C. Optical sections from the middle of the nucleus were obtained, with an image size of 256 by 256 pixels and a resolution of 100 nm/pixel. A 514-nm laser line and a 560- to 600-nm-band-pass filter were used to acquire the EYFP signal. In half-nucleus FRAP (hnFRAP) experiments, seven successive iterations of high laser intensity (100% of 25 mV) were used to bleach approximately half of the nucleus. Images were acquired with low laser intensity (0.5 to 1%) and an appropriate time interval (0.5 to 2 frames per s [fps]). The optimal imaging rate was determined in preliminary studies and was 4 fps for K406M-deYFP and EYFP, 1 fps for E444Q-deYFP and R510A-deYFP, and 0.5 fps for NS1-deYFP, R508A-deYFP, and E445Q-deYFP. Image data were processed as described for fixed samples and were analyzed with ImageJ and spreadsheet software. FRAP data were normalized as follows (49): FREL = (Bt/B0)/(Nt/N0). Relative fluorescence (FREL) was obtained from the fluorescence of the bleached area at time point t (Bt), the average fluorescence of the bleached area before bleaching (B0), total nuclear fluorescence at time point t (Nt), and average nuclear fluorescence before bleaching (N0). Background fluorescence was measured from outside the cell and subtracted from all values. Half-time recovery (t[1/2]) was the median value between the lowest and highest values after bleaching. Student's t test was used to evaluate the significance of differences in half-time values.
ivFRAP.
In vitro FRAP (ivFRAP) experiments were conducted essentially as described before (36). Cells were cultivated on glass-bottom dishes and transfected with an appropriate construct ∼20 h before the experiment. Samples were prepared by washing cells with permeabilization buffer (20 mM HEPES, 110 mM potassium acetate, 2 mM magnesium acetate, pH 7.5) and were permeabilized with 100 μg/ml digitonin in permeabilization buffer for 6 min at room temperature. Imaging was conducted in pH 6.8 permeabilization buffer or the same buffer containing 1 mM ATP, 1 mM AMP-PNP, or 1 mM ATP-γS (Sigma) from a freshly prepared stock solution. Imaging settings were the same as in live-cell FRAP experiments, but the frame rate was 4 fps for all samples. Normalization was done as in FRAP experiments, except that the minimum value after bleaching was reduced from the values for all time points after bleaching to balance differences in bleaching efficiency.
P38 transactivation studies.
Transactivation was studied using both NS1-EYFP and pIC series of mutant constructs. Constructs were transfected into NLFK cells. Total cell lysates were collected at 24 h p.t. (NS1-EYFP) or at 48 h p.t. (pIC), and samples were analyzed with SDS-PAGE and Western blotting. In NS1-EYFP series, EYFP and NS1-EYFP were detected with anti-GFP Ab (Sigma). In pIC series, NS1 and VP2 were detected from replica Western blots, using anti-NS1 Ab and anti-capsid Ab, respectively. Horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG and a SuperSignal (Pierce Biotechnology, Rockford, IL) chemiluminescence detection kit were used for detection. Signal was collected quantitatively with a Chemidoc XRS (Bio-Rad, United Kingdom) hood equipped with a CoolSNAP HQ2-CCD camera. Exposure time was adjusted to avoid saturation of the signal. Intensities of individual bands were analyzed with ImageJ, and transactivation activity was analyzed according to the following equation: transactivation = (PMut/NS1Mut)/(PNS1/NS1NS1), where PMut is the intensity of the product under the control of the P38 promoter in the mutated construct (EYFP or VP2), NS1Mut is the corresponding intensity of the mutated NS1 protein in the sample (NS1-EYFP or NS1), and PNS1 and NS1NS1 are the intensities of nonmutated NS1 under the control of the P38 promoter (EYFP or VP2) and of the nonmutated NS1 protein (NS1-EYFP or NS1), respectively. This equation gives proportional transactivation activities of individual mutants. Five individual experiments were analyzed.
Virtual cell simulations.
The half-nucleus FRAP experiments were simulated with Virtual Cell software (56). The reaction map for simulations is shown in Fig. S5 in the supplemental material. The nucleus was assumed to have an ellipsoidal shape, with a width of 10 μm and a height of 15 μm. The time step of the simulation was set to 10 ms, and images were collected with the same frame rate as that in the FRAP studies. The bleaching reaction was modeled by a laser light-induced general reaction. The bleaching region of interest (ROI) was set to correspond to a half-nucleus, as in the hnFRAP experiments. The length of the bleach pulse was adjusted to 500 ms, and the first recovery image was collected 50 ms after the bleach phase to simulate image acquisition by confocal microscopy. In addition, bleaching due to the imaging laser was modeled by a second continuous bleaching reaction in the nucleus. The binding reactions were modeled as a mass action reaction in which the free ligand (diffusing NS1) reacted with the substrate (immobile binding partner) and formed the complex (immobile NS1). The concentration of free binding sites was set at 1 μM. In this situation, the pseudo-on rate (k*on) is the product of the binding site concentration and the actual on rate. The diffusion coefficient for EYFP (molecular mass = 26.9 kDa) was first modeled from hnFRAP data (30 μm2/s). This was used to calculate a theoretical diffusion coefficient for NS1-deYFP (molecular mass = 105 kDa), using the following mass scaling equation: DNS1-deYFP = DEYFP(mEYFP/mNS1-deYFP)^(1/3). In this equation, DNS1-deYFP is the diffusion coefficient and mNS1-deYFP is the theoretical mass of the NS1-deYFP molecule. DEYFP is the diffusion coefficient and mEYFP is the theoretical mass of EYFP. The calculated diffusion coefficient for monomeric NS1-deYFP was 18.8 μm2/s. The binding reaction kon and koff rates were changed in the modeling process until the model fit with the measured data.
RESULTS
NS1 modeling.
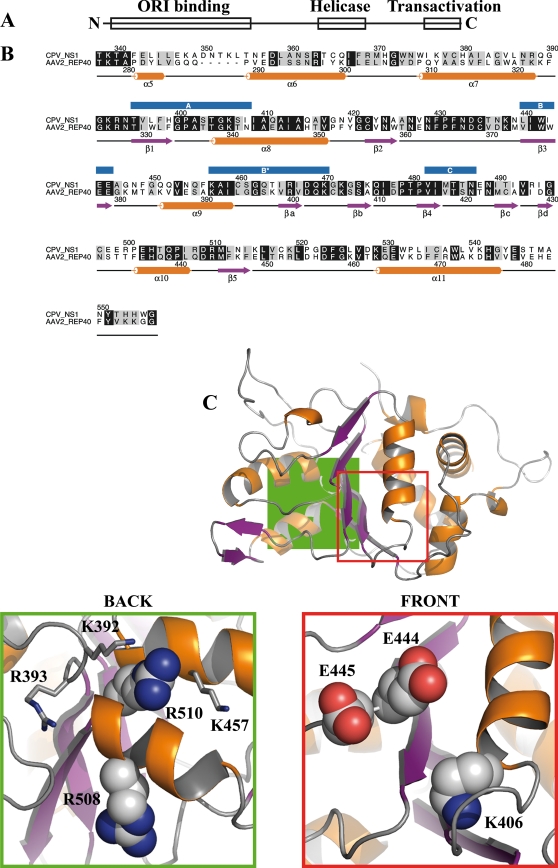
Our aim was to determine the effect of ATP binding and hydrolysis on the nuclear dynamics of NS1. To determine the positions of critical amino acid residues in the helicase domain (Fig. 1A and B), we built a comparative model of NS1. We used the REP40 protein structure from another parvovirus, AAV2, as our template. The identity of the helicase domain amino acid sequences was ∼38% (Fig. 1B). Visual inspection of the NS1 model did not show any biochemical anomalies or clashes between amino acid side chains. The conserved regions between REP40 and NS1 were located in the ATP-binding pocket and in the region that was near the arginine finger at the opposite side of the helicase domain. To further clarify their function in CPV NS1, five individual amino acid residues were selected and mutated to neutralize their charge. Three of them were positioned in the ATP-binding pocket: lysine 406 of the A motif was mutated to methionine, and two glutamates, at positions 444 and 445 of the B motif, were changed to glutamine (Fig. 1C, front). Two arginines, R508 and R510, at the other side of the helicase domain, are conserved in parvoviruses (e.g., minute virus of mice [MVM], H1 virus, and B19 virus [data not shown]), and the latter is also conserved in REP40 (Fig. 1C, back). These arginines lay in close proximity to a region that contained other positively charged amino acids (K392, R393, and K457) (Fig. 1C, back) and were mutated to alanine to neutralize their charge.
