Abstract
A large number of human papillomavirus (HPV) types, distributed over five papillomavirus genera, are detectable in the skin. HPV types belonging to the alpha, gamma, and mu genera have been detected in cutaneous warts. A state-of-the-art HPV genotyping assay for these cutaneous wart-associated HPV types does not exist although warts constitute a highly prevalent skin condition, especially in children (33%) and organ transplant recipients (45%). Cutaneous warts are again the focus of attention as their clinical relevance rises with the increasing number of chronically immunosuppressed patients. The objective of this study was to develop and evaluate a DNA-based genotyping system for all known cutaneous wart-related HPV types using PCR and Luminex xMAP technology. The broad-spectrum PCR amplified DNA of all known wart-associated HPV types from the genera alpha (HPVs 2, 3, 7, 10, 27, 28, 29, 40, 43, 57, 77, 91, and 94), gamma (HPVs 4, 65, 95, 48, 50, 60, and 88), mu (HPVs 1 and 63), and nu (HPV41). The probes were evaluated using plasmid HPV DNA and a panel of 45 previously characterized cutaneous wart biopsy specimens showing high specificity. HPV was also identified in 96% of 100 swabs from nongenital cutaneous warts. HPV types 1, 2, 27, and 57 were the most prevalent HPV types detected in 89% of the swabs. In conclusion, this Luminex-based genotyping system identifies all known cutaneous wart HPV types including phylogenetically related types, is highly HPV type specific, and is suitable for large-scale epidemiological studies.
Papillomaviruses (PV) constitute a group of viruses associated with benign and malignant proliferative lesions of cutaneous and mucosal epithelia. The taxonomic family of PV includes 16 genera (5). More than 100 human papillomavirus (HPV) types have been fully sequenced. The number of HPV types that are not yet fully characterized is probably much larger. HPV types, distributed over five genera (alpha, beta, gamma, mu, and nu) and 16 species, infect the skin (5). HPV types belonging to species of three genera (alpha, gamma, and mu) have most frequently been detected in hyperkeratotic skin lesions (warts).
The alphapapillomavirus (alpha-PV) types infecting the genital mucosa (e.g., HPV16, HPV18, HPV6, and HPV11) are the best understood. HPV16 and HPV18 are the most prevalent types involved in the pathogenesis of anogenital (e.g., cervical) cancer, and HPV types 6 and 11 cause genital warts and laryngeal papillomas. The alpha-PV types belonging to species 2, 4, and 8, which have been detected in cutaneous warts, have been studied less thoroughly. At present the most frequently detected alpha-PV types in cutaneous warts are HPV types 2, 3, 10, 27, and 57 (3, 10, 12-16, 18, 19). Gamma-PV types, including HPV4, HPV60, and HPV65, have also regularly been detected in cutaneous warts (4, 10, 12, 15). Mu-PV types, HPV1 and HPV63, have also been detected in cutaneous warts (7). HPV types included in the beta and nu genera have rarely been detected in cutaneous warts. Only Harwood and coworkers (13) have described the presence of beta-PV types in cutaneous warts of immunosuppressed patients. The nu-PV genus consists of only one HPV type (HPV41), which was originally isolated from a facial wart (11).
In general, cutaneous warts from immunocompetent and immunocompromised patients appear to show the same genotype distributions (16, 19). This has to be confirmed by a large-scale study comparing the distributions in the two populations with the same techniques. The number of detected types per lesion appeared to differ between immunocompetent and immunosuppressed patients (13). Currently, cutaneous warts are again the focus of attention with the increasing number of chronically immunosuppressed patients (9, 20). Significant physical and psychological morbidity in these patients may result from the existence of confluent plaques of warts present on cosmetically sensitive areas such as the face and hands and the presence of large warts occurring on pressure-bearing areas such as the feet.
