Abstract
Rhodococcus equi and Dietzia spp. are closely related actinomycetes that show similar phenotypic properties. In humans, R. equi is an opportunistic pathogen associated with severe immunodeficiency. Dietzia spp. are environmental bacteria that have been isolated recently from clinical material and are presumptively associated with human infections. During the last 5 years, 15 bacterial isolates from human clinical samples collected at the Hospital Marqués de Valdecilla, Santander, Spain, were identified as R. equi by the API Coryne test. 16S rRNA gene sequencing confirmed seven isolates to be true R. equi strains, whereas the other eight were identified as members of the genus Dietzia, including Dietzia maris (four isolates), Dietzia natronolimnaea (two isolates), and Dietzia timorensis and Dietzia sp. (one isolate each). The eight Dietzia isolates were highly sensitive to 12 antimicrobial compounds.
The nocardioform actinomycete Rhodococcus equi is a multihost pathogen that causes pyogranulomatous infections in a variety of animal species, with special significance in horses and humans (23). In immunocompromised persons, R. equi causes tuberculosis-like pneumonia associated with a high case-fatality rate, particularly in human immunodeficiency virus (HIV)-infected patients (22). The number of cases of infection associated with R. equi in humans increased with the expansion of the AIDS pandemia but in recent years has diminished due to the control of HIV spread.
R. equi infections are diagnosed by culture and subsequent phenotypic analysis of samples by means of classical morphological and biochemical tests (9). In clinical microbiology laboratories, R. equi is usually identified using the API Coryne system (bio-Mérieux, Marcy l'Étoile, France), a commercial multisubstrate kit that includes R. equi in its database. However, its reliability for the identification of rhodococcal isolates is limited, resulting in misidentification of R. equi as other rhodococcal species or even other actinomycetes (9). Several molecular methods based on amplification of DNA sequences by PCR for detection and identification of this pathogen have been proposed. Since R. equi is particularly relevant in equine medicine, most target the plasmid-borne vapA gene, encoding a virulence factor associated with horse pathogenesis. However, these methods are not useful for human isolates, because they generally do not carry vapA-type virulence plasmids but alternative types, like the pig-associated vapB plasmid and a recently identified new bovine type, or are plasmid-less (16). We therefore developed a PCR method for species-specific R. equi identification based on the amplification of the choE gene (11, 20), a chromosomal locus encoding a secreted cholesterol oxidase (14). ChoE is the cytolytic factor responsible for the synergistic hemolysis (CAMP-like) reaction elicited by R. equi in the presence of sphingomyelinase C-producing bacteria, such as Listeria ivanovii, Bacillus cereus, and Staphylococcus aureus (14). We use this CAMP-like reaction as a phenotypic marker for the rapid presumptive identification of R. equi. However, this functional test may miss R. equi isolates not expressing cholesterol oxidase despite having the choE gene or give false-positive results for other extracellular cholesterol oxidase-producing actinomycetes.
During the last 5 years, 15 bacterial isolates with phenotypic properties resembling those of rhodococci were collected from human specimens and isolated at the Clinical Microbiology Laboratory of the Hospital Marqués de Valdecilla, Santander, Spain. All of them were identified as R. equi by the API Coryne test. However, several of these isolates showed colony morphology and pigmentation characteristics that differed from those typical of R. equi strains. The 15 isolates were additionally tested with the CAMP assay and the R. equi-specific choE PCR assay, and their 16S rRNA genes were sequenced. Analysis of 16S rRNA gene sequences provides reliable identification at the species level for most of the clinically significant bacteria (6).
CAMP tests were performed on sheep blood agar plates with Columbia base medium (bioMérieux) and L. ivanovii ATCC 19119 as the indicator strain, as described previously (14). Highly purified genomic DNA samples from all the strains under study were prepared by following a previously described protocol for R. equi genomic DNA extraction (14). The R. equi-specific choE PCR was carried out with COX primers as described in detail elsewhere (11). The universal primers PA (5′-AGAGTTTGATCCTGGCTCAG-3′) and PL06 (5′-GGTTAAGTCCCGCAACGAGCGA-3′) for the forward strand and PH (5′-AAGGAGGTGATCCAGCCGCA-3′) and PLO6-R (5′-GCGCTCGTTGCGGGACTTAACC-3′) for the reverse strand were used to generate overlapping DNA amplicons from the 16S rRNA genes. These amplicons were sequenced by primer walking using dye terminator chemistry in an Applied Biosystems model 377 apparatus. Homology searches were performed with the BLAST tool (1) at the website of the National Center for Biotechnology Information (Bethesda, MD; http://www.ncbi.nlm.nih.gov). The 16S rRNA gene sequences from the eight isolates identified as Dietzia species (Table 1) were registered in the GenBank database. A phylogenetic tree was inferred from the aligned sequences by the neighbor-joining method (21).