FIG. 1.
Sequence alignment and NS1 model. (A) Organization of functional domains in CPV NS1, including the N-terminal origin of replication (amino acids 16 to 275), the SF3 helicase domain (amino acids 299 to 486), and the C-terminal transactivation domain (amino acids 600 to 667). (B) Alignment of CPV NS1 and AAV2 REP40 helicase domain protein sequences. Numbers above the alignment refer to the CPV NS1 sequence, and those below refer to the AAV2 REP40 sequence. Positions of conserved domains (A, B, B′, and C) are indicated with blue boxes above the alignment. Secondary structures according to REP40 are shown below the alignment (34). α-Helices are shown in purple, and β-sheets are shown in orange. (C) Comparative model of CPV NS1 helicase domain. α-Helices (orange) and β-sheets (purple) are shown according to the alignment. (Back) Close-up of the area denoted in green, as seen from behind the structure. R508 and R510 are shown as space-filling models, and adjacent positively charged amino acids are shown in a stick model. (Front) Close-up of the ATP-binding pocket, denoted as a red square in the model. K406 from the A motif and E444 and E445 from the B motif are shown as space-filling models.
SF3 helicases are thought to function as ring-shaped homomultimers. We used the hexameric structures of SV40 LTag and Haemophilus influenzae proteasome-chaperone complex (62) to build hexameric models of NS1. The models revealed slightly different orientations of the subunits in the hexameric ring (see Fig. S1A and C in the supplemental material). Both models indicated that the conserved regions between REP40 and NS1 lay at the proposed intersubunit interfaces. In addition, our model placed a region of positively charged amino acids, including R508 and R510, in close proximity to the ATP-binding pocket of a neighboring subunit (see Fig. S1B and D in the supplemental material).
Characterization of fluorescent NS1 constructs.
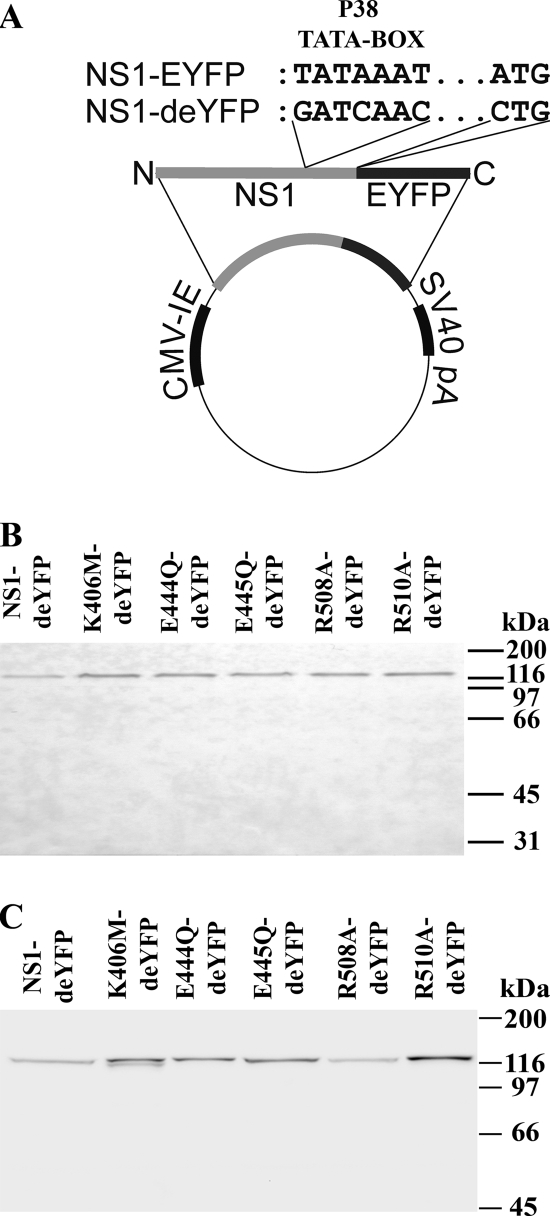
After identifying critical amino acids, we assessed their effect on NS1 function. A series of fluorescent fusion constructs were made by cloning NS1 upstream of the EYFP gene in a mammalian expression plasmid (Fig. 2A, NS1-EYFP). Because the viral capsid promoter P38 is embedded in the NS1 gene, transfection of this construct into mammalian cells lead to expression of NS1-EYFP fusion proteins under the control of the cytomegalovirus (CMV) promoter and of free EYFP proteins under the control of the P38 promoter (Fig. 3D). To remove the EYFP background, NS1-deYFP protein constructs were generated (Fig. 2A, NS1-deYFP). Next, analysis of the total cellular extracts from NS1-deYFP-transfected cells confirmed the presence of a single 120-kDa product that was recognized by both anti-GFP (Fig. 2B) and anti-NS1 (Fig. 2C) antibodies. The theoretical size of wild-type (wt) NS1 during viral infection is 76.6 kDa, and it is detected as a 95-kDa band by Western blotting. The sizes of NS1-deYFP proteins were consistent with this observation, as proteins were detected at ∼20 kDa more than expected by their theoretical size of 105 kDa. Immunofluorescence microscopy studies demonstrated good colocalization and correlation between the transfected NS1-deYFP mutant constructs with antibodies against GFP and NS1 (not shown). Collectively, these results confirm that NS1-deYFP transfectants express a single product in which NS1 is fused to EYFP.
FIG. 2.
NS1 fluorescent fusion constructs. (A) Schematic presentation of NS1-EYFP and NS1-deYFP constructs. CPV NS1 with an N-terminal EYFP fusion is under the control of the CMV immediate-early (IE) promoter. NS1-deYFP constructs have mutations in the P38 promoter area and in the start codon of EYFP. Western blots are shown for whole-cell lysates of NLFK cells transfected with NS1-deYFP constructs and detected with anti-GFP (B) and anti-NS1 (C) antibodies.
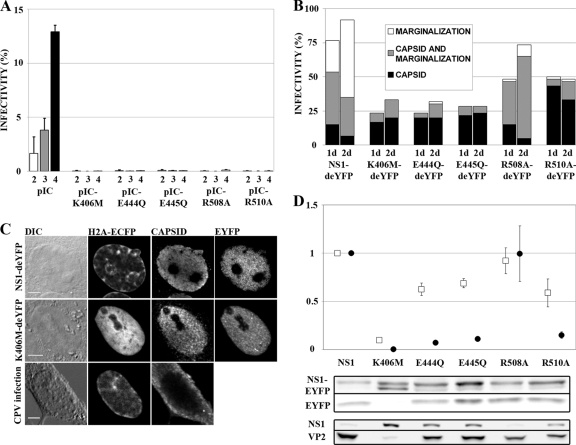
FIG. 3.
Functions of mutated NS1 constructs. (A) Secondary infections with concentrated medium collected at 2, 3, and 4 days p.t. are shown with white, gray, and black bars, respectively. Infectivity is the percentage of NS1-positive nuclei in the sample (n > 5,100). Error bars indicate standard deviations. (B) Percentages of transfected cells with signs of infection (n = 60). Chromatin marginalization is indicated in white, nuclear capsid antibody staining in black, and cells with both markers in gray. (C) NS1-deYFP-expressing infected cell with marginalized chromatin and nuclear capsid accumulation, K406M-deYFP-expressing infected cell with nuclear capsid accumulation but normal chromatin distribution, and CPV-infected cell, fixed at 48 h p.i., with marginalized chromatin and cytoplasmic capsid localization. Bars, 5 μm. DIC, differential interference contrast. (D) P38 transactivation activities of mutated NS1 proteins. Average transactivation values from EYFP (white boxes) and pIC (black circles) experiments are shown (n = 5). Transactivation values are average values for the product under the P38 promoter (EYFP or VP2) divided by the value for the corresponding NS1 protein (NS1-EYFP or NS1) and normalized to the value for nonmutated NS1. Proteins were detected in Western blots with anti-GFP (NS1-EYFP and EYFP), anti-NS1 (NS1), or anti-capsid (VP2) antibody. Error bars indicate standard errors of the means.