Although cutaneous warts constitute a highly prevalent skin condition, especially in children (33%) (22) and organ transplant recipients (45%) (2), only a few large-scale epidemiological studies including more than 100 lesions have investigated the distribution of HPV types in cutaneous warts. These studies utilized different detection methods, such as general primer-mediated PCR followed by HPV typing with direct sequencing (12, 15) or by restriction enzyme cleavage of the PCR product (18). Other methods employed were multiple-type-specific PCR combined with direct sequencing (3), in situ hybridization (8), and (Southern) blot hybridization (6, 10). Although these methods are in general type specific, they are time-consuming and therefore less suitable for large scale-epidemiological studies. In the case of multiple infections, reliable typing might also be difficult with direct sequencing of the PCR amplimer without prior molecular cloning. Therefore, we have evaluated a novel broad-spectrum PCR-multiplex genotyping (MPG) assay called the hyperkeratotic skin lesion (HSL) PCR/MPG for identification of established and candidate cutaneous wart HPV types. These types are included in the alpha genus species 2 (HPVs 3, 10, 28, 29, 77, and 94), species 4 (HPVs 2, 27, and 57), and species 8 (HPVs 7, 40, 43, and 91); gamma genus species 1 (HPVs 4, 65, and 95), species 2 (HPV48), species 3 (HPV50), species 4 (HPV60), and species 5 (HPV88); mu genus species 1 (HPV1) and species 2 (HPV63); and nu genus species 1 (HPV41). In this study the sensitivity, the HPV type specificity, and the clinical performance of the HSL-PCR/MPG broad-spectrum assay have been investigated.
MATERIALS AND METHODS
Clinical materials.
Two panels of clinical samples were available. The first panel consisted of purified DNA obtained from 45 nongenital cutaneous warts that were taken from immunocompetent patients, immunosuppressed organ transplant patients, and HIV-positive patients. These samples had been previously HPV genotyped by DNA sequencing using a purified PCR product from two rounds of nested PCR utilizing the primer pair HVP2-B5 for the first round and the pairs CN1F-CN1R, CN2F-CN2R, and CN3F-CN3R for the second round (sequencing) or the single-round primer pair C4F-C4R (PCR and sequencing), as described by Harwood and coworkers (13). The use of separate reaction mixtures with different primer pairs allowed the identification of multiple HPV types. In some cases DNA sequencing was not carried out with all of the primer pairs (Table 1).
TABLE 1.
Comparison of HSL-PCR/MPG assay results with direct DNA sequencing on PCR amplimers obtained from purified DNA from 45 warts
| Sample no. | HPV type(s) by: |
Comparison of resultsf | |
|---|---|---|---|
| HSL-PCR/MPG | DNA sequencing | ||
| 1 | 27 | 27 | I |
| 2 | 27 | 27 | I |
| 3 | 27 | 27, 94 | C |
| 4 | 27 | 27, 94 | C |
| 5 | 27 | 27 | I |
| 6 | 28 | 28 | I |
| 7 | 27 | 27 | I |
| 8 | 2 | 2 | I |
| 9 | Negative | Negative | I |
| 10 | 27 | 27 | I |
| 11 | (3)a, 7 | 3 | C |
| 12 | (3)a, 7 | 3 | C |
| 13 | 27 | 27 | I |
| 14 | 2 | 2 | I |
| 15 | 57 | 57 | I |
| 16 | 57 | 57 | I |
| 17 | 57 | 28, 57 | C |
| 18 | 57 | 57 | I |
| 19 | 57 | 57 | I |
| 20b | 57 | 72 | D |
| 21b | Negative | 72 | I |
| 22c | 4 | 27, 28 | D |
| 23 | 27 | 27, 28 | C |
| 24 | 57 | 57 | I |
| 25 | 27 | 27 | I |
| 26 | 27 | 27 | I |
| 27 | 27, 57, 95 | 27 | C |
| 28 | 27, 57 | 27 | C |
| 29 | 57 | 57 | I |
| 30 | 57 | 57 | I |
| 31 | 1, 3, 57 | 3 | C |
| 32 | 3, 57 | 57 | C |
| 33 | 57 | 57 | I |
| 34 | 4 | 4 | I |
| 35 | 2 | 2 | I |
| 36 | 2 | 2 | I |
| 37d | 7 | 10 | D |
| 38 | 4, 7, 10 | 7, 10, 27 | C |
| 39 | 4, 7, 10 | 7, 10, 27 | C |
| 40 | 7 | 7, 10 | C |
| 41 | 7, 10 | 10, 27 | C |
| 42 | 10, 27 | 27 | C |
| 43e | 63 | 27 | D |
| 44 | 27 | 27 | I |
| 45 | 4 | 4 | I |
Elevated signal but below cutoff for HPV3 with the HSL-PCR/MPG. There was no sequencing result for HPV7; HPV7 was confirmed by a third assay.
