Abstract
Drug resistance mutations in HIV-1 protease selectively alter inhibitor binding without significantly affecting substrate recognition and cleavage. This alteration in molecular recognition led us to develop the substrate-envelope hypothesis which predicts that HIV-1 protease inhibitors that fit within the overlapping consensus volume of the substrates are less likely to be susceptible to drug-resistant mutations, as a mutation impacting such inhibitors would simultaneously impact the processing of substrates. To evaluate this hypothesis, over 130 HIV-1 protease inhibitors were designed and synthesized using three different approaches with and without substrate-envelope constraints. A subset of 16 representative inhibitors with binding affinities to wild-type protease ranging from 58 nM to 0.8 pM was chosen for crystallographic analysis. The inhibitor-protease complexes revealed that tightly binding inhibitors (at the picomolar level of affinity) appear to “lock” into the protease active site by forming hydrogen bonds to particular active-site residues. Both this hydrogen bonding pattern and subtle variations in protein-ligand van der Waals interactions distinguish nanomolar from picomolar inhibitors. In general, inhibitors that fit within the substrate envelope, regardless of whether they are picomolar or nanomolar, have flatter profiles with respect to drug-resistant protease variants than inhibitors that protrude beyond the substrate envelope; this provides a strong rationale for incorporating substrate-envelope constraints into structure-based design strategies to develop new HIV-1 protease inhibitors.
Human immunodeficiency virus type 1 (HIV-1) infects an estimated three million people every year worldwide (12). The viral life cycle is critically influenced by the activity of one enzyme, HIV-1 protease, which processes the Gag and Gag-Pol polyproteins into structural and functional proteins essential for proper virion assembly and maturation (7). Inhibition of HIV-1 protease results in immature, noninfectious viral particles. Thus, HIV-1 protease is a prime target for the rational design of anti-HIV-1 therapeutics.
To date, the U.S. Food and Drug Administration (FDA) has approved nine HIV-1 protease inhibitors (PIs): saquinavir (SQV), indinavir (IDV), ritonavir (RTV), nelfinavir (NFV), amprenavir (APV), lopinavir (LPV), atazanavir (ATV), tipranavir (TPV), and darunavir (DRV) (8, 9, 13-15, 17, 22-26). The development of these PIs is considered a major success of structure-based drug design, since they have dramatically reduced mortality and morbidity rates for AIDS patients. However, this success has not ended the need for new PIs, as the existing inhibitors are becoming increasingly ineffective against rapidly emerging drug-resistant HIV-1 mutants (5, 6, 19). Therefore, new inhibitors need to be designed with broad specificity not only for existing drug-resistant variants of HIV-1 but also for drug-resistant mutants that may emerge in the future.
All HIV-1 PIs in clinical use are competitive inhibitors that compete with protease substrates by binding at the active site of the enzyme. Because of drug-resistant mutations in protease, it is no longer being efficiently inhibited by PIs, but it still recognizes its substrates and cleaves them into the individual proteins necessary for viral maturation (10). To understand the mechanism by which protease recognizes the viral substrates, we analyzed the crystal structures of six substrates in complex with an inactive (D25N) protease variant and found that the volumes of the substrates overlapped in the active site of the protease (21). This consensus volume, or conserved shape, which we defined as the substrate envelope, was hypothesized to determine substrate specificity for HIV-1 protease. Comparison of this substrate envelope with the crystal structures of FDA-approved PIs in complex with wild-type protease revealed that some inhibitor atoms protrude beyond the envelope (16). The protruding inhibitor atoms contacted protease residues that mutate in HIV-1-infected patients to develop drug resistance to PI therapy. These protease residues are important for inhibitor binding but not for substrate binding. The two observations referred to above led to the substrate-envelope hypothesis: HIV-1 protease inhibitors that fit completely within the substrate envelope are less likely to be susceptible to drug resistance mutations (16, 21).
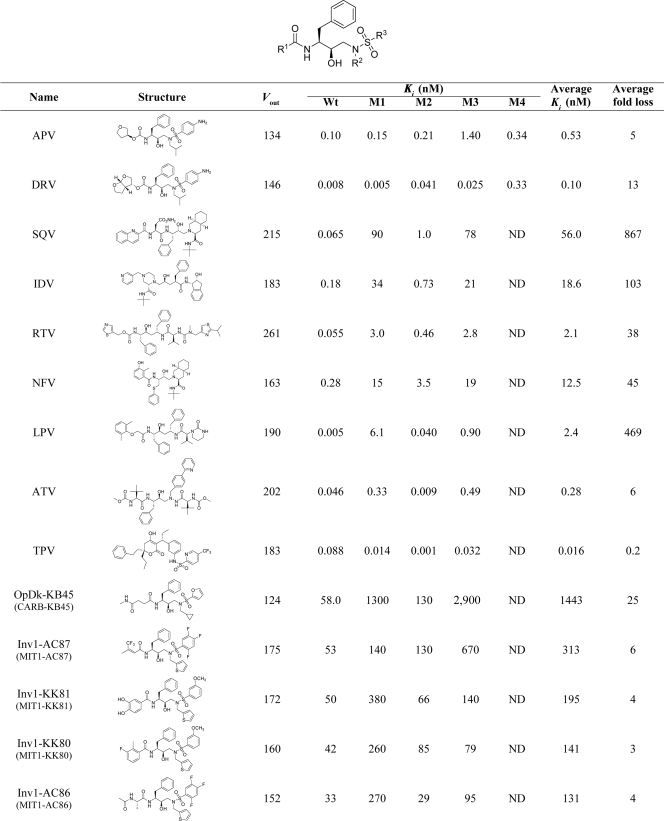
The substrate-envelope hypothesis can be used to design new inhibitors that fit within the substrate envelope, thus possibly eluding drug resistance, because mutations that decreased inhibitor binding would also affect substrate processing. To evaluate the substrate-envelope hypothesis, new protease inhibitors were designed based on the (R)-(hydroxyethylamino)sulfonamide isostere (Table 1), the scaffold present in the clinical inhibitors APV and DRV (1, 2, 4, 20). This scaffold fits predominately within the substrate envelope and has three sites to which various substituent groups can be easily attached to manipulate functional characteristics. By using various substituent groups at these three sites (R1, R2, and R3) on the (R)-(hydroxyethylamino)sulfonamide isostere, more than 130 new inhibitors were designed and synthesized. The binding affinities of these inhibitors to wild-type HIV-1 protease were measured as previously described (1, 18). Representative compounds from the designed inhibitors were also tested against a panel of three to four drug-resistant protease variants. These drug-resistant protease variants are prototypes of commonly found drug resistance patterns seen with PI-treated patients.
TABLE 1.
Vout values and the experimental Ki values of binding against the wild-type and resistant variants of HIV-1 protease for the 9 FDA approved and 16 designed protease inhibitors a
Wt, wild-type; M1, L10I, G48V, I54V, L63P, V82A; M2, D30N, L63P, N88D; M3, L10I, L63P, A71V, G73S, I84V, L90M; M4, I50V, A71V; average fold loss, average ratio between the Ki values against mutants and the wild-type protease; ND, not determined. The labeling of the Inv, OpDk, and SAR compounds is different in the previously published papers (references 1, 2, and 4) and is given in parentheses for each inhibitor. The binding affinity data were previously published (see references 1, 2, and 4). *, updated values; †, new data.