TABLE 1.
Identification of eight Dietzia isolates initially classified as R. equi or Rhodococcus sp.
| Isolatea | Result of CAMP test (using L. ivanovii) | API Coryne identification | Result of choE-specific PCR assay | Length (bp) of 16S rRNA gene sequence | Identification based on 16S rRNA gene sequence (% identity to best match) | GenBank accession no. for 16S rRNA gene |
|---|---|---|---|---|---|---|
| CA138 | + | R. equi | + | 1,424 | D. maris DSM 43672 (99.8) | GU247959 |
| CA149 | + | Rhodococcus sp. | − | 1,467 | D. schimae YIM 65001 (94.6) | GQ870422 |
| CA150 | + | Rhodococcus sp. | − | 1,468 | D. maris DSM 43672 (99.7) | GQ870423 |
| CA155 | − | Gordonia, Dietzia, or Nocardia species or R. equi | − | 1,472 | D. timorensis ID05-A0528 (99.6) | GQ870424 |
| CA160 | − | Rhodococcus sp. | − | 1,465 | D. maris DSM 43672 (99.9) | GQ870425 |
| CA161 | − | Rhodococcus sp. | − | 1,433 | D. natronolimnaea CV46 (99.5) | GQ870426 |
| CA165 | − | R. equi | − | 1,451 | D. natronolimnaea CV46 (100) | GQ870427 |
| CA167 | − | R. equi | − | 1,373 | D. maris DSM 43672 (100) | GQ870428 |
| ATCC 616106 | − | − | D. maris | |||
| ATCC 6939 | + | + | R. equi |
ATCC 616106 and ATCC 6939 are reference type strains of D. maris and R. equi, respectively.
Ten of the 15 isolates gave a positive CAMP test result with L. ivanovii, whereas PCR amplification of the genomic DNA samples with COX primers gave positive results for only 8 isolates. Sequencing of the 16S rRNA genes from all isolates revealed that for eight isolates, the best match was a sequence from the genus Dietzia. The genus Dietzia has been established very recently and includes environmental isolates with great resemblance in morphology and colony appearance to R. equi (13). Several authors have reported the isolation of Dietzia strains, notably Dietzia maris (3, 18) but also Dietzia papillomatosis (12) and Dietzia cinnamea (25), from human clinical material. The 16S rRNA genes of isolates CA138, CA150, CA160, and CA167 showed >99% identity to the corresponding gene from D. maris strain DSM 43672 (Table 1), the type strain of the genus Dietzia (15). D. maris has been isolated from a variety of environments, including soils, deep sea mud, and the dinoflagellate Pyrodinium bahamense (2). It has also been recovered from the skin of healthy humans (8), and in four instances, it has been associated with human infections. Bemer-Melchior et al. isolated D. maris from the blood of an immunocompromised patient suffering septic shock and carrying a long-term central line in situ (3). This D. maris isolate was susceptible to β-lactam agents, aminoglycosides, macrolides, pristinamycin, rifampin, trimethoprim-sulfamethoxazole, and vancomycin. D. maris was also identified as the causal agent of a prosthetic hip infection (18). This isolate was susceptible to amoxicillin, imipenem, gentamicin, pristinamycin, trimethoprim-sulfamethoxazole, rifampin, clindamycin, and vancomycin. Reyes et al. isolated D. maris from the pericardial liquid and the aortic wall of a patient with aortitis (19). The isolate was susceptible to teicoplanin, tobramycin, linezolid, rifampin, imipenem, ciprofloxacin, gentamicin, vancomycin, clarithromycin, clindamycin, and amoxicillin-clavulanic acid. Finally, Broseta et al. isolated D. maris from the blood of a patient with cardiac and respiratory insufficiency (5).
The 16S rRNA genes of two isolates, CA161 and CA165, were >99% identical to that of Dietzia natronolimnaea CV46. D. natronolimnaea strains have been isolated from salt lakes, alkaline waters, and the guts of Japanese horned beetle (Trypoxylus dichotomus) larvae (13). Morphologically, colonies from isolate CA155 did not resemble those from the other five isolates. The CA155 16S rRNA gene was 99% identical to that from Dietzia timorensis ID05-A0528. This species was proposed to classify a soil isolate from West Timor, Indonesia, and has recently been validly recognized as a novel Dietzia species (24). This is the first reported isolation of the alkaliphilic D. natronolimnaea and the recently established species D. timorensis from human specimens. Finally, the 16S rRNA gene of isolate CA149 was most similar (94.6%) to that of Dietzia schimae YIM 65001. However, since the levels of identity of the CA149 16S rRNA gene to genes from D. schimae YIM 65001 and other Dietzia spp. registered in GenBank were lower than 99%, CA149 was assigned the identification Dietzia sp.