In live-cell confocal microscopy, all mutants were seen to localize predominantly to the nucleus in NLFK transfectants, excluding the nucleolus (Fig. 4, “live” column). The nuclear distributions of NS1-deYFP and R508A-deYFP were heterogeneous and concentrated in small foci. A more homogenous distribution and some focal accumulation were observed with the two ATP-binding-pocket mutants, E444Q-deYFP and E445Q-deYFP, and the arginine finger mutant, R510A-deYFP. Notably, K406M-deYFP was distributed throughout the nucleus but was excluded from the nucleolus. In conclusion, the mutations did not affect nuclear localization of NS1 but had patent effects on intranuclear distribution.
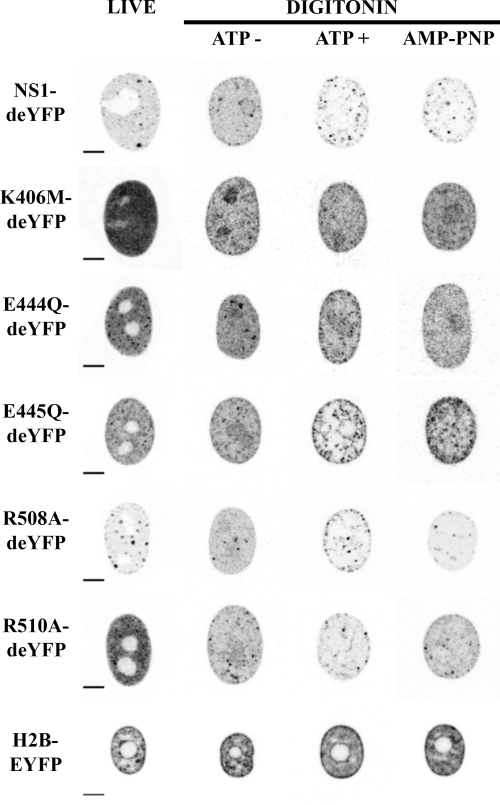
FIG. 4.
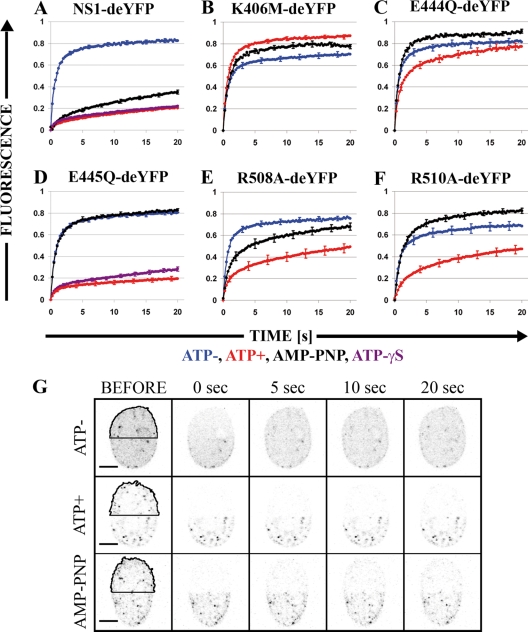
Nuclear distribution of NS1-deYFP constructs and H2B-EYFP under different imaging conditions. (Live) NS1-deYFP constructs under live-cell imaging conditions. (Digitonin) Permeabilization experiments are divided into three columns depending on the imaging conditions: ATP−, imaged in buffer without ATP; ATP+, imaged with 1 mM ATP; and AMP-PNP, imaged with 1 mM AMP-PNP. Bars, 5 μm. Images are shown with inverted gray-scale coloring.
Analysis of replication efficacy.
NS1 is an important component and marker of parvovirus infection-induced replication bodies (20, 32, 72). Effects of ATP-binding-pocket mutations on the replication efficacy of CPV were studied by use of a secondary infection series (Fig. 3A). Medium of pIC transfectants collected at 24, 48, or 72 h p.t. was used to inoculate new cells. Immunofluorescence analysis demonstrated that only the nonmutated infectious clone of CPV (pIC) showed a time-dependent increase in secondary infection (Fig. 3A). Infectivity percentages of the pIC media collected at 24, 48, and 72 h p.t. were 1.7% ± 1.5%, 3.8% ± 1.1%, and 12.9% ± 0.6%, respectively (mean ± standard deviation) (Fig. 3A). All mutants were essentially unable to produce infectious viruses. The highest infectivity rate (0.1% ± 0.07%) was observed for medium collected from pIC-R508A transfectants at 72 h p.t., and it remained below 0.1% for all other mutants and time points.
Next, the progression of the wt CPV infection was studied in the presence of NS1-deYFP constructs in cells that stably expressed the chromosomal marker H2B-ECFP. Cells were fixed at 24 h or 48 h p.t./p.i. and were immunolabeled with CPV capsid antibody. Sixty transfectants for each construct at each time point were analyzed. Nuclear labeling with capsid antibody and host cell chromatin marginalization (CM) were indicative of ongoing CPV infection (Fig. 3C) (31).
At 24 h p.t./p.i., 77% of NS1-deYFP-expressing cells were positive for one or both markers (Fig. 3B). The proportion of infected cells increased at 48 h p.t./p.i., when >90% of the cells were infected. A marked decline in infectivity was observed in mutant NS1-deYFP-expressing cells. Three of the constructs that had mutations in the ATP-binding-pocket area (K406M-deYFP, E444Q-deYFP, and E445Q-deYFP) had the lowest infectivities. Only 20% to 30% of these cells were infected at 24 h p.t./p.i., and <35% were infected at 48 h p.t./p.i. An infection rate of 50% at 24 h p.t./p.i. was observed for R508A-deYFP and R510A-deYFP. The proportion of infected cells rose to over 70% for R508A-deYFP-expressing cells at 48 h p.t./p.i. but remained 50% for R510A-deYFP-positive cells.
Further analysis revealed differences between constructs in the ratio of the two infection markers. For infected NS1-deYFP-expressing cells, the proportion that had the CM phenotype increased from 80% at 24 h p.t./p.i. to 93% at 48 h p.t./p.i. (Fig. 3B, white and gray portions of bars). Notably, the only mutant that experienced a similar trend was R508A-deYFP, for which the proportion of CM cells climbed from 69% to 93%. In infected cells that expressed any of the remaining mutants, the proportion of CM cells remained below 40%, even at 48 h p.t./p.i. The percentage of cells with nuclear virus capsid label (Fig. 3B, gray and black portions of bars) decreased from 70% to 38% of all infected NS1-deYFP-expressing cells. Again, a similar trend was observed only in R508A-deYFP-expressing cells, where a decrease from 97% to 89% was observed between 24 h p.t./p.i. and 48 h p.t./p.i. In cells that expressed any of the other mutants, the proportion of infected cells with nuclear virus capsid antibody staining remained over 94% of total infected cells.
These data demonstrated that all mutations rendered infectious clones unable to produce viruses. Moreover, the expression of mutated NS1 proteins inhibited and modified the progression of the CPV infection. Although mutant NS1-deYFP-expressing infected cells produced viral capsids inside the nucleus, only R508A-deYFP affected the proportion of CM phenotype cells similarly to NS1-deYFP.
P38 transactivation.
The ability of the NS1 mutants to transactivate the capsid promoter, P38, was studied by transfecting cells with either the NS1-EYFP or pIC series of mutants, followed by monitoring the production of the proteins under the control of the P38 promoter. NS1-EYFP constructs expressed NS1-EYFP under the CMV promoter and EYFP under the P38 promoter, while pIC constructs expressed NS1 under the viral P4 promoter and VP2 under the P38 promoter. The CPV P38 promoter is constitutively active in mammalian cells (14). Accordingly, the amount of protein produced under the P38 promoter in a cell culture depends on transfection efficiency, P38 basal activity, and P38 transactivation by NS1. In our approach, data were normalized to the amount of produced NS1 to correct for transfection efficiency and the amount of transactivating agent. This experimental setup allowed us to measure the levels of P38 promoter transactivation induced by individual NS1 mutants (NS1-EYFP or NS1).