HPV72 not included in the HSL-PCR/MPG assay.
No sequencing result for HPV4 available.
Repeated DNA sequencing negative.
Repeated DNA sequencing did not confirm HPV27.
I, both methods yielded completely identical genotyping results (concordant); C, both methods showed one or more of the same genotype(s) (compatible); D, no similarity between genotypes detected by both methods (discordant).
A second panel comprised swabs taken from 100 cutaneous warts derived from 100 individuals (age of >4 years) with one or more cutaneous warts who self-presented to their general practitioners. The warts were present mainly on hands and feet. The swabs were taken from one wart from each patient by firmly rubbing a prewetted cotton-tipped stick five times over the surface of the lesion. Next, the swabs were put in 1 ml of saline solution, and 10 μl was used directly in the novel HSL-PCR/MPG assay.
Plasmids.
For the evaluation of the analytical type specificity of the HSL-PCR/MPG assay (Labo Biomedical Products BV, Rijswijk, Netherlands), amplimers were generated using plasmid clones containing sequences of the following types: HPV types 1, 2, 3, 4, 7, 10, 27, 28, 29, 40, 41, 43, 48, 50, 57, 60, 63, 65, 77, 88, 91, 94, and 95. Plasmids containing HPV genomic DNA were kindly provided by E.-M. de Villiers (HPVs 1, 4, 7, 41, 48, and 63), O. Forslund (HPV88), M. Favre (HPV10), T. Matsukura (HPV60), R. Ostrow (HPV3), G. Orth (HPVs 28, 29, and 50), and A. Lorincz (HPV43). For HPV types 2, 27, 40, 57, 65, 77, 91, 94, and 95, the relevant HPV sequence was synthesized, cloned, and sequenced (GenScript USA, Inc.). In order to confirm amplification of an HPV-specific fragment, gel electrophoresis was performed with 2.2% FlashGel agarose gels (Lonza, Basel, Switzerland). To determine the analytical sensitivity of the assay, DNA concentrations of the plasmids were determined using a NanoDrop 2000 instrument (Thermo Fisher Scientific Inc.), and 10-fold serial dilutions were made in a background of 12 ng/μl human genomic DNA. This background human DNA is equivalent to approximately 24,000 cells per 10 μl of plasmid sample.
HSL-PCR/MPG assay.
The HSL-PCR/MPG assay comprises the HSL-PCR generating a biotinylated amplimer of 76 to 84 bp from the L1 region and a genotyping assay with bead-based xMAP suspension array technology, which is able to simultaneously identify 23 HPV genotypes. Within the L1 gene of HPV a region suitable for broad-spectrum PCR and genotyping was used (Fig. 1). The primer set consisted of 27 nondegenerate primers (13 forward and 14 reverse) without inosines. All PCRs were carried out with all precautions to avoid contamination, as described by the manufacturer (Labo Biomedical Products BV, Rijswijk, Netherlands).
FIG. 1.
Nucleotide sequence alignment of the target region for the HSL-PCR for 23 HPV types. The nucleotides identical to the top sequence (HPV3) are indicated as dots. Within the alpha genus the HPV types are ordered per species, with species 2 consisting of HPV types 3, 10, 28, 29, 77, and 94; species 4 consisting of HPV types 2, 2 variant, 27, and 57; and species 8 consisting of HPV types 7, 40, 43, and 91.