All the inhibitors were designed using three design approaches: optimized docking (OpDk) (previously CARB) (4), inverse design (Inv) (previously MIT) (2), and structure activity relationship (SAR) (1). Both the OpDk and Inv libraries were designed by computational methods, using the substrate envelope as an added constraint. Briefly, OpDk used a two-step procedure: first, available substituents at the R1, R2, and R3 positions were adjusted one position at a time with the other two substituents held constant, generating 150 optimal candidate substituents at each position, based on their fit to the substrate envelope and a simple energy-based affinity-scoring function; second, a genetic algorithm was used to discover optimal combinations of the most promising substituents. This OpDk approach led to the design and synthesis of 26 inhibitors with binding affinities ranging from >10 μM to 24 nM (reference 4 and unpublished results).
The second computational design method, Inv, is complementary to standard docking approaches. This procedure used a library of scaffold molecules containing (R)-(hydroxyethylamino)sulfonamide isostere and placed them discretely within the substrate envelope. A combinatorial search of candidate substituents was then performed to identify molecules that do not extend beyond the substrate envelope. This design resulted in a rank-ordered list of inhibitors based on estimated binding affinities. In the first round of design (with the Inv1 library), 15 inhibitors were synthesized with binding affinities to wild-type protease ranging from 26 μM to 30 nM, values which represent affinities that are weaker than the binding affinities of clinically approved PIs. Based on retrospective analysis of the Inv1 library, a second library (the Inv2 library) was designed by reoptimizing the computational algorithm, using successful substituent R groups from the first round and the crystal structure of a DRV-protease complex (1T3R) instead of the crystal structure of a substrate-protease complex (1KJG). In the second round of Inv, 36 inhibitors were synthesized with binding affinities to wild-type protease ranging from 4.2 nM to 14 pM (3 orders of magnitude better than the binding affinities seen with the first-round inhibitors) (2). Compounds in the third library (SAR) were designed based on the same consensus scaffold, (R)-(hydroxyethylamino)sulfonamide isostere, without using any explicit substrate-envelope constraint. A heterocyclic moiety with multiple polar atoms, N-phenyloxazolidinone-5-carboxamide, was used as an R1 group to mimic the critical hydrogen bonds formed by tetrahydrofuranyl (THF)/bis-THF moieties in APV and DRV. Substituents were adjusted on the phenyl group at the nitrogen of the oxazolidinone ring to get different R1 groups. The SAR library inhibitors bound to wild-type protease with affinities ranging from 250 nM to 0.8 pM (1).
A set of 16 representative inhibitors was chosen from the high-affinity compounds generated by these three design schemes, and their crystal structures in complex with the wild-type HIV-1 protease were determined. In the current report, the crystal structures of these 16 protease-inhibitor complexes are analyzed in detail. The 16 inhibitors bound to wild-type protease with affinities ranging from 58 nM to 0.8 pM and comprised the 6 best inhibitors from the OpDk and Inv1 libraries and 10 inhibitors from Inv2 and SAR libraries. Analyses of the van der Waals (vdW) contacts and hydrogen bonds in all of the inhibitor-protease complexes showed that the nanomolar and picomolar inhibitors interacted differently with some active-site residues of the protease. The extent to which these inhibitors protrude beyond the substrate envelope generally correlates with the loss of their inhibitory activity against the mutant proteases, providing a rationale for using substrate-envelope constraints in designing new protease inhibitors.
MATERIALS AND METHODS
Nomenclature for inhibitors.
Wild-type protease-inhibitor complexes are distinguished from inhibitors by subscript notations. For example, APVWT denotes the complex of APV with wild-type protease, whereas APV denotes the inhibitor itself.
Structural analysis. (i) Estimation of van der Waals contact energy.
The van der Waals interaction energy can be calculated as a “6-12” or Lennard-Jones potential, with a long-range shallow attractive interaction and a short-range repulsive one. While this potential works well in computing interactions in a molecular dynamics simulation, the repulsive term is too restrictive in assessing particular interactions of an experimentally determined crystal structure, where slight changes in position can cause a favorable interaction to be considered unfavorable.
Protease-inhibitor van der Waals contacts were computed using a simplified “6-12” or Lennard-Jones potential, V(r), and the relation 4ɛ[(σ/r)12 − (σ/r)6, where r represents the protease-inhibitor interatomic distance and ɛ and σ represent the depth of the potential well and the collision diameter, respectively, for each protease inhibitor atom pair. V(r) was computed for all possible protease-inhibitor atom pairs within 5 Å, and potentials for nonbonded pairs separated by less than the distance at the minimum of the potential were equated to −ɛ. Using this simplified potential value for each nonbonded protease-inhibitor atom pair, the total van der Waals contact energy, ΣV(r), was then computed for each protease residue and the inhibitor molecule.
(ii) Crystal structure of APV with wild-type protease.
The crystal structure of wild-type protease complexed with APV (3EKV) was determined to a resolution of 1.8 Å. It crystallized in the P212121 space group with one protease dimer per asymmetric unit. The inhibitor has one orientation with clear electron density for all the atoms. The APVWT in the Protein Data Bank (PDB) (1HPV) was determined to a resolution of 1.9 Å in the P61 space group. The conformations of APV in the 1HPV and 3EKV structures are different, especially at the nitrogen atom attached to the sulfone group. In principle, two geometries are possible at the sulfonamide nitrogen atom site of APV because of the possibility of umbrella inversion. The two geometries are observed in the two crystal structures of APV (1HPV and 3EKV). The orientations of the isobutyl group of APV in the two structures are different, maybe because of the umbrella inversion at the nitrogen atom. The observed orientations of the isobutyl group and the geometries at the nitrogen atom of APV in 3EKV are similar to those of all DRV structures, which have the same molecular core as APV. Since all the newly designed inhibitors crystallized in the P212121 space group with dimensions similar to those of the new APVWT (3EKV), this structure was used for comparative analysis.
RESULTS
Overall structural determination.
All complexes of the 16 designed inhibitors with the wild-type protease crystallized in the P212121 space group, with similar cell dimensions and one protease dimer per asymmetric unit. The crystallographic and refinement statistics for all structures are shown (Table 2). The structures were refined to resolutions from 2.1 to 1.7 Å. The electron densities for all inhibitor and protease atoms were well ordered in each structure. One conformation was modeled for the inhibitor in all structures except for inhibitor Inv2-AD86, which was modeled with two conformations. All crystal structures had some residues that were modeled in multiple conformations, though most are outside the active site. In eight complexes, two conformations of V82 were observed within the active site, and in the complex of inhibitor OpDk-KB45 with the wild-type protease (OpDk-KB45WT; for the labeling conventions used for complexes, see Materials and Methods), multiple conformations were observed for I50 and G51. The crystal structures of the 16 new inhibitor-protease complexes were compared with the structure of the APVWT complex. The quality and similarities of the crystallographic parameters for these structures permit the inhibitor-protease complexes to be compared in detail.
TABLE 2.