All mutants except for the R508A mutant reduced P38 transactivation (Fig. 3D). The largest reduction was observed for K406M-EYFP, which retained only 10% ± 1.1% and 0.5% ± 0.3% of nonmutated NS1 activity in EYFP- and pIC-based assays, respectively (in both EYFP and pIC series, P < 0.001 and n = 5; errors are standard errors of means for all experiments). Significant reductions were seen for the E444Q (in EYFP series, 63% ± 6.5% [P < 0.01]; in pIC series, 7.2% ± 1.2% [P < 0.001]), E445Q (in EYFP series, 69% ± 4.7% [P < 0.01]; in pIC series, 11% ± 2.2% [P < 0.001]), and R510A (in EYFP series, 59% ± 14.6% [P < 0.05]; in pIC series, 15% ± 3.3% [P < 0.001]) mutants compared with the corresponding NS1 results. The R508A mutant retained NS1-like P38 transactivation activity in both assays (in EYFP series, 92% ± 13.3% [P > 0.59]; in pIC series, 100% ± 29% [P > 0.99]).
In vitro fluorescence recovery after photobleaching.
ATP binding and hydrolysis properties of CPV NS1 have not been characterized. Thus, we used a recently described in vitro FRAP (ivFRAP) method (65) to study these properties. Cells that were transfected with NS1-deYFP constructs were permeabilized with digitonin to remove endogenous ATP (53). Half of the nucleus was exposed to a high laser intensity under a confocal microscope to irreversibly bleach the fluorescent molecules in that area. Redistribution of the remaining fluorophores was monitored with low-laser-intensity scanning. Studies were conducted in buffer (Fig. 4, ATP− column), buffer with 1 mM ATP (ATP+ column), or buffer with 1 mM nonhydrolyzable ATP analog (AMP-PNP column). NS1-deYFP and E445Q-deYFP constructs were also studied in a buffer containing 1 mM ATP-γS. At least 20 cells were analyzed for each construct under all conditions.
All NS1-deYFP constructs remained nuclear in digitonin-treated cells (Fig. 4). Intranuclear distribution, however, varied depending on the experimental conditions. The expression of all constructs was more homogenous in buffer without ATP than that in nonpermeabilized live cells (Fig. 4). Some constructs (NS1-deYFP, E445Q-deYFP, R508A-deYFP, and R510A-deYFP) appeared as specks in the presence of ATP and AMP-PNP. Notably, digitonin permeabilization induced the accumulation of all NS1-deYFP constructs in the nucleoli. Nucleolar exclusion was partially restored for NS1-deYFP and E445Q-deYFP in the presence of ATP.
ivFRAP recovery was monitored for 20 s after bleaching (Fig. 5G), and normalized results were analyzed (Fig. 5A to F). The recovery of NS1-deYFP was slower under ATP-containing (Fig. 5A, red line) than under non-ATP (Fig. 5A, blue line) conditions; E445Q-deYFP gave a similar result (Fig. 5D). The shapes of the recovery curves for these constructs were nearly linear with ATP and logarithmic without ATP. Compared with NS1-deYFP, changes in recovery on introduction of ATP were not as large for E444Q-deYFP, R508A-deYFP, and R510A-deYFP. Also, recovery in ATP was slowest in these mutants, but the shapes of the curves with and without ATP were similar (Fig. 5C, E, and F). Recovery of the K406M-deYFP mutant was faster in ATP-containing buffer than without ATP, but the recovery curves were similarly shaped (Fig. 5B).
FIG. 5.
In vitro FRAP experiments with NS1-deYFP constructs. (A) NS1-deYFP. (B) K406M-deYFP. (C) E445Q-deYFP. (D) E444Q-deYFP. (E) R508A-deYFP. (F) R510A-deYFP. Average recovery in buffer without ATP is shown in blue, that in buffer with 1 mM ATP is shown in red, that in buffer with 1 mM AMP-PNP is shown in black, and that in buffer with 1 mM ATP-γS is shown in purple (only in panels A and D). The y axis is normalized average fluorescence, and the x axis is the time after bleaching, in seconds. Error bars show standard errors (SE). Twenty or more cells were analyzed within each condition. (G) Inverted gray-scale images of NS1-deYFP recovery in ivFRAP experiments under different imaging conditions. The bleached area is highlighted with a black line. Bar, 5 μm.
Next, we studied the effect of the nonhydrolyzable ATP analog AMP-PNP. Only NS1-deYFP recovered considerably slower in AMP-PNP than in buffer without ATP (Fig. 5A, black line). In addition, the shape of the AMP-PNP recovery curve for NS1-deYFP was similar to that with ATP. For all other constructs, the recovery of AMP-PNP resembled that in buffer without ATP. Notably, the fastest recovery for R510A-deYFP and E444Q-deYFP was observed in AMP-PNP. ivFRAP recovery was also measured for NS1-deYFP and E445Q-deYFP in buffer that contained ATP-γS. Recovery of both NS1-deYFP and E445Q-deYFP was considerably slower with ATP-γS than that without ATP but similar to the recovery with ATP (Fig. 5A and D, purple lines).
FRAP analysis of NS1 mutants.
The binding properties of NS1-deYFP constructs were assessed with FRAP. Half of each nucleus from transfected cells was bleached, and the recovery was followed until it reached a plateau (Fig. 6 and Fig. S2 in the supplemental material). First, we estimated the contribution of diffusion to the recovery by visualizing gradient smoothing during recovery (see Fig. S3 in the supplemental material). We compared images that were taken immediately after the bleach pulse and at the point of approximate half-time recovery (t[1/2]) for individual constructs. After normalization, the intensity profiles for all constructs shifted, except for that of E445Q-deYFP, indicating that the dynamics of all other constructs depend on diffusion (6) and that the binding kinetics of E445Q-deYFP is considerably slower than those in other constructs.
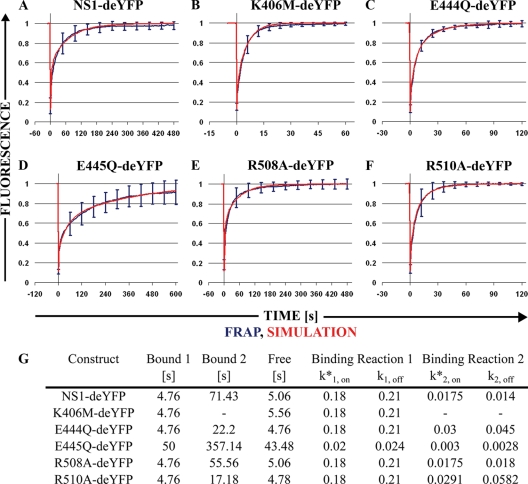
FIG. 6.
Half-nucleus FRAP experiments and virtual cell simulations. The blue line denotes normalized half-nucleus recovery of the construct. Error bars are SE. The red line shows the virtual cell simulation reconstruction of recovery. (A) NS1-deYFP. (B) K406M-deYFP. (C) E444Q-deYFP. (D) E445Q-deYFP. (E) R508A-deYFP. (F) R510A-deYFP. The y axis is normalized average fluorescence, and the x axis is time, in seconds. (G) Average binding times for both binding sites (Bound 1 and Bound 2) and average free diffusion times (free), in seconds. Binding reaction pseudo-on rates (k*1, on and k*2, on) and off rates (k1, off and k2, off) were obtained from simulations.