Briefly, HSL-PCR was performed in a final reaction volume of 50 μl, containing 10 μl of the isolated DNA, 2.0 mM MgCl2, 1× GeneAmp PCR Buffer II, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1.5 U of AmpliTaq Gold DNA polymerase, and 10 μl of the primer mix. The PCR was performed by a 9-min preheating step at 94°C, followed by 35 cycles of amplification comprising 30 s at 94°C, 45 s at 55°C, and 45 s at 72°C. A final elongation step at 72°C for 5 min ended the PCR.
The HSL-PCR/MPG assay permitted the simultaneous identification of multiple HPV genotypes in a single hybridization step. Twenty-four genotyping probes were selected from the PCR target region (Fig. 1) and were immobilized on color-coded beads. Since the HPV genotypes included in the assay often differ by only a few nucleotides in the probe region, well-controlled assay conditions and probe selection were required. The HSL-PCR/MPG assay was performed according to the manufacturer's instructions in a Luminex 100 IS System (Luminex Corporation, Austin, TX) that was calibrated weekly. Briefly, 3B buffer was added to the provided bead mix to minimize background in the final Luminex readout. Subsequently, HSL-PCR products were added. Next, heat denaturation, hybridization under stringent conditions, and incubation with streptavidin-conjugated R-phycoerythrin detection conjugate, followed by readout according to the specified instrument settings, resulted in median fluorescence intensity (MFI) levels per HPV type for each specimen. No distinction was made between samples positive for HPV type 2 variant (HPV2var) and HPV2 as they are both scored as HPV2 positive.
Sequence analysis.
For sequence analysis of the HSL amplimers from the plasmid clones generated with use of type-specific primers, amplimers were treated with ExoSAP-IT (USB, OH) to remove unconsumed dNTPs and primers. Purified amplimers were directly sequenced according to the manual of the Big Dye Terminator Cycle Sequencing kit using forward and reverse HSL-PCR primers. The sequence products were subsequently read using a 3100-Avant Genetic Analyzer. The resulting DNA sequences were analyzed with Vector NTI Advance, version 9.0, software and compared with all known HPV types present in the National Center for Biotechnology Information (NCBI) database utilizing nucleotide-nucleotide BLAST (BLASTN) searches (1) (http://www.ncbi.nlm.nih.gov/BLAST/).
HPV sequences from GenBank.
The following are accession numbers of HPV sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) and used as references for the corresponding HPV genotypes: HPV type 3, X74462; HPV type 10, X74465; HPV type 28, U31783; HPV type 29, U31784; HPV type 77, Y15175; HPV type 94, AJ620211; HPV2var, DQ080000; HPV type 2, X55964; HPV type 27, X73373; HPV type 57, X55965; HPV type 7, X74463; HPV type 40, X74478; HPV type 43, AJ620205; HPV type 91, AF419318; HPV type 4, X70827; HPV type 48, U31789; HPV type 50, U31790; HPV type 60, U31792; HPV type 65, X70829; HPV type 88, EF467176; HPV type 95, AJ620210; HPV type 1, V01116; HPV type 63, X70828; HPV type 41, X56147.
RESULTS
Analytical type specificity of the HSL-PCR/MPG.
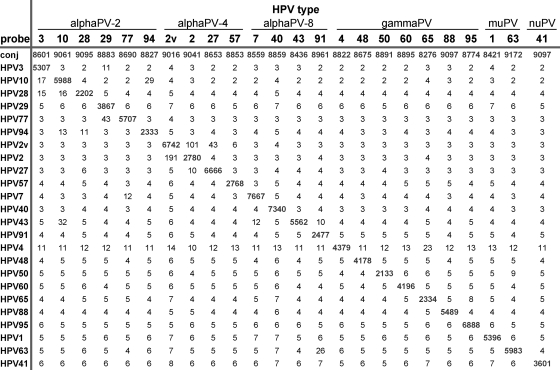
DNA from 24 plasmids containing partial or complete HPV genomic sequences representing established types from alpha-, gamma-, mu-, and nu-PV genera was analyzed by HSL-PCR/MPG. Amplimers were identified on agarose gels, and a fragment of the expected size (76 to 84 bp) was present (data not shown). Subsequent sequence analysis (data not shown) confirmed that the correct genotypes were amplified. For all HPV sequences the genotyping assay showed type-specific hybridization, with MFI values ranging from 2,133 to 7,667 for targeted HPV types (Fig. 2).