Crystallographic and refinement statistics
| Parametera | Value for indicatedprotease inhibitor |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APV | OpDk (KB45) | Inv1 |
OpDk (AD37) | Inv2 |
SAR |
||||||||||||
| AC87 | KK81 | KK80 | AC86 | AD94 | AD86 | AD93 | KB98 | KC08 | KB60 | KB62 | AD78 | KB19 | AD81 | ||||
| >Resolution (Å) | 1.75 | 1.75 | 1.85 | 2.00 | 2.10 | 1.90 | 1.93 | 1.95 | 1.85 | 1.80 | 1.85 | 1.85 | 1.85 | 1.80 | 1.85 | 1.80 | 1.95 |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| a (Å) | 50.72 | 50.69 | 50.74 | 50.83 | 50.91 | 50.67 | 50.65 | 50.88 | 50.80 | 50.86 | 50.91 | 50.88 | 50.81 | 50.83 | 50.80 | 50.77 | 50.78 |
| b (Å) | 57.40 | 57.59 | 57.94 | 58.06 | 58.11 | 57.98 | 57.64 | 58.37 | 58.04 | 58.18 | 58.26 | 58.23 | 58.13 | 58.34 | 58.02 | 57.65 | 58.31 |
| c (Å) | 61.74 | 61.84 | 61.70 | 61.76 | 61.63 | 61.54 | 61.78 | 61.84 | 61.76 | 61.80 | 61.81 | 61.81 | 61.57 | 61.63 | 61.67 | 61.89 | 61.69 |
| Z | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Rmerge (%) | 2.9 | 4.5 | 6.5 | 5.4 | 5.1 | 4.7 | 6.5 | 6.6 | 5.8 | 4.2 | 4.4 | 4.2 | 4.3 | 5.0 | 4.1 | 5.0 | 4.7 |
| Completeness (%) | 79.7 | 98.4 | 95.4 | 99.7 | 99.0 | 98.6 | 99.4 | 98.2 | 99.9 | 94.5 | 92.5 | 95.8 | 99.7 | 98.4 | 95.7 | 95.6 | 95.3 |
| Total no. of reflections | 49,469 | 114,039 | 86,034 | 85,715 | 76,441 | 98,469 | 92,428 | 94,050 | 89,209 | 106,905 | 75,550 | 97,446 | 10,702 | 10,357 | 10,999 | 10,706 | 65,161 |
| No. of unique reflections | 14,987 | 18,590 | 15,443 | 12,846 | 11,126 | 14,666 | 14,044 | 13,691 | 16,171 | 16,632 | 15,105 | 15,597 | 16,121 | 17,324 | 15,595 | 16,632 | 13,238 |
| Rfree (%) | 22.6 | 20.8 | 20.5 | 20.5 | 23.0 | 19.7 | 19.8 | 21.0 | 20.3 | 20.3 | 21.2 | 21.0 | 22.1 | 19.4 | 19.6 | 18.5 | 21.6 |
| Rfactor (%) | 19.0 | 16.5 | 16.6 | 15.6 | 17.0 | 15.9 | 16.1 | 17.4 | 17.4 | 17.3 | 17.0 | 17.3 | 16.9 | 17.0 | 16.4 | 16.0 | 16.6 |
| RMSD | |||||||||||||||||
| Bond length (Å) | 0.007 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.007 | 0.007 | 0.008 | 0.008 | 0.008 | 0.007 | 0.008 |
| Bond angle (°) | 1.414 | 1.266 | 1.227 | 1.211 | 1.267 | 1.240 | 1.221 | 1.160 | 1.215 | 1.193 | 1.253 | 1.298 | 1.403 | 1.356 | 1.283 | 1.232 | 1.294 |
| Temperature (°C) | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 | −80 |
| PDB no. | 3EKV | 2PSU | 2QHZ | 2QI1 | 2QI0 | 2QHY | 2PSV | 2QI3 | 2QI7 | 2QI4 | 2QI6 | 2QI5 | 3GI4 | 3GI5 | 3GI6 | 2I0A | 2I0D |
Z, number of molecules in the unit cell; RMSD, root mean square deviation; PDB, Protein Data Bank.
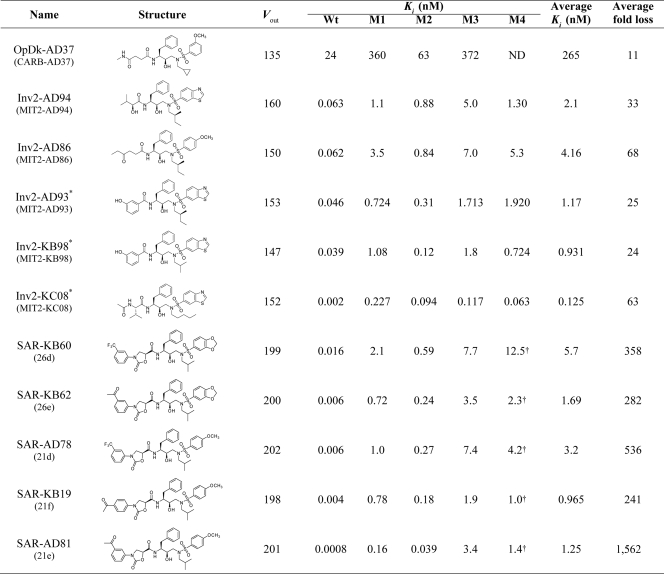
To compare the overall structures of the complexes, their backbones were superimposed. The alpha carbon backbone of each complex differed from that of the APVWT complex by a root mean square deviation (RMSD) of 0.18 to 0.39Å. Slight variations (RMSD of 0.70) were observed in the flaps for 6 nanomolar inhibitors, whereas 8 of the 10 pM inhibitor-protease complexes superimposed very closely, with a maximum RMSD of 0.35 (Fig. 1). The two exceptions, the complexes of Inv2-KC08 and Inv2-AD94, have benzothiazole as an R3 group on one side and different small R1 groups on the other, a finding that deviates from the tight conformational clustering seen with the other picomolar complexes. While much of the core protease structures exhibited extensive overlap, the variations in flap conformation were not significant enough to distinguish the picomolar inhibitor complexes from the nanomolar inhibitor complexes.
FIG. 1.
Superposition of the crystal structures of HIV-1 protease in complex with nanomolar inhibitors Inv1-AC86 (green) and OpDk-AD37 (orange) and picomolar inhibitors APV (cyan), Inv2-AD93 (purple), and SAR-KB19 (yellow). (Top panel) An α-carbon trace of all the 16 complexes shows that most of the core protease structure extensively overlaps. A slight variation in the flaps is observed among nanomolar inhibitors, whereas the flaps tightly superpose among picomolar inhibitors. The variation is not significant for differentiating the nanomolar and picomolar inhibitors. (Bottom panel) View down the flaps (inset).
Differential van der Waals contacts between inhibitors.
The interactions within a protease-inhibitor complex can be assessed by analyzing the interactions within the complex, i.e., hydrogen bonds and van der Waals (vdW) contacts. Until recently, vdW contacts were calculated using a simple distance criterion; however, that approach does not accurately distribute the energetic contributions. To overcome this problem and to quantitatively estimate vdW contacts, we have developed a simplified vdW calculation in which the attractive potential is retained but the repulsive potential is removed. This method was used to calculate the distribution of vdW contacts for APV and the 16 new inhibitors for all protease active-site residues.