Next, we analyzed the recovery from half-nucleus FRAP experiments in NLFK cells. EYFP was used as a free diffusion marker and recovered quickly, as expected (mean t[1/2] ± standard error of the mean, 0.46 ± 0.03 s; n = 12) (see Fig. S4 in the supplemental material). The recovery of all NS1-deYFP constructs was considerably slower. The fastest recovery was observed for the A motif mutant K406M-deYFP, which equilibrated in approximately 60 s (t[1/2] = 3.04 ± 0.20 s; n = 20) (Fig. 6B); its recovery, however, was much slower than what is expected for a freely diffusing molecule (e.g., EYFP). NS1-deYFP reached a plateau in approximately 240 s (t[1/2] = 17.90 ± 1.97 s; n = 20) (Fig. 6A), while R508A-deYFP recovered more quickly (t[1/2] = 15.35 ± 1.73 s; n = 20) (Fig. 6E). The shapes of the recovery curves were nearly identical for these constructs, and there was no significant difference in t[1/2] values (see Fig. S4G in the supplemental material). Both the B motif mutant E444Q-deYFP (t[1/2] = 5.70 ± 0.27 s; n = 20) (Fig. 6C) and the arginine finger mutant R510A-deYFP (t[1/2] = 5.82 ± 0.62 s; n = 22) (Fig. 6F) equilibrated in 120 s. The recoveries of these mutants were nearly identical in these experiments, and t[1/2] values did not differ significantly (see Fig. S4G in the supplemental material). Finally, considerably slower dynamics was observed for the B motif mutant E445Q-deYFP (t[1/2] = 40.19 ± 6.90 s; n = 21) (Fig. 6D), which leveled off in approximately 600 s; despite this pattern, approximately 10% of the fraction was not recovered.
Virtual cell modeling of FRAP results.
Half-nucleus FRAP experiments were reconstituted in a virtual cell modeling environment (59) to glean more information on the binding kinetics of the NS1-deYFP constructs. Free EYFP FRAP data yielded a diffusion constant of 30 μm2/s in simulations, which was used to calculate a theoretical diffusion constant of 18.8 μm2/s for NS1-deYFP monomers by use of mass scaling.
A reasonable fit was obtained for K406M-deYFP FRAP data by using the calculated diffusion constant and assuming a single binding reaction with a pseudo-on rate (k*1, on) of 0.18 and an off rate (k1, off) of 0.21 (Fig. 6B and G). Under these parameters, protein was bound for an average of 4.76 s (1/koff) and diffused freely for 5.56 s (1/k*on). Other constructs, however, did not fit the single-binding-site model (data not shown). Further simulations were conducted, assuming a second independent binding reaction. The first binding reaction was assumed to remain the same as in the K406M-deYFP simulation. The values for the second binding reaction (k*2, on and k2, off) were adjusted to fit the FRAP data (Fig. 6G). Recovery data for all remaining mutants, except for E445Q-deYFP, were reproduced using the second binding reaction with adjusted parameters. The second binding reaction had slower kinetics for all mutants than the first one. NS1-deYFP bound to the second binding site for 71.43 s and diffused freely for 5.06 s. Slightly faster dissociation for R508A-deYFP resulted in a binding time of 55.56 s for the second binding reaction, and the free diffusion time was 5.06 s, the same as that for NS1-deYFP; this mutant differed from NS1-deYFP only by its slightly higher k2, off rate. The kinetics of the second binding site was faster for both E444Q-deYFP and R510A-deYFP, yielding binding times of 22.2 s and 17.18 s, respectively. Free diffusion times were 4.76 s for E444Q-deYFP and 4.78 s for R510A-deYFP. The slow recovery of E445Q-deYFP did not fit the same model as that for the other mutants; its recovery did not contain a highly dynamic component that was comparable with K406M-deYFP and the other constructs, neither was its recovery reproducible with the single-binding-site model. We had to alter our model to contain two binding sites to successfully reproduce the FRAP data. Binding times for E445Q-deYFP were 50 s and 357 s for the first and second binding reactions, respectively. Association rate constants were much lower than those for other constructs, and the free diffusion time was 43.5 s. Recovery of this mutant was incomplete in the hnFRAP experiments, but the full recovery time predicted by the simulation was 1,120 s (Fig. 6D).
DISCUSSION
After nuclear import, the single-stranded-DNA (ssDNA) genome of parvovirus is converted to double-stranded DNA (dsDNA), and production of the viral NS1 protein initiates. All ensuing steps of the infection appear to depend on this multifunctional protein, whose functions rely on ATP. In this study, we mutated amino acids that are critical for ATP binding and monitored the changes in the function and dynamics of canine parvovirus NS1 in living cells.
The ATP-binding site of parvoviral NS1 proteins is located in the conserved superfamily 3 (SF3) helicase domain (27) and is likely to be the only binding site, because mutations therein abolish ATP-dependent functions of NS1 (35, 40). The general mechanisms of ATP binding and hydrolysis of AAA+ fold proteins, including SF3 helicases, are well established (reviewed in references 23 and 61). Our comparative model of the helicase domain shows an α/β fold where the conserved ATP-binding A and B motifs meet on one of the facets. These properties are conserved in all SF3 helicase domain structures (30). Some helicases control ATP binding and hydrolysis though an arginine finger, a trans-acting, positively charged amino acid (57). Based on sequence alignment, CPV NS1 is one of these helicases, and R510 is a prominent arginine finger residue. In our comparative model, this residue lies in a region of positively charged amino acids opposite the A and B motifs. A similar positively charged region has been shown to be critical for oligomerization of LTAg, an SF3 helicase from simian virus 40 (29), and an analogous role can be inferred for other members of this family. Note that a mutation in this area of the MVM parvovirus renders it unable to replicate the viral genome, thus suggesting its importance for parvovirus NS1 function (35). This region is close to the neighboring subunit in our hexameric models, further supporting its role in multimerization of parvovirus NS1.
In our comparative model, we identified the locations of ATP-binding amino acid residues in CPV NS1. We created a series of fluorescent fusion constructs with mutations in the identified residues to study the effects of ATP binding and hydrolysis on the dynamics of NS1. After expression in mammalian cells, these proteins localized to the nucleus, confirming that neither the C-terminal fluorescent fusion nor any of the mutations affected nuclear transport. This result was expected, because a bipartite nuclear localization signal resides before the helicase domain in the NS1 sequence (46). At the early stages of parvoviral infection, characteristic intranuclear replication structures, called autonomous parvovirus replication bodies, are formed (20). We have reported that CPV NS1 expressed in uninfected cells as a fluorescent fusion protein forms interchromosomal foci (32). The role or constituents of these foci are unknown, but similar structures are formed by inert multimerizing proteins, arguing that this is a property of multimerizing proteins (28). Focus formation similar to that of nonmutated NS1 was observed for the R508A mutant and both motif B mutants, the E444Q and E445Q mutants. These structures were, however, less pronounced for the R510A mutant and absent from K406M mutant-expressing cells. Notably, K406 is a crucial amino acid for ATP-binding-associated functions in many SF3 helicases, including NS1 of the parvovirus MVM (35) and multimerization in AAV Rep (42). Also, according to sequence analysis, R510 is the arginine finger residue in CPV NS1 and hence regulates ATP binding in multimeric conformation (34). These observations suggest a link between focus formation and multimerization of NS1. ATP binding per se, however, does not appear to be critical, because these structures also exist in ATP-depleted cells.
ivFRAP is a new method for estimation of the ATP dependency of a binding reaction (36). With this method, we demonstrated that NS1 binding dynamics is highly dependent on ATP. Moreover, results with the nonhydrolyzable ATP analog AMP-PNP initially suggested that ATP hydrolysis is unnecessary. The motif B E445Q mutant retains significant ATP-binding ability, but ATP hydrolysis is severely impaired (35). Binding of the E445Q mutant was ATP dependent in the ivFRAP experiments, further suggesting that hydrolysis is unnecessary for NS1 binding. Yet binding was not induced by AMP-PNP in the ivFRAP experiments. These findings should be interpreted with some caution, however, since proteins complexed with a nonhydrolyzable ATP analogue might not be fully functionally equivalent to corresponding ATP-bound forms (24). Proteins also vary in their ability to recognize and hydrolyze ATP analogs, and this could explain the difference between NS1 and the E445Q mutant in AMP-PNP experiments (43, 48, 64). Indeed, we could observe binding for the E445Q mutant in the presence of another nonhydrolyzable ATP analog, ATP-γS. Accordingly, we propose that some degree of ATP hydrolysis is required for NS1 binding, as has been shown for MCM helicase (54). Alternatively, the changes could be due to differences in the binding of ATP analogues, as has been reported for the SecA protein of the translocase complex (24). Notably, a mechanism has been reported for E1, an SF3 helicase from bovine papillomavirus for which there are two states in the ATP hydrolysis cycle, with opposing binding properties (9). When bound to ATP, E1 readily forms multimers with a high affinity and fast binding kinetics for both ss- and dsDNA. Incorporation of ADP or AMP-PNP, however, decreases the DNA binding affinity and kinetics, indicating that ATP hydrolysis is required to dissociate E1. ivFRAP results indicate that a similar mechanism exists for CPV NS1, in which affinity for DNA cycles between high-affinity (ATP-bound) and low-affinity (hydrolyzed) stages. The motif A K406M mutant did not alter its binding at detectable levels in the presence of ATP or AMP-PNP. This result was expected, because many studies have reported that the amino acid at this position is critical for mediating ATP-dependent functions (35, 40, 46). Binding of constructs in the presence of ATP decreased gradually from the wt to the K406M mutant (NS1 ∼ E445Q > R510A > R508A > E444Q > K406M), indicating that we correctly identified the amino acids that are important for ATP binding.