FIG. 2.
Analytical type specificity of the HSL-PCR/MPG. Indicated are the MFI readouts (without background subtraction) of HSL-PCR amplimers generated from 6 × 108 to 7 × 107 copies of different HPV plasmids (listed on top) in relation to the bead-bound capture probes (listed on the left). The conjugate control (conj) serves as the positive control for correct incubation with the detection conjugate for each separate specimen. HPV2 and HPV2var (2v) are both scored as HPV2 positive.
Analytical sensitivity of the HSL-PCR/MPG.
Sensitivity tests were performed on 10-fold serial dilutions of plasmid clones of HPV types 1, 2, 3, 4, 7, 10, 27, 28, 29, 40, 41, 43, 48, 50, 57, 60, 63, 65, 77, 88, 91, 94, and 95 made in a background of 12 ng/μl human DNA. The analytical sensitivity of the HSL-PCR/MPG assay ranged from 1 to 10 copies per PCR for HPV types 1 and 2; from 10 to 100 copies per PCR for HPV types 2 variant, 4, 7, 10, 43, 48, 63, and 95; from 100 to 1,000 copies per PCR for HPV types 3, 27, 29, 57, 60, 65, and 77; and from 1,000 to 10,000 copies per PCR for HPV types 28, 40, 41, 50, 88, 91, and 94 (data not shown).
Clinical performance of the HSL-PCR/MPG.
The HSL-PCR/MPG yielded an HPV genotyping result identical to that of sequencing in 26 of the 45 nongenital cutaneous wart DNA samples derived from immunocompetent patients, immunosuppressed organ transplant patients, and HIV-positive patients. In 15 of 45 cases the result was compatible (i.e., an additional HPV type was detected by either method [21]) (Table 1). In four of 45 cases the results were discordant. In two of these four cases the result with one or both assays was positive for an HPV type that was not included in the other assay, and these two samples were excluded. Two other discordant results remain unresolved since repeated sequencing did not confirm the first result.
Second, the performance of the HSL-PCR/MPG assay was evaluated on cutaneous wart swabs. The positivity rate was 96% in swabs taken from nongenital cutaneous warts mainly present on hands and feet of 100 patients (see Materials and Methods). The most prevalent HPV types were HPV1 (genus mu species 1) and HPVs 2, 27, and 57 (genus alpha species 4). Together, these HPV types were detected in 89% of the cutaneous wart swabs. Other HPV types detected were HPVs 3, 4, 7, 63, 65, and 95 (Table 2). In one case the generated amplimer of the expected size, as visualized by agarose gel electrophoresis, did not hybridize with the available genotyping probes. In three other cases the HSL-PCR/MPG did not generate an amplimer of the expected size and did not give a positive signal with the genotyping assay. Within these 100 samples, 11 multiple infections were observed; of these 10 contained two HPV types and 1 was positive for three types.
TABLE 2.
HPV type distribution in 100 swabs taken from a single wart of 100 persons
| Genus | Species no. | Genotype | % Positivitya |
|---|---|---|---|
| Alpha | 2 | HPV3 | 2 |
| 4 | HPV2 | 20 | |
| HPV27 | 31 | ||
| HPV57 | 15 | ||
| 8 | HPV7 | 1 | |
| Gamma | 1 | HPV4 | 5 |
| HPV65 | 1 | ||
| HPV95 | 1 | ||
| Mu | 1 | HPV1 | 31 |
| 2 | HPV63b | 1 |
Eleven samples showed multiple infections; 10 of these samples were positive for two types and 1 was positive for three types. Four samples were negative with the genotyping assay.
Only detected once in a multiple infection with HPV1.