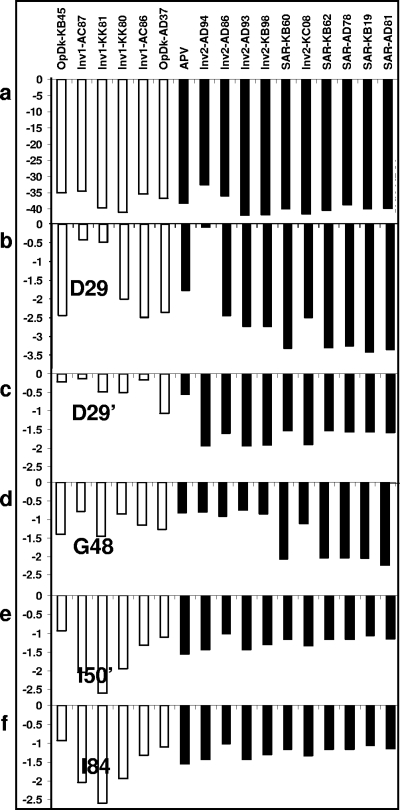
The total vdW contact energy values for APV and each of the 16 inhibitors in complex with the wild-type protease are shown (Fig. 2a). The binding affinity of APV was 100 pM, six inhibitors had binding affinities of 20 to 60 nM, and the other 10 inhibitors had affinities of 0.8 to 63 pM. Only one conformation of Inv2-AD86 was used in the analysis, as its protease contacts (except for D30 and I47) are fairly similar to those of the other conformation. The total vdW energy did not correlate with inhibitor binding affinity to the protease: many high-affinity inhibitors as well as some low-affinity inhibitors made extensive vdW contacts with protease.
FIG. 2.
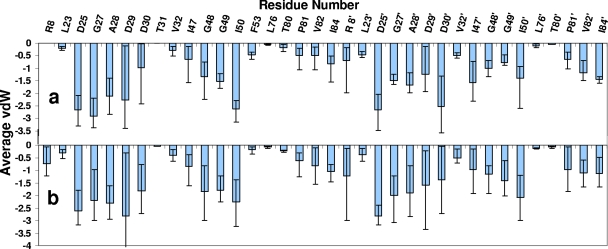
Approximate vdW contacts between inhibitors and wild-type protease calculated using the simplified force field. The nanomolar inhibitors are shown in white, and the picomolar inhibitors are shown in black. (a) vdW contacts to all protease residues for each inhibitor. (b to f) vdW contacts to residues D29, D29′, G48, I50′, and I84 calculated for each inhibitor. The vertical scale represents an approximation of van der Waals energy, shown as kilocalories/mole.
To better understand the differences in binding between nanomolar and picomolar inhibitors, the vdW contact values for all inhibitors were averaged and analyzed on a residue-by-residue basis (Fig. 3a) and compared with those of the FDA-approved PIs (Fig. 3b). All the inhibitors interacted with a subset of 34 residues in the protease active site: L23, D25, G27, A28, D29, D30, V32, I47, G48, G49, I50, F53, L76, T80, P81, V82, I84, R8′, L23′, D25′, G27′, A28′, D29′, D30′, V32′, I47′, G48′, G49′, I50′, L76′, T80′, P81′, V82′, and I84′. Some of these residues (D25, G27, A28, D29, I50, D25′, A28′, and D30′) consistently made extensive contacts with inhibitors, while others made minimal contacts.
FIG. 3.
Average vdW contacts to each active site residue calculated using the simplified force field for 16 newly designed inhibitors (a) and all FDA-approved inhibitors (b). “Error” bars represent the minimum and maximum vdW energy values for contacts between the inhibitors and a particular residue. The vertical scale is an approximation of van der Waals energy (shown as kilocalories/mole).
Although the variability for some contacts did not directly correlate with binding affinity, the extent of contact for a subset of residues does seem to distinguish nanomolar from picomolar inhibitors. In most cases, picomolar inhibitors made extensive contacts with D29 and D29′ whereas fewer such contacts were formed by nanomolar inhibitors (Fig. 2b and c). All SAR compounds made extensive contacts with G48 (Fig. 2d). Interestingly, D29 is an invariant residue in the floor of the active site, and G48 formed a conserved hydrogen bond with protease substrates. In contrast, more extensive contacts with I84 (in the P-loop region) and I50′ (in the flap region) were strongly associated with inhibitors that bound poorly to protease (Fig. 2e to f). It is also of interest that mutations at I84 and I50 were associated with multidrug resistance; thus, high-affinity inhibitors that do not make extensive contacts with these resistance-prone residues could be designed.
Each active site protease residue interacted with the inhibitors to a varying degree. Figure 3 shows average vdW contact values for each active site residue for 16 newly designed inhibitors and all FDA-approved inhibitors. The “error” bars represent the variations in contacts and the minimum and maximum vdW energy values for contacts between the inhibitors and a particular residue. Small variations for a given residue, e.g., I84′, indicate that it was contacted by all the inhibitors to similar extents, whereas large variations reflect the finding that a particular residue, e.g., D29, contacted some inhibitors more than others. Interestingly, the variations in contact values for each residue for the new inhibitors (Fig. 3a) were quite different from the variations in contact values seen for FDA-approved inhibitors (Fig. 3b).
Conservation and variation of hydrogen bonds.
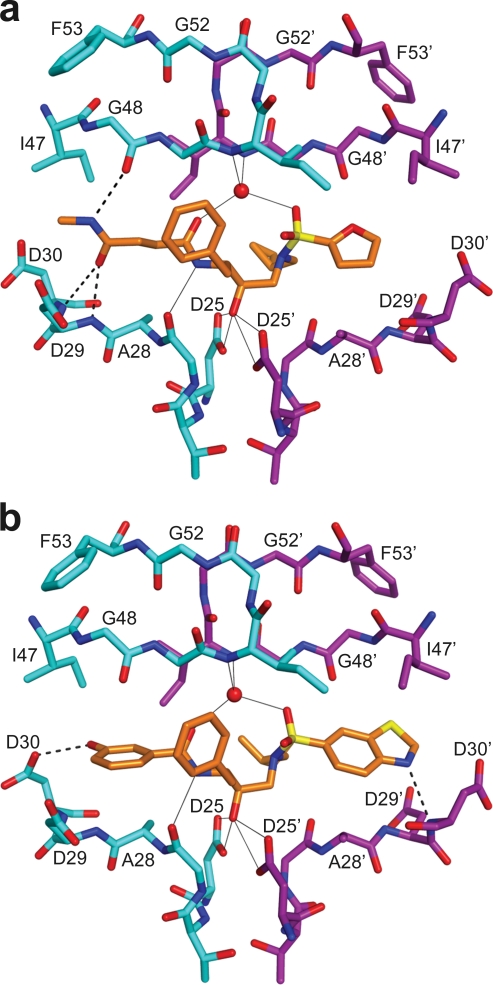
Hydrogen bonds confer specificity in protease-ligand interactions. APV and the 16 new inhibitors formed both direct and water-mediated hydrogen bonds to the protease (Table 3). All inhibitors formed seven hydrogen bonds (five direct and two water-mediated; see solid lines in Fig. 4) from the central (R)-(hydroxyethylamino)sulfonamide isostere, the scaffold used for design of the inhibitors. The secondary hydroxyl group in the center of this inhibitor core forms four hydrogen bonds to the two catalytic aspartic acid residues. The amide nitrogen on the inhibitor core forms a direct hydrogen bond to the carbonyl oxygen of the G27 residue. A water molecule mediates hydrogen bonds between the two protease flaps and the carbonyl oxygen and one sulfone oxygen atom on the inhibitor. In fact, this water-mediated hydrogen bond is a conserved hydrogen bond present in all protease-substrate and peptidomimetic inhibitor complexes.