To determine the effects of the mutations on the transactivation activity of NS1, we studied both NS1 fusion proteins (with EYFP) and infectious clones (pIC). All mutants, with the exception of the R508A mutant, had reduced P38 transactivation. Interestingly, the levels of transactivation in all mutants, except the R508A mutant, were lower in EYFP assays than in pIC assays. This difference could be explained either by the effect of the EYFP fusion or by the differences in the levels of NS1 in these assays. The possible influence of the EYFP fusion on NS1 function is most likely independent of the mutations in the ATP-binding pocket and thus should not affect the relative transactivation values of different mutants. In pIC assays, NS1 and R508A mutant expression levels were constantly lower than others. This led to lower relative transactivation values for the other constructs, due to normalization. The reason for reduced NS1 expression is not clear but could arise from its cytotoxicity (37). Interestingly, these data suggest that a small amount of functional NS1 is sufficient for high-level transactivation of the P38 promoter. The P38 promoter could be saturated by an excess of NS1 in the EYFP assay leading to underestimation of the transactivation potential. Despite the differences between the assays, the only mutant that retained the NS1-like transactivation was the R508A mutant, and the lowest activity was observed for the K406M mutant, a mutant that did not react to ATP in ivFRAP. However, the E445Q and E444Q mutants, which had strong and weak reactivities to ATP in the ivFRAP experiments, respectively, were indistinguishable in promoter transactivation. Overall, it appears that although ATP binding is important for transactivation, it does not correlate with differences in the overall binding properties of mutants.
Viral replication efficacy was analyzed in secondary infection experiments. A time-dependent increase in the amount of infectious viruses was observed for the nonmutated infectious clone. This was not observed in any of the mutants, which were essentially unable to produce infectious viruses. Additionally, infectivity was drastically reduced in cells that expressed constructs that had a mutation in the A or B motif and slightly decreased with arginine mutants. Because these cells also expressed wt NS1, these reductions in infectivity were likely a consequence of dominant-negative inhibition by the mutant proteins. What is the mechanism of this dominant-negative effect? NS1 is inhibited in a dominant-negative manner by coexpression of a nonfunctional protein that has an intact oligomerization domain (22) and in a competitive manner by peptides that mimic the oligomerization domain sequence (50). These observations highlight the importance of multimerization for NS1 function. In many helicases, multimerization is an ATP-dependent process (42). Although there is no direct link between ATP binding and the multimerization of autonomous parvovirus NS1 proteins, some indirect evidence exists (46). Site-specific nicking (13) and capsid promoter transactivation depend on ATP in MVM NS1 (10), and multimerization is speculated to occur during both events. Moreover, forced multimerization of NS1 by cross-linking antibodies is sufficient for transactivation (10). Both nicking and transactivation are likely to be independent of energy input (10, 35, 44), further suggesting that the function of ATP binding is mediated through multimerization of NS1. Notably, the two mutants that induced nearly identical reductions in infectivity, the K406M and E445Q mutants, had disparate effects on P38 transactivation. In general, defects in transactivation do not appear to explain the observed dominant-negative effect. An intriguing feature of CPV infection is the marginalization of host chromatin toward the nuclear envelope and the nucleolus (31). Marginalization was not observed in cells that expressed fluorescent fusions of the viral proteins NS1, NS2, and VP2, suggesting that viral genome replication is required, either directly or indirectly (data not shown). One of the obligatory roles of NS1 in genomic replication is helicase activity (15). The current opinion on the function of SF3 helicases suggests a model with “all sites sequential” (23). In this model, ssDNA makes contact directly with consecutive subunits of the multimeric protein ring while being escorted though a central pore. The corollary of this model is that a single nonfunctional monomer can slow or halt the helicase activity of the entire complex. Consequently, NS1 helicase activity is prone to malfunctioning in the presence of even minute amounts of mutated subunits. We observed a substantial decrease in the proportion of cells with the chromatin-marginalized phenotype in the infected cells that expressed any of the mutants, except for the R508A mutant. Also, the reduction in infectivity was substantially lower in this mutant. Notably, in some infected cells, viral capsids were mostly cytoplasmic. We suggest that this phenotype represents cells where production of capsid proteins is ceased while nuclear export of capsids is still ongoing. At later time points, the majority of the NS1-expressing cells, in contrast to only a few R508A mutant-expressing cells, belonged to this category. It is tempting to speculate that the difference in capsid localization is the explanation for the mutant's inability to produce infectious viruses. In conclusion, our data suggest that defects in helicase activity explain the dominant-negative effect of mutant NS1 proteins.
To measure the binding properties of individual ATP-binding mutants, we studied their dynamics in living cells by use of FRAP. To our knowledge, this is the first study to use FRAP to systematically characterize ATP binding in a protein. This approach has the benefit of monitoring the steady state of all interactions in the cellular environment, obviating the need to concentrate on a particular predefined interaction. This is particularly advantageous in the case of a multifunctional protein, e.g., NS1, which has many ATP-dependent functions. FRAP analysis was performed in noninfected cells due to the dominant-negative-effect-induced poor infectivity of mutant protein-expressing cells. The recovery of all constructs was slower than that of a similarly sized freely diffusing protein, indicating that all mutants retained some binding properties. The fastest dynamics was observed for the K406M mutant, and simulation data supported the single-binding-site model. Based on ivFRAP results, we propose that this binding is ATP independent. The single-binding-site model does not explain the recovery of the remaining constructs, which we concluded to have two binding modes with distinct binding kinetics. We further assumed that one of the reactions represented ATP-independent binding and thus retained the same binding parameters as those for the K406M mutant. This assumption yielded a good fit to the experimental data for all of the mutants except the E445Q mutant. The binding kinetics of the second reaction was slower for all constructs than that for the K406M mutant. Notably, these constructs showed ATP-dependent binding in the ivFRAP experiments, indicating that the second binding reaction was ATP dependent. Hence, the differences in the kinetics of the second binding reaction reflect ATP-dependent binding properties of the constructs. Recently, we reported two binding modes for NS1 in infected cells, with binding times of 83 s and 10 s and a free diffusion time of 8.1 s (31). Notably, very similar values for average binding times (71 s and 4.8 s) and for free diffusion time (5.1 s) were observed in this study for noninfected cells. Evidently, this result suggests that the interactions that define NS1 dynamics in infected and noninfected cells are the same, despite the drastic changes that occur in the nuclear environment. As anticipated from recovery profile analysis of the E445Q mutant, patently slower binding kinetics, without a fast component comparable to that of the K406M mutant, was observed. A homologous mutant has been reported to hydrolyze ATP slowly in the context of MVM NS1 protein (35) and other AAA+ fold-containing proteins (4). These data, with ivFRAP analysis, imply that ATP hydrolysis is required for the dissociation of NS1. While other constructs readily hydrolyze bound ATP molecules and return to low-affinity states or dissociate from DNA, the E445Q mutant remains bound to DNA. What are the probable targets for NS1 binding in the noninfected cell nucleus? We have recently shown that NS1 protein stays in the nucleus, even after permeabilization of the nuclear membrane, but that its binding is sensitive to the removal of DNA (31). Also, only binding to an immobile target, e.g., DNA, leads to large changes in recovery dynamics; thus, it seems plausible that the dynamics of NS1 in living cells is dictated by its ability to bind DNA. Of the several instances of NS1-DNA interactions that have been reported, some are likely to have a major effect on the overall dynamics only during infection. Nicking activity is controlled by specific interactions between NS1 and endogenous proteins on the viral DNA sequence, and the helicase activity of NS1 requires single-stranded or nicked DNA for initiation in vitro (11, 15, 45). These interactions are thus unlikely to take place in noninfected cells. NS1 binds ACCA repeat sequences in dsDNA, and recently, two binding modes were observed for MVM NS1 in vitro (16); one mode was highly dependent on ATP, while the effect of ATP was negligible in the other. Consensus binding sequences were found to be noncomplex, and thus candidate binding sequences are likely to be found in the endogenous genome. Similarly, a recent study proposed two DNA-binding modes for the AAV Rep protein (42), one of which was ATP dependent and cooperative between the N-terminal and helicase domains, while the other was independent of ATP and maintained by the N-terminal domain.