DISCUSSION
In this study, we have described the evaluation of a novel, sensitive, single-step broad-spectrum PCR targeting the HPV L1 region combined with a Luminex-based hybridization assay for the detection and genotyping of HPV types associated with nongenital cutaneous warts. The HSL-PCR was able to amplify HPV DNA from various sources and showed an analytical sensitivity of 1 to 10,000 HPV copies per PCR in a human genomic DNA background. The analytical HPV type specificity was 100%, as shown by analyzing amplimers generated from all 23 included HPV types at a concentration ranging from 6 × 108 to 7 × 107 viral genome copies per PCR. When the novel HSL-PCR/MPG method was compared with an established DNA sequencing method (13) targeting a different part of the L1 open reading frame, the results were highly concordant. Forty-one of the 43 included samples gave identical or compatible results (95%), and two gave discordant results, confirming the high analytical specificity of the novel genotyping method.
The reproducibility of the Luminex system which was used for the HSL-PCR/MPG assay has been extensively investigated by an analogue, broad-spectrum PCR genotyping assay (Digene HPV Genotyping LQ Test package insert) and was higher than 95%.
The median MFI values, representing the general background signals in all 130 analyzed clinical samples consisting of swabs and biopsy specimens, were very low, ranging from 1 to 9 MFI for the different probes. The observed cross-reactions in clinical samples were all confirmed by experiments with plasmid dilutions using an input of up to 108 viral genome copies per PCR. Furthermore, DNA sequence alignments between the probes and cross-hybridizing HPV types (data not shown) indicated that a small number of mismatches explained the cross-hybridization reactions with a maximum signal of 131 MFI. Whereas the general background signals were between 1 and 9 MFI, theoretically allowing for a low cutoff, the detected cross-hybridization reactions with some probes resulted in the use of a conventional cutoff of 200 MFI for all types. Using this high cutoff for all HPV types included in the assay could cause an underestimation of the prevalence of certain HPV types.
Amplimers of HPV27 gave cross-hybridization reactions with the probe for HPV2var that resulted in MFI signals higher than 200 MFI. The highest observed cross-reactive signal was detected in purified DNA from a cutaneous wart positive for HPV27, with an MFI value of 9,226. The cross-reactive signal for HPV2var was 534 MFI in this case. Therefore, the cutoff for HPV2var positivity was set at 700 MFI in multiple infections with HPV27 signals above 7,500 MFI.
Similarly, amplimers of HPV10 and HPV28 gave cross-hybridization reactions with the probe for HPV94 that resulted in MFI signals higher than 200 MFI, with a maximum of 221 MFI. Consequently, we used a cutoff for HPV94 positivity of 500 MFI in a multiple infection with HPV10 or HPV28 when these types had higher signals than 4,000 or 1,500 MFI, respectively.
In the absence of these types, the assay's conventional cutoff of 200 MFI was used. This cutoff value is well above the background signals and was chosen as a precaution because cutaneous warts could produce very high concentrations of viral DNA, possibly challenging the specificity of the genotyping system. The downside of this precaution is reflected by the analytical sensitivity of the assay, which ranges from 1 to 10,000 copies per PCR. However, in the panel of 100 swabs of cutaneous warts, 96 (96%) of the samples were HPV positive, indicating a high enough analytical sensitivity of the assay for this kind of sample.
Current methods for the determination the HPV genotypes in cutaneous warts are generally based on Southern blot hybridization or on general primer-mediated PCR or type-specific PCR followed by direct DNA sequencing. Although these methods can provide accurate results, they are laborious and not suitable for use in large epidemiological studies. The assay format of the novel HSL-PCR/MPG method encompasses a broad-spectrum PCR amplifying all known cutaneous wart-associated HPV types, followed by a multiplex genotyping assay in a high-throughput setting, which allowed genotyping of a panel of PCR products derived from 96 samples in about 2 h.