TABLE 3.
Number of hydrogen bonds in APV and 16 designed inhibitors
| Inhibitor | No. of direct H bonds | No. of water-mediated H bonds | Total no. of H bonds |
|---|---|---|---|
| APV | 9 | 2 | 11 |
| OpDk-KB45 | 8 | 2 | 10 |
| Inv1-AC87 | 5 | 2 | 7 |
| Inv1-KK81 | 7 | 5 | 12 |
| Inv1-KK80 | 5 | 4 | 9 |
| Inv1-AC86 | 7 | 3 | 10 |
| OpDk-AD37 | 8 | 2 | 10 |
| Inv2-AD94 | 7 | 4 | 11 |
| Inv2-AD86 | 7 | 3 | 10 |
| Inv2-AD93 | 8 | 2 | 10 |
| Inv2-KB98 | 8 | 2 | 10 |
| Inv2-KC08 | 9 | 3 | 12 |
| SAR-KB60 | 11 | 7 | 18 |
| SAR-KB62 | 11 | 7 | 18 |
| SAR-AD78 | 10 | 5 | 15 |
| SAR-KB19 | 11 | 5 | 16 |
| SAR-AD81 | 10 | 5 | 15 |
FIG. 4.
Hydrogen bonding interactions between wild-type HIV-1 protease and OpDk-KB45, a nanomolar inhibitor (a), and Inv2-AD93, a picomolar inhibitor (b). The two protease monomers are shown in cyan and magenta. Oxygen, nitrogen, and sulfur atoms are shown in red, blue, and yellow, respectively. The hydrogen bonds formed by the scaffold, (R)-(hydroxyethylamino)sulfonamide, in all the 16 inhibitor-protease structures are shown by continuous lines. Hydrogen bonds specific to the inhibitors are shown by dashed lines. All the picomolar inhibitors seemed to be locked into the active site by making at least one hydrogen bond to residues D29 and/or D30 in the floor of the active-site in both monomers, whereas the nanomolar inhibitors either formed no hydrogen bonds to residues D29 and/or D30 or formed bonds to these residues in only in one of the monomers. The hydrogen bonds formed between the inhibitor and protease flap residues do not seem to influence inhibitor activity.
The variations in hydrogen bonding among the 16 inhibitors depend only on the substituents at R1 and R3, as the R2 groups do not have hydrogen bond donors or acceptors. Comparing the specifics of hydrogen bonding between different inhibitors, the protease may distinguish which hydrogen bonds are responsible for affinity and/or specificity. Apart from residues D25, G27, and I50, which form hydrogen bonds with all the inhibitors, the residues that might form hydrogen bonds with the inhibitors are D29 and D30 in the floor of the active site and G48 in the flap region. APV formed 11 hydrogen bonds with the protease, including the 7 hydrogen bonds mentioned above (Table 3). All the inhibitors designed using the SAR approach formed at least four additional hydrogen bonds. The remaining inhibitors each formed 9 to 12 hydrogen bonds to the protease, except for inhibitor Inv1-AC87, which formed only 7 hydrogen bonds. The nanomolar inhibitors (except Inv1-AC87) formed either direct or water-mediated hydrogen bonds to one or both G48 residues in the protease flaps. These nanomolar inhibitors also formed hydrogen bonds (direct or water mediated) to D29 and/or D30, but only in one protease monomer, not both. In contrast, the picomolar inhibitors formed a less-conserved hydrogen bond (direct or water mediated) to G48. However, all picomolar inhibitors formed hydrogen bonds (direct or water mediated) to both sides of the floor of the active site at D29 and/or D30. The pattern seen for picomolar inhibitors is conserved among all FDA-approved peptidomimetic inhibitors, except for LPV. Thus, tightly binding picomolar inhibitors appear to “lock” into the active site by making hydrogen bonds to particular sites on both sides of the floor of the active site. The exact number of hydrogen bonds does not distinguish between nanomolar and picomolar binding affinity of the inhibitors, but certain combinations of hydrogen bonds are associated with high affinity.
Impact of plasticity of HIV-1 protease on binding ligands.
Although the 16 inhibitors analyzed in this study had the same molecular scaffold, their various R1, R2, and R3 substituents generated a range of inhibitor sizes and shapes and a range of affinities for the wild-type protease. The conformation of the protease responded to each compound somewhat differently, and much of the conformational variation was localized in the flaps and P-1 loops. For a few protease residues, the variations in vdW contacts distinguished nanomolar from picomolar inhibitors. However, these residues formed a small subset of all vdW contacts; the remaining residues either contacted the inhibitors in a uniform manner or differed with the particular functional group, without an obvious correlation to affinity. Except for the conformations of a few residues that may specify protease affinity to the inhibitor, most protease residues seem to adapt to the shape of the inhibitor by a combination of backbone and side chain rearrangements throughout the enzyme. This finding implies that the active site of HIV-1 protease is a highly plastic and interdependent binding site, where changes in one region can propagate throughout the enzyme and influence inhibitor affinity and specificity, sometimes unexpectedly.
The plasticity of the binding site has implications for computational inhibitor design. Thus, the Inv1 inhibitors which were designed using the substrate-bound protease structure (1KJG [micromolar affinity]) yielded compounds with micromolar affinities, and retrospective calculations were unable to distinguish high- from low-affinity inhibitors (2). In contrast, when the crystal structure of protease bound to the picomolar affinity inhibitor DRV (1T3R) was used for designing PIs, high-affinity inhibitors were obtained and the correlation between calculations and experiments was improved. Comparison of the two protease crystal structures (1KJG and 1T3R) shows significant differences of approximately 1 Å in the backbone conformation, chiefly localized in the flap and P-1 loop regions but also found throughout the enzyme. The protease structure was not allowed to relax or change throughout the inhibitor design process. These results suggest that it is preferable to base inhibitor design on a target structure solved with an existing high-affinity inhibitor, if available. Perhaps part of the explanation for this finding is that the protease conformations induced by higher-affinity ligands are more intrinsically stable. This concept may well generalize to other therapeutic targets.
Fit of inhibitors in the substrate envelope.