In conclusion, we propose two binding modes for autonomous parvovirus NS1 proteins in living cells. These binding modes are likely to reflect the ATP-independent fast kinetics for binding of the N-terminal domain and the ATP-dependent slower binding kinetics with both the N-terminal and helicase domains. Furthermore, our data suggest that DNA binding of NS1 depends on both binding and hydrolysis of ATP.
Supplementary Material
Acknowledgments
Past and present M.V.-R. group members are thanked for creating an inspiring working environment. First-class experimental support by Irene Helkala is gratefully acknowledged.
This work was supported by the Academy of Finland (http://www.aka.fi/en-gb/A/) (grants 107311 and 129774). E.A.N. and O.K. were supported by the National Graduate School in Informational and Structural Biology (http://web.abo.fi/isb/). E.A.N. was also supported by Emil Aaltonen's foundation and the Finnish cultural foundation. T.O.I. was supported by the National Graduate School in Nanoscience (NGS-NANO).
Footnotes
Published ahead of print on 10 March 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Ahn, J. K., Z. W. Pitluk, and D. C. Ward. 1992. The GC box and TATA transcription control elements in the P38 promoter of the minute virus of mice are necessary and sufficient for transactivation by the nonstructural protein NS1. J. Virol. 66:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, G. J. 1993. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 6:37-40. [DOI] [PubMed] [Google Scholar]
- 6.Beaudouin, J., F. Mora-Bermudez, T. Klee, N. Daigle, and J. Ellenberg. 2006. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys. J. 90:1878-1894. doi: 10.1529/biophysj.105.071241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman, H., K. Henrick, and H. Nakamura. 2003. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 8.Bleker, S., M. Pawlita, and J. A. Kleinschmidt. 2006. Impact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. J. Virol. 80:810-820. doi: 10.1128/JVI.80.2.810-820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castella, S., D. Burgin, and C. M. Sanders. 2006. Role of ATP hydrolysis in the DNA translocase activity of the bovine papillomavirus (BPV-1) E1 helicase. Nucleic Acids Res. 34:3731-3741. doi: 10.1093/nar/gkl554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, J., S. F. Cotmore, and P. Tattersall. 1995. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 69:5422-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, J., S. F. Cotmore, and P. Tattersall. 1997. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J. Virol. 71:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, J., S. F. Cotmore, and P. Tattersall. 1997. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J. Virol. 71:5733-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, J., S. F. Cotmore, and P. Tattersall. 2001. Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur. J. Virol. 75:7009-7017. doi: 10.1128/JVI.75.15.7009-7017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen, J., T. Storgaard, B. Viuff, B. Aasted, and S. Alexandersen. 1993. Comparison of promoter activity in Aleutian mink disease parvovirus, minute virus of mice, and canine parvovirus: possible role of weak promoters in the pathogenesis of Aleutian mink disease parvovirus infection. J. Virol. 67:1877-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., R. L. Gottlieb, and P. Tattersall. 2007. Replication initiator protein NS1 of the parvovirus minute virus of mice binds to modular divergent sites distributed throughout duplex viral DNA. J. Virol. 81:13015-13027. doi: 10.1128/JVI.01703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotmore, S. F., and P. Tattersall. 1988. The NS-1 polypeptide of minute virus of mice is covalently attached to the 5′ termini of duplex replicative-form DNA and progeny single strands. J. Virol. 62:851-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotmore, S. F., and P. Tattersall. 1998. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J. Virol. 72:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotmore, S. F., and P. Tattersall. 2005. Encapsidation of minute virus of mice DNA: aspects of the translocation mechanism revealed by the structure of partially packaged genomes. Virology 336:100-112. doi: 10.1016/j.virol.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Cziepluch, C., S. Lampel, A. Grewenig, C. Grund, P. Lichter, and J. Rommelaere. 2000. H-1 parvovirus-associated replication bodies: a distinct virus-induced nuclear structure. J. Virol. 74:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLano, W. L. 2002. The PyMOL molecular graphics system. http://www.pymol.org/.
- 22.Deleu, L., A. Pujol, J. P. Nuesch, and J. Rommelaere. 2001. Inhibition of transcription-regulating properties of nonstructural protein 1 (NS1) of parvovirus minute virus of mice by a dominant-negative mutant form of NS1. J. Gen. Virol. 82:1929-1934. [DOI] [PubMed] [Google Scholar]
- 23.Enemark, E. J., and L. Joshua-Tor. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270-275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 24.Fak, J. J., A. Itkin, D. D. Ciobanu, E. C. Lin, X. J. Song, Y. T. Chou, L. M. Gierasch, and J. F. Hunt. 2004. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry 43:7307-7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- 25.Gai, D., R. Zhao, D. Li, C. V. Finkielstein, and X. S. Chen. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119:47-60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparison and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429.