The HSL-PCR was designed to amplify all HPV types previously found in cutaneous warts as well as related types from the same species (5). Complementing established types with the HPV types present in the same species is based on the assumption that these types might also be involved in cutaneous wart etiology analogous to the cervical cancer risk associated with not only HPV16 but also other types included in the same alpha-PV species 9. In this study swabs from 100 cutaneous warts were analyzed from individuals attending their general practitioners for treatment. Therefore, this panel was probably representative for warts in the general (Dutch) population. We confirmed the presence of similar HPV types as reported in the literature, but the other types present in the same HPV species were not often detected in this panel. Whether these other types occur less frequently or only in specific patient groups (e.g., HIV-infected persons) will be the subject of further investigations.
The common and easily accessible nature of cutaneous warts led early on to prolific clinical research in PV (17). Currently, these skin lesions are again the focus of attention as the clinical relevance of cutaneous warts rises with the increasing number of chronically immunosuppressed patients (9, 20). Significant physical and psychological morbidity in these patients may result from the existence of confluent plaques of warts present on cosmetically sensitive areas such as the face and hands and the presence of large warts occurring on pressure-bearing areas such as the feet. The impact of these problems and the difficulty in treating them should not be underestimated. Indeed, in some cases they may prompt the use of lower levels of immunosuppression, thus potentially endangering graft survival. In addition, the recently available preventive vaccines for HPV types causing cervical cancer pave the way for preventive and possibly therapeutic vaccines against cutaneous warts, particularly in the case of organ transplant recipients and patients with HIV. The development of these vaccines will, however, require a greater understanding of the HPV types that cause cutaneous warts and their natural history. Easy-to-use, high-throughput genotyping systems such as that described in this study will be invaluable for this future research.
Acknowledgments
We thank J. Lindeman and Labo Biomedical Products B.V. (Rijswijk, Netherlands) for providing the HSL-PCR/MPG assay.
M.C.W.F. is supported by the Netherlands Organization for Health Research and Development (ZonMW Clinical Fellowship 907-00-150).
Footnotes
Published ahead of print on 17 March 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bouwes Bavinck, J. N., S. Euvrard, L. Naldi, I. Nindl, C. M. Proby, R. Neale, D. Abeni, G. P. Tessari, M. C. Feltkamp, A. Claudy, E. Stockfleth, and C. A. Harwood. 2007. Keratotic skin lesions and other risk factors are associated with skin cancer in organ-transplant recipients: a case-control study in the Netherlands, United Kingdom, Germany, France, and Italy. J. Invest. Dermatol. 127:1647-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, S. Y., S. H. Chew, K. Egawa, E. I. Grussendorf-Conen, Y. Honda, A. Rubben, K. C. Tan, and H. U. Bernard. 1997. Phylogenetic analysis of the human papillomavirus type 2 (HPV-2), HPV-27, and HPV-57 group, which is associated with common warts. Virology 239:296-302. [DOI] [PubMed] [Google Scholar]
- 4.Chen, S. L., Y. P. Tsao, J. W. Lee, W. C. Sheu, and Y. T. Liu. 1993. Characterization and analysis of human papillomaviruses of skin warts. Arch. Dermatol. Res. 285:460-465. [DOI] [PubMed] [Google Scholar]
- 5.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 324:17-27. [DOI] [PubMed] [Google Scholar]
- 6.Egawa, K. 1994. New types of human papillomaviruses and intracytoplasmic inclusion bodies: a classification of inclusion warts according to clinical features, histology and associated HPV types. Br. J. Dermatol. 130:158-166. [DOI] [PubMed] [Google Scholar]