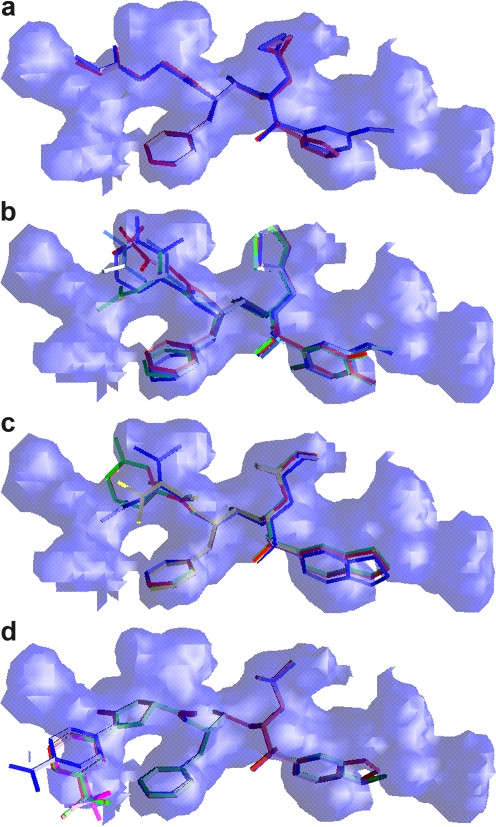
The validity of the substrate-envelope hypothesis was tested by comparing inhibitors designed with and without application of this criterion. The fit of the 16 inhibitors to the substrate envelope was assessed quantitatively using Vout (3), which represents the volume of the inhibitor outside the substrate envelope. Each inhibitor was also examined qualitatively by looking at the extent to which its atoms protruded beyond the envelope. These assessments produce complementary interpretations of how the 16 inhibitors fit within the active site and within the substrate envelope or substrate-binding region (Table 1 and Fig. 5). The Vout values for APV and the 16 inhibitors range from 124 to 202 (where smaller Vout values signify a better fit of the inhibitor in the substrate envelope). APV, which has one of the best fits, has a Vout of 134. The molecular core atoms in APV that protrude beyond the envelope are the oxygen of the hydroxyl group that forms hydrogen bonds with the catalytic aspartic acids, D25 and D25′, and one oxygen on the sulfone group. These two oxygen atoms, which are part of the conserved APV molecular core, also protrude beyond the substrate envelope in all 16 novel inhibitor complexes. The range of fits for similar inhibitors allows comparison of the different design schemes.
FIG. 5.
Superposition of the inhibitors onto the substrate envelope, showing the fit of the inhibitors in the substrate envelope designed by three methodologies: optimized docking (OpDk [a]), inverse design (Inv1 [b] and Inv2 [c]), and structure activity relationship (SAR [d]).
The two compounds designed using the optimized docking scheme, OpDk-AD37 and OpDk-KB45, had the same R1 and R2 groups but different R3 groups. All three R-groups of the two OpDk inhibitors fit well within the substrate envelope, in agreement with the computational design (Fig. 5a). The fit of the OpDk inhibitors within the substrate envelope also correlates with their Vout values. The Vout for OpDk-KB45 is 135, a value similar to that determined for APV, while the OpDk-AD37 Vout is only 124. Although these two inhibitors fit well within the substrate envelope, they exhibited only nanomolar-level binding affinity, suggesting that the relative weighting values for the substrate envelope and docking interactions were not sufficiently optimized to produce high-affinity compounds.
The second set of computationally designed inhibitors was made using the Inv scheme. The nine compounds based on this method had Vout values from 147 to 175, all greater than that of APV. The four nanomolar inhibitors designed in the first round had a thiophene ring as their R2 group, different R1 and R3 groups, and high Vout values (152 to 175). The orientations of the thiophene ring were similar in all four crystal structures, with 3 atoms of the 5-membered thiophene ring protruding beyond the envelope (Fig. 5b). Occasionally, one or two additional atoms at R1 and R3 also protruded beyond the substrate envelope. The five inhibitors from the second round of the inverse design all bound with picomolar affinity and had Vout values from 147 to 160 (Fig. 5c). Four of the 5 second-round Inv inhibitors had a benzothiazole group at R3. In all four structures containing this R3 group, three atoms in the 5-membered ring of the benzothiazole group laid outside the substrate envelope. The fifth compound, Inv2-AD86, had a 4-methoxyphenyl group at R3, and the two atoms of the methoxy group protruded beyond the envelope. The R2 groups of all five inhibitors fit well within the substrate envelope. The R1 group of Inv2-KC08 fits within the envelope, while one atom of the R1 groups of Inv2-KB98, Inv2-AD93, and Inv2-AD94 protruded beyond the envelope. The R1 group of the fifth inhibitor, Inv2-AD86, had two conformations in the crystal structure. In one conformation, R1 fits within the envelope, whereas in the second conformation, most of the R1 group laid outside the envelope. Interestingly, the average of these two conformations corresponds to the R1 group of the predicted structure, which fits within the envelope. Whereas all nine inhibitors were designed using the Inv scheme, the five inhibitors from the second round had a much better affinity and fit in the substrate envelope than the first-round inhibitors, likely due to reoptimization of the design method.
The inhibitors designed using the SAR approach without any substrate envelope constraints had the highest Vout values (∼200) among all of the inhibitors, indicating that they protrude the most from the substrate envelope. All five SAR inhibitors have a phenyloxazolidinone moiety at R1 with different substituents on the phenyl ring. Two atoms of the phenyl ring and its substituents protruded beyond the substrate envelope in all five inhibitors (Fig. 5d). All five inhibitors had an isobutyl group at R2, as seen with APV. As with the APV group, this R2 group fits within the envelope in all structures. All these inhibitors had either a 4-methoxyphenyl or 1,3-benzodioxolane group at R3. In these compounds, two atoms of R3 protruded from the substrate envelope. Overall, the fit of the five SAR inhibitors in the substrate envelope was not as good as the fit of those designed using the OpDk and Inv schemes, although they had the best binding affinities against the wild-type protease compared to the OpDk and Inv inhibitors.
Binding to drug-resistant HIV-1 protease variants.
The correlation between inhibitor fit in the substrate envelope and the susceptibility of the inhibitor to drug resistance was assessed by measuring the binding affinities of all 16 inhibitors against four drug-resistant protease variants (Table 1). These four protease variants (M1, L10I, G48V, I54V, L63P, and V82A; M2, D30N, L63P, and N88D; M3, L10I, L63P, A71V, G73S, I84V, and L90M; and M4, I50V and A71V) were selected from previously identified sets of coevolving mutations in drug-resistant clinical isolates (11, 27). APV, the molecular core from which all the inhibitors were designed, has a Vout value of 134 and exhibited an average of a 5-fold loss of affinity against the drug-resistant mutants tested. The two inhibitors designed with the OpDk (OpDk-KB45 and OpDk-AD37) had Vout values better than and similar to APV, respectively, indicating that they fit well in the substrate envelope. OpDk-KB45 and OpDk-AD37 exhibited an average of a 25- and 11-fold loss of affinity, respectively, against three drug-resistant variants. In addition, the four first-round Inv inhibitors had Vout values between 152 to 175, which, although higher than the APV values, are lower than most of the other FDA-approved inhibitors, exhibiting an average of only a 3- to 6-fold loss of affinity with the mutants. Thus, five of the six nanomolar inhibitors designed using either scheme (OpDk and first-round Inv inhibitors) exhibited low binding affinity but relatively flat binding profiles to the drug-resistant variants.
The Inv2 inhibitors have binding affinities higher than that of APV and better than the first-round Inv1 inhibitors, but their Vout values (147 to 160) are somewhat larger than that of APV. Inv2-KC08, Inv2-KB98, and Inv2-AD93 had the best drug resistance profiles of all the computationally designed inhibitors; these inhibitors had Vout values of 152, 147, and 153, respectively, and retained relatively high affinity (average Ki values of 0.125, 0.93, and 1.17 nM) for all four mutant proteases. Thus, an inhibitor can retain high affinity, remain within the substrate envelope, and have a relatively flat binding profile against the drug-resistant variants.