- 27.Gorbalenya, A. E., E. V. Koonin, and Y. I. Wolf. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145-148. [DOI] [PubMed] [Google Scholar]
- 28.Gorisch, S. M., M. Wachsmuth, C. Ittrich, C. P. Bacher, K. Rippe, and P. Lichter. 2004. Nuclear body movement is determined by chromatin accessibility and dynamics. Proc. Natl. Acad. Sci. U. S. A. 101:13221-13226. doi: 10.1073/pnas.0402958101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenleaf, W. B., J. Shen, D. Gai, and X. S. Chen. 2008. Systematic study of the functions for the residues around the nucleotide pocket in simian virus 40 AAA+ hexameric helicase. J. Virol. 82:6017-6023. doi: 10.1128/JVI.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickman, A. B., and F. Dyda. 2005. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15:77-85. doi: 10.1016/j.sbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Ihalainen, T. O., E. A. Niskanen, J. Jylhava, O. Paloheimo, N. Dross, H. Smolander, J. Langowski, J. Timonen, and M. Vihinen-Ranta. 2009. Parvovirus induced alterations in nuclear architecture and dynamics. PLoS One 4:e5948. doi: 10.1371/journal.pone.0005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ihalainen, T. O., E. A. Niskanen, J. Jylhava, T. Turpeinen, J. Rinne, J. Timonen, and M. Vihinen-Ranta. 2007. Dynamics and interactions of parvoviral NS1 protein in the nucleus. Cell. Microbiol. 9:1946-1959. doi: 10.1111/j.1462-5822.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- 33.James, J. A., A. K. Aggarwal, R. M. Linden, and C. R. Escalante. 2004. Structure of adeno-associated virus type 2 Rep40-ADP complex: insight into nucleotide recognition and catalysis by superfamily 3 helicases. Proc. Natl. Acad. Sci. U. S. A. 101:12455-12460. doi: 10.1073/pnas.0403454101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James, J. A., C. R. Escalante, M. Yoon-Robarts, T. A. Edwards, R. M. Linden, and A. K. Aggarwal. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025-1035. [DOI] [PubMed] [Google Scholar]
- 35.Jindal, H. K., C. B. Yong, G. M. Wilson, P. Tam, and C. R. Astell. 1994. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J. Biol. Chem. 269:3283-3289. [PubMed] [Google Scholar]
- 36.Kota, K. P., S. R. Wagner, E. Huerta, J. M. Underwood, and J. A. Nickerson. 2008. Binding of ATP to UAP56 is necessary for mRNA export. J. Cell Sci. 121:1526-1537. doi: 10.1242/jcs.021055. [DOI] [PubMed] [Google Scholar]
- 37.Legendre, D., and J. Rommelaere. 1992. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J. Virol. 66:5705-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehtonen, J. V., D. J. Still, V. V. Rantanen, J. Ekholm, D. Bjorklund, Z. Iftikhar, M. Huhtala, S. Repo, A. Jussila, J. Jaakkola, O. Pentikainen, T. Nyronen, T. Salminen, M. Gyllenberg, and M. S. Johnson. 2004. BODIL: a molecular modeling environment for structure-function analysis and drug design. J. Comput. Aided Mol. Des. 18:401-419. [DOI] [PubMed] [Google Scholar]
- 39.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512-518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 40.Li, X., and S. L. Rhode III. 1990. Mutation of lysine 405 to serine in the parvovirus H-1 NS1 abolishes its functions for viral DNA replication, late promoter trans activation, and cytotoxicity. J. Virol. 64:4654-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorson, C., L. R. Burger, M. Mouw, and D. J. Pintel. 1996. Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J. Virol. 70:834-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansilla-Soto, J., M. Yoon-Robarts, W. J. Rice, S. Arya, C. R. Escalante, and R. M. Linden. 2009. DNA structure modulates the oligomerization properties of the AAV initiator protein Rep68. PLoS Pathog. 5:e1000513. doi: 10.1371/journal.ppat.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newnham, C. M., and C. C. Query. 2001. The ATP requirement for U2 snRNP addition is linked to the pre-mRNA region 5′ to the branch site. RNA 7:1298-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuesch, J. P., J. Christensen, and J. Rommelaere. 2001. Initiation of minute virus of mice DNA replication is regulated at the level of origin unwinding by atypical protein kinase C phosphorylation of NS1. J. Virol. 75:5730-5739. doi: 10.1128/JVI.75.13.5730-5739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nuesch, J. P., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122-135. doi: 10.1006/viro.1995.1236. [DOI] [PubMed] [Google Scholar]
- 46.Nuesch, J. P., and P. Tattersall. 1993. Nuclear targeting of the parvoviral replicator molecule NS1: evidence for self-association prior to nuclear transport. Virology 196:637-651. doi: 10.1006/viro.1993.1520. [DOI] [PubMed] [Google Scholar]
- 47.Parrish, C. R. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183:195-205. [DOI] [PubMed] [Google Scholar]
- 48.Peck, M. L., and D. Herschlag. 2003. Adenosine 5′-O-(3-thio)triphosphate (ATPgammaS) is a substrate for the nucleotide hydrolysis and RNA unwinding activities of eukaryotic translation initiation factor eIF4A. RNA 9:1180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 50.Pujol, A., L. Deleu, J. P. Nuesch, C. Cziepluch, J. C. Jauniaux, and J. Rommelaere. 1997. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J. Virol. 71:7393-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reed, A. P., E. V. Jones, and T. J. Miller. 1988. Nucleotide sequence and genome organization of canine parvovirus. J. Virol. 62:266-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhode, S. L., III. 1985. trans-Activation of parvovirus P38 promoter by the 76K noncapsid protein. J. Virol. 55:886-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ribbeck, K., U. Kutay, E. Paraskeva, and D. Gorlich. 1999. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr. Biol. 9:47-50. [DOI] [PubMed] [Google Scholar]
- 54.Sakakibara, N., F. P. Schwarz, and Z. Kelman. 2009. ATP hydrolysis and DNA binding confer thermostability on the MCM helicase (dagger). Biochemistry 48:2330-2339. doi: 10.1021/bi801921j. [DOI] [PubMed] [Google Scholar]
- 55.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 56.Schaff, J., C. C. Fink, B. Slepchenko, J. H. Carson, and L. M. Loew. 1997. A general computational framework for modeling cellular structure and function. Biophys. J. 73:1135-1146. doi: 10.1016/S0006-3495(97)78146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheffzek, K., M. R. Ahmadian, W. Kabsch, L. Wiesmuller, A. Lautwein, F. Schmitz, and A. Wittinghofer. 1997. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277:333-338. [DOI] [PubMed] [Google Scholar]
- 58.Singleton, M. R., M. S. Dillingham, and D. B. Wigley. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76:23-50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 59.Slepchenko, B. M., J. C. Schaff, I. Macara, and L. M. Loew. 2003. Quantitative cell biology with the virtual cell. Trends Cell Biol. 13:570-576. [DOI] [PubMed] [Google Scholar]
- 60.Smith, P. C., N. Karpowich, L. Millen, J. E. Moody, J. Rosen, P. J. Thomas, and J. F. Hunt. 2002. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snider, J., and W. A. Houry. 2008. AAA+ proteins: diversity in function, similarity in structure. Biochem. Soc. Trans. 36:72-77. doi: 10.1042/BST0360072. [DOI] [PubMed] [Google Scholar]
- 62.Sousa, M. C., C. B. Trame, H. Tsuruta, S. M. Wilbanks, V. S. Reddy, and D. B. McKay. 2000. Crystal and solution structures of an HslUV protease-chaperone complex. Cell 103:633-643. [DOI] [PubMed] [Google Scholar]
- 63.Suikkanen, S., K. Saajarvi, J. Hirsimaki, O. Valilehto, H. Reunanen, M. Vihinen-Ranta, and M. Vuento. 2002. Role of recycling endosomes and lysosomes in dynein-dependent entry of canine parvovirus. J. Virol. 76:4401-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor, J. S. 1981. Sarcoplasmic reticulum ATPase catalyzes hydrolysis of adenyl-5′-yl imidodiphosphate. J. Biol. Chem. 256:9793-9795. [PubMed] [Google Scholar]
- 65.Wagner, S., S. Chiosea, M. Ivshina, and J. A. Nickerson. 2004. In vitro FRAP reveals the ATP-dependent nuclear mobilization of the exon junction complex protein SRm160. J. Cell Biol. 164:843-850. doi: 10.1083/jcb.200307002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, D., W. Yuan, I. Davis, and C. R. Parrish. 1998. Nonstructural protein-2 and the replication of canine parvovirus. Virology 240:273-281. doi: 10.1006/viro.1997.8946. [DOI] [PubMed] [Google Scholar]
- 67.Weidemann, T., M. Wachsmuth, T. A. Knoch, G. Muller, W. Waldeck, and J. Langowski. 2003. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J. Mol. Biol. 334:229-240. [DOI] [PubMed] [Google Scholar]
- 68.Willwand, K., A. Q. Baldauf, L. Deleu, E. Mumtsidu, E. Costello, P. Beard, and J. Rommelaere. 1997. The minute virus of mice (MVM) nonstructural protein NS1 induces nicking of MVM DNA at a unique site of the right-end telomere in both hairpin and duplex conformations in vitro. J. Gen. Virol. 78:2647-2655. [DOI] [PubMed] [Google Scholar]
- 69.Wilson, G. M., H. K. Jindal, D. E. Yeung, W. Chen, and C. R. Astell. 1991. Expression of minute virus of mice major nonstructural protein in insect cells: purification and identification of ATPase and helicase activities. Virology 185:90-98. [DOI] [PubMed] [Google Scholar]
- 70.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 A resolution. J. Mol. Biol. 264:497-520. doi: 10.1006/jmbi.1996.0657. [DOI] [PubMed] [Google Scholar]
- 71.Yeung, D. E., G. W. Brown, P. Tam, R. H. Russnak, G. Wilson, I. Clark-Lewis, and C. R. Astell. 1991. Monoclonal antibodies to the major nonstructural nuclear protein of minute virus of mice. Virology 181:35-45. [DOI] [PubMed] [Google Scholar]
- 72.Young, P. J., K. T. Jensen, L. R. Burger, D. J. Pintel, and C. L. Lorson. 2002. Minute virus of mice NS1 interacts with the SMN protein, and they colocalize in novel nuclear bodies induced by parvovirus infection. J. Virol. 76:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.