- 7.Egawa, K., H. Delius, T. Matsukura, M. Kawashima, and E. M. de Villiers. 1993. Two novel types of human papillomavirus, HPV 63 and HPV 65: comparisons of their clinical and histological features and DNA sequences to other HPV types. Virology 194:789-799. [DOI] [PubMed] [Google Scholar]
- 8.Egawa, K., Y. Honda, Y. Inaba, and T. Ono. 1998. Pigmented viral warts: a clinical and histopathological study including human papillomavirus typing. Br. J. Dermatol. 138:381-389. [DOI] [PubMed] [Google Scholar]
- 9.Euvrard, S., J. Kanitakis, and A. Claudy. 2003. Skin cancers after organ transplantation. N. Engl. J. Med. 348:1681-1691. [DOI] [PubMed] [Google Scholar]
- 10.Gassenmaier, A., P. Fuchs, H. Schell, and H. Pfister. 1986. Papillomavirus DNA in warts of immunosuppressed renal allograft recipients. Arch. Dermatol. Res. 278:219-223. [DOI] [PubMed] [Google Scholar]
- 11.Grimmel, M., E. M. de Villiers, C. Neumann, M. Pawlita, and H. zur Hausen. 1988. Characterization of a new human papillomavirus (HPV 41) from disseminated warts and detection of its DNA in some skin carcinomas. Int. J. Cancer 41:5-9. [DOI] [PubMed] [Google Scholar]
- 12.Hagiwara, K., H. Uezato, H. Arakaki, S. Nonaka, K. Nonaka, H. Nonaka, T. Asato, M. Oshiro, K. Kariya, and A. Hattori. 2005. A genotype distribution of human papillomaviruses detected by polymerase chain reaction and direct sequencing analysis in a large sample of common warts in Japan. J. Med. Virol. 77:107-112. [DOI] [PubMed] [Google Scholar]
- 13.Harwood, C. A., P. J. Spink, T. Surentheran, I. M. Leigh, E. M. de Villiers, J. M. McGregor, C. M. Proby, and J. Breuer. 1999. Degenerate and nested PCR: a highly sensitive and specific method for detection of human papillomavirus infection in cutaneous warts. J. Clin. Microbiol. 37:3545-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai, J. Y., R. J. Doyle, J. M. Bluhm, and J. C. Johnson. 2006. Multiplexed PCR genotyping of HPVs from plantaris verrucae. J. Clin. Virol. 35:435-441. [DOI] [PubMed] [Google Scholar]
- 15.Pfister, H. 2003. Chapter 8: human papillomavirus and skin cancer. J. Natl. Cancer Inst. Monogr. 31:52-56. [DOI] [PubMed] [Google Scholar]
- 16.Porro, A. M., M. M. Alchorne, G. R. Mota, N. Michalany, A. C. Pignatari, and I. E. Souza. 2003. Detection and typing of human papillomavirus in cutaneous warts of patients infected with human immunodeficiency virus type 1. Br. J. Dermatol. 149:1192-1199. [DOI] [PubMed] [Google Scholar]
- 17.Rowson, K. E., and B. W. Mahy. 1967. Human papova (wart) virus. Bacteriol. Rev. 31:110-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rübben, A., K. Kalka, B. Spelten, and E. I. Grussendorf-Conen. 1997. Clinical features and age distribution of patients with HPV 2/27/57-induced common warts. Arch. Dermatol. Res. 289:337-340. [DOI] [PubMed] [Google Scholar]
- 19.Rübben, A., R. Krones, B. Schwetschenau, and E. I. Grussendorf-Conen. 1993. Common warts from immunocompetent patients show the same distribution of human papillomavirus types as common warts from immunocompromised patients. Br. J. Dermatol. 128:264-270. [DOI] [PubMed] [Google Scholar]
- 20.Ulrich, C., M. Hackethal, T. Meyer, A. Geusau, I. Nindl, M. Ulrich, T. Forschner, W. Sterry, and E. Stockfleth. 2008. Skin infections in organ transplant recipients. J. Dtsch. Dermatol. Ges. 6:98-105. [DOI] [PubMed] [Google Scholar]
- 21.van Doorn, L. J., W. Quint, B. Kleter, A. Molijn, B. Colau, M. T. Martin, I. Kravang, N. Torrez-Martinez, C. L. Peyton, and C. M. Wheeler. 2002. Genotyping of human papillomavirus in liquid cytology cervical specimens by the PGMY line blot assay and the SPF10 line probe assay. J. Clin. Microbiol. 40:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Haalen, F. M., S. C. Bruggink, J. Gussekloo, W. J. Assendelft, and J. A. Eekhof. 2009. Warts in primary schoolchildren: prevalence and relation with environmental factors. Br. J. Dermatol. 161:148-152. [DOI] [PubMed] [Google Scholar]