The five SAR-designed inhibitors had the largest Vout values (larger than those of the OpDk and Inv inhibitors). Although the SAR inhibitors exhibited very high binding affinities to wild-type protease (0.8 to 16.0 pM), they exhibited significant losses of affinity (an average of 241- to 1,562-fold) to the drug-resistant variants. All five SAR inhibitors exhibited a 120- to 200-fold loss of affinity against the protease variants containing the G48V mutation (L10I, G48V, I54V, L63P, and V82A), probably due in part to a vdW clash with V48. These inhibitors exhibited an even greater loss of affinity for the multidrug-resistant variant (L10I, L63P, A71V, G73S, I84V, and L90M), perhaps indicating strong interactions with I84. Overall, the SAR-derived inhibitors possessed high binding affinity to wild-type protease, but they also exhibited a significantly greater loss of affinity to drug-resistant variants than OpDk and Inv inhibitors, although, even with the large loss, they still retained nanomolar affinity for resistant variants.
Comparing methodologies.
The 16 inhibitors were designed using three different methodologies. The two computational methods, OpDk and Inv, use different methodologies to implement the substrate-envelope constraint. The third methodology uses a SAR approach without the substrate-envelope constraint but also starts with the same scaffold, which is amenable to fitting within the substrate envelope. Crystallographic analysis showed that the OpDk inhibitors fit the substrate envelope and had midnanomolar binding affinity values. The two OpDk inhibitors exhibited an average of an 11- to 25-fold loss of affinity to the drug-resistant variants. The four first-round Inv inhibitors exhibited an average of only a 3- to 6-fold loss of affinity, but their thiophene ring had a conformation different from the predicted structure and protruded slightly from the substrate envelope. The difference in conformation of the thiophene ring might account for the low affinity of the first-round inhibitors. When the Inv design scheme was optimized in the second round, the predicted conformations of most inhibitors were not only similar to the observed conformations but also had picomolar levels of affinity and fit within the substrate envelope. Introducing the phenyloxazolidinone ring on the (R)-(hydroxyethylamino)sulfonamide in the SAR approach led to the design of high-affinity inhibitors, but the lack of substrate-envelope constraint resulted in low adaptability of the inhibitors to various mutants. Thus, while a variety of methodologies can lead to highly potent HIV-1 protease inhibitors, incorporation of the substrate envelope constraints into the design methodology would likely lead to inhibitors that retain high-affinity even under evolutionary pressure.
DISCUSSION
The major challenge in treating HIV-1-infected patients is not only to develop potent inhibitors against wild-type and drug-resistant viruses but also to prevent the virus from evolving resistant mutations to those inhibitors. The substrate-envelope hypothesis provides a rational approach to designing such protease inhibitors. Analysis of the 16 protease-inhibitor complexes has given insights into the interactions important for binding and indicates that protrusion of inhibitors from the substrate envelope generally correlates with their loss of affinity to mutant proteases. These inhibitors and two FDA-approved inhibitors, APV and DRV, share the same (R)-(hydroxyethylamino)sulfonamide molecular core and have different affinities to the protease, allowing for detailed comparisons. With this series of well-resolved crystal structures of similar inhibitors, ranging in affinity from the nanomolar to the picomolar level, systematic comparisons of the subtle changes in their interactions were performed, including the use of a quantitative method for assessing vdW interactions. By performing a detailed comparison of the structures, small differences in the crystal structures of nanomolar and picomolar inhibitors can be elucidated. By quantitatively analyzing the interactions of the various inhibitors with HIV-1 protease, the interdependent adaptability of HIV-1 protease, the specifics of the hydrogen bonding pattern, and the exact alterations in the extent and pattern of vdW contacts were elucidated.
Attention to the details of such interactions should significantly aid in future inhibitor design, as much of structure-based drug design involves altering local interactions of specific substituent groups. As these inhibitors were designed utilizing different methodologies and philosophies, the validity of the substrate-envelope hypothesis has been assessed. The results of this study indicate that inhibitors are more likely to be robust and retain binding affinity to drug-resistant variants when they fit within the substrate envelope, whereas they may exhibit a relative loss of binding affinity when they protrude from the substrate envelope. Although some of the potent SAR compounds, while losing substantial affinity, retained a nanomolar level of affinity for resistant variants, the substrate-envelope hypothesis allows a larger percentage of the designed inhibitors to retain a flat or robust binding profile for resistant variants.
Many instances of drug resistance result from mutations in a macromolecular drug target that allow it to retain function while no longer being efficiently inhibited by the drug. Traditional drug design is largely geared toward disrupting the activity of the drug target and often ignores the molecular basis for its function, resulting in the rapid evolution of resistance to the drug. As a consequence, many inhibitors found by high-throughput screening and structure-based drug design can contact residues within the target that could mutate and confer resistance without significantly impairing its function. Current drug design may therefore inadvertently facilitate drug resistance. We developed the concept of the substrate-envelope hypothesis by elucidating the molecular details of substrate recognition by HIV protease, allowing the development of inhibitors less likely to be susceptible to resistance. This study indicates that inhibitors designed using substrate-envelope constraints are more likely to retain similar levels of affinity across a panel of drug-resistant protease variants. Thus, for quickly evolving therapeutic targets, combining the substrate-envelope hypothesis with structure-based drug design may result in the creation of inhibitors that are less susceptible to drug resistance.
Acknowledgments
This work was made possible by a grant from the National Institute of General Medical Sciences, National Institutes of Health (P01-GM66524). V.K. thanks Fundação para a Ciência e a Tecnologia (Portugal) for grant SFRH/BPD/41787/2007.
Footnotes
Published ahead of print on 17 March 2010.
REFERENCES
- 1.Ali, A., G. S. K. K. Reddy, H. Cao, S. G. Anjum, M. N. L. Nalam, C. A. Schiffer, and T. M. Rana. 2006. Discovery of HIV-1 protease inhibitors with picomolar affinities incorporating N-aryl-oxazolidinone-5-carboxamides as novel P2 ligands. J. Med. Chem. 49:7342-7356. [DOI] [PubMed] [Google Scholar]
- 2.Altman, M. D., A. Ali, G. S. K. K. Reddy, M. N. L. Nalam, S. G. Anjum, H. Cao, S. Chellappan, V. Kairys, M. X. Fernandes, M. K. Gilson, C. A. Schiffer, T. M. Rana, and B. Tidor. 2008. HIV-1 Protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants. J. Am. Chem. Soc. 130:6099-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chellappan, S., V. Kairys, M. X. Fernandes, C. Schiffer, and M. K. Gilson. 2007. Evaluation of the substrate envelope hypothesis for inhibitors of HIV-1 protease. Proteins 68:561-567. [DOI] [PubMed] [Google Scholar]
- 4.Chellappan, S., G. S. K. K. Reddy, A. Ali, M. N. L. Nalam, S. G. Anjum, H. Cao, V. Kairys, M. X. Fernandes, M. D. Altman, B. Tidor, T. M. Rana, C. A. Schiffer, and M. K. Gilson. 2007. Design of mutation-resistant HIV protease inhibitors with the substrate envelope hypothesis. Chem. Biol. Drug Des. 69:298-313. [DOI] [PubMed] [Google Scholar]
- 5.Clavel, F., and A. J. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023-1035. [DOI] [PubMed] [Google Scholar]
- 6.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Tepplert, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 7.Debouck, C. 1992. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res. Hum. Retrovir. 8:153-164. [DOI] [PubMed] [Google Scholar]
- 8.De Meyer, S., H. Azijn, D. Surleraux, D. Jochmans, A. Tahri, R. Pauwels, P. Wigerinck, and M. P. de Bethune. 2005. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 49:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorsey, B. D., R. B. Levin, S. L. McDaniel, J. P. Vacca, J. P. Guare, P. L. Darke, J. A. Zugay, E. A. Emini, W. A. Schleif, J. C. Quintero, J. H. Lin, I.-W. Chen, M. K. Holloway, P. M. D. Fitzgerald, M. G. Axel, D. Ostovic, P. S. Anderson, and J. R. Huff. 1994. L-735,524: The design of a potent and orally bioavailable HIV protease inhibitor. J. Med. Chem. 37:3443-3451. [DOI] [PubMed] [Google Scholar]
- 10.Erickson, J. W. 1995. The not-so-great escape. Nat. Struct. Mol. Biol. 2:523-529. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman, N. G., C. A. Schiffer, and R. Swanstrom. 2003. Covariation of amino acid positions in HIV-1 protease. Virology 314:536-548. [DOI] [PubMed] [Google Scholar]
- 12.Joint United Nations Program on HIV/AIDS. 2008. 2008 report on the global AIDS epidemic. Joint United Nations Program on HIV/AIDS (UNAIDS), Geneva, Switzerland.
- 13.Kaldor, S. W., V. J. Kalish, J. F. Davies II, B. V. Shetty, J. E. Fritz, K. Appelt, J. A. Burgess, K. M. Campanale, N. Y. Chirgadze, D. K. Clawson, B. A. Dressman, S. D. Hatch, D. A. Khalil, M. B. Kosa, P. P. Lubbehusen, M. A. Muesing, A. K. Patick, S. H. Reich, K. S. Su, and J. H. Tatlock. 1997. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J. Med. Chem. 40:3979-3985. [DOI] [PubMed] [Google Scholar]
- 14.Kempf, D. J., K. C. Marsh, J. F. Denissen, E. McDonald, S. Vasavanonda, C. A. Flentge, B. E. Green, L. Fino, C. H. Park, X. P. Kong, N. E. Wideburg, A. Saldivar, L. Ruiz, W. M. Kati, H. L. Sham, T. Robins, K. D. Stewart, A. Hsu, J. J. Plattner, J. M. Leonard, and D. W. Norbeck. 1995. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Natl. Acad. Sci. U. S. A. 92:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, E. E., C. T. Baker, M. D. Dwyer, M. A. Murcko, B. G. Rao, R. D. Tung, and M. A. Navia. 1995. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J. Am. Chem. Soc. 117:1181-1182. [Google Scholar]
- 16.King, N. M., M. Prabu-Jeyabalan, E. A. Nalivaika, and C. A. Schiffer. 2004. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem. Biol. 11:1333-1338. [DOI] [PubMed] [Google Scholar]
- 17.Koh, Y., H. Nakata, K. Maeda, H. Ogata, G. Bilcer, T. Devasamudram, J. F. Kincaid, P. Boross, Y. F. Wang, Y. Tie, P. Volarath, L. Gaddis, R. W. Harrison, I. T. Weber, A. K. Ghosh, and H. Mitsuya. 2003. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 47:3123-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matayoshi, E. D., G. T. Wang, G. A. Krafft, and J. Erickson. 1990. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 247:954-958. [DOI] [PubMed] [Google Scholar]
- 19.Menendez-Arias, L. 2010. Molecular basis of human immunodeficiency virus drug resistance: an update. Antiviral Res. 85:210-231. [DOI] [PubMed] [Google Scholar]
- 20.Nalam, M. N. L., and C. A. Schiffer. 2008. New approaches to HIV protease inhibitor drug design II: testing the substrate envelope hypothesis to avoid drug resistance and discover robust inhibitors. Curr. Opin. HIV AIDS 3:642-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabu-Jeyabalan, M., E. Nalivaika, and C. A. Schiffer. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369-381. [DOI] [PubMed] [Google Scholar]
- 22.Roberts, N. A., J. A. Martin, D. Kinchington, A. V. Broadhurst, J. C. Craig, I. B. Duncan, S. A. Galpin, B. K. Handa, J. Kay, A. Krohn, R. W. Lambert, J. H. Merrett, J. S. Mills, K. E. B. Parkes, S. Redshaw, A. J. Ritchie, D. L. Taylor, G. J. Thomas, and P. J. Machin. 1990. Rational design of peptide-based HIV proteinase inhibitors. Science 248:358-361. [DOI] [PubMed] [Google Scholar]
- 23.Robinson, B. S., K. A. Riccardi, Y. F. Gong, Q. Guo, D. A. Stock, W. S. Blair, B. J. Terry, C. A. Deminie, F. Djang, R. J. Colonno, and P. F. Lin. 2000. BMS-232632, a highly potent human immunodeficiency virus protease inhibitor that can be used in combination with other available antiretroviral agents. Antimicrob. Agents Chemother. 44:2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sham, H. L., D. J. Kempf, A. Molla, K. C. Marsh, G. N. Kumar, C. M. Chen, W. Kati, K. Stewart, R. Lal, A. Hsu, D. Betebenner, M. Korneyeva, S. Vasavanonda, E. McDonald, A. Saldivar, N. Wideburg, X. Chen, P. Niu, C. Park, V. Jayanti, B. Grabowski, G. R. Granneman, E. Sun, A. J. Japour, J. M. Leonard, J. J. Plattner, and D. W. Norbeck. 1998. ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob. Agents Chemother. 42:3218-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Surleraux, D. L., A. Tahri, W. G. Verschueren, G. M. Pille, H. A. de Kock, T. H. Jonckers, A. Peeters, S. De Meyer, H. Azijn, R. Pauwels, M. P. de Bethune, N. M. King, M. Prabu-Jeyabalan, C. A. Schiffer, and P. B. Wigerinck. 2005. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J. Med. Chem. 48:1813-1822. [DOI] [PubMed] [Google Scholar]
- 26.Turner, S. R., J. W. Strohbach, R. A. Tommasi, P. A. Aristoff, P. D. Johnson, H. I. Skulnick, L. A. Dolak, E. P. Seest, P. K. Tomich, M. J. Bohanon, M. M. Horng, J. C. Lynn, K. T. Chong, R. R. Hinshaw, K. D. Watenpaugh, M. N. Janakiraman, and S. Thaisrivongs. 1998. Tipranavir (PNU-140690): a potent, orally bioavailable nonpeptidic HIV protease inhibitor of the 5,6-dihydro-4-hydroxy-2-pyrone sulfonamide class. J. Med. Chem. 41:3467-3476. [DOI] [PubMed] [Google Scholar]
- 27.Wu, T. D., C. A. Schiffer, M. J. Gonzales, J. Taylor, R. Kantor, S. Chou, D. Israelski, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 77:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]