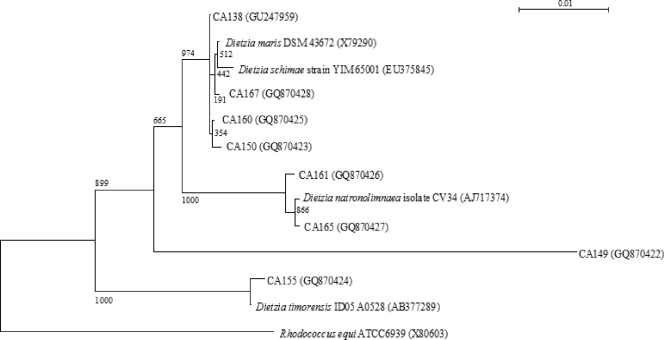
Figure 1 shows a phylogenetic tree based on 16S rRNA genes depicting the relationships among the eight Dietzia isolates identified in this study and representative strains of Dietzia and R. equi. Isolate CA149 forms a detached branch in the tree, whereas the other isolates group with a representative Dietzia strain.
FIG. 1.
Neighbor-joining tree based on 16S rRNA gene sequences showing the phylogenetic relationships among isolates identified as Dietzia and representative strains of Dietzia and R. equi. Bar, 1% sequence variation. The bifurcations are supported by the indicated bootstrap values.
Previous reports by other authors and our own data suggest that dietziae have the potential to act as opportunistic pathogens. To date, no comprehensive investigation of the susceptibilities of human Dietzia isolates to antibiotics has been carried out. The in vitro susceptibilities of the eight identified Dietzia bacteria to antimicrobial agents were evaluated by the Etest diffusion gradient method according to the instructions of the Etest manufacturer (AB Biodisk, Solna, Sweden). The guidelines of the CLSI for testing the antibiotic susceptibilities of mycobacteria, nocardiae, and other aerobic actinomycetes (7) were followed. The antibiotics recommended by the CLSI, including amikacin, amoxicillin-clavulanic acid, ceftriaxone, ciprofloxacin, clarithromycin, trimethoprim-sulfamethoxazole (cotrimoxazole), imipenem, linezolid, minocycline, and tobramycin, were tested. Ampicillin and vancomycin were also tested. Escherichia coli ATCC 25922 and S. aureus ATCC 25923 served as controls. Resistance or susceptibility to the antibiotics was interpreted as suggested by the CLSI standards (7). The results of susceptibility tests are given in Table 2. The eight Dietzia isolates and ATCC 616106, a collection strain representative of D. maris, were susceptible to all antibiotics tested. D. maris has been reported previously to be susceptible to aztreonam, ciprofloxacin, mezlocillin, oxacillin, penicillin G, perfloxacin, and ticarcillin and to be resistant to sulfamethoxazole by disk diffusion testing (4).
TABLE 2.
Antibiotic susceptibilities of the eight Dietzia isolates identified in this study
| Isolate | MICa (μg/ml) of: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | AMC | AMP | CRO | CIP | CLR | COT | IPM | LZD | MIN | TOB | VAN | |
| D. maris isolate CA138 | 0.25 | 0.016 | 0.047 | 0.06 | 0.032 | 0.016 | 1 | 0.125 | 0.125 | 0.064 | 0.19 | 0.19 |
| Dietzia sp. isolate CA149 | 0.50 | 0.016 | 0.500 | 0.750 | 0.016 | <0.016 | 0.094 | 0.064 | 0.250 | 0.032 | 0.500 | 0.500 |
| D. maris isolate CA150 | 0.75 | 0.032 | 0.250 | 0.750 | 0.032 | <0.016 | 0.094 | 0.047 | 0.190 | 0.047 | 0.750 | 0.125 |
| D. maris isolate CA160 | 0.50 | 0.032 | 0.250 | 0.250 | 0.047 | <0.016 | 0.032 | 0.064 | 0.380 | 0.125 | 0.190 | 0.500 |
| D. maris isolate CA167 | 0.125 | <0.016 | 0.047 | 0.750 | 0.008 | <0.016 | 0.094 | <0.002 | 0.016 | <0.016 | 0.094 | 0.190 |
| D. natronolimnaea isolate CA161 | 1.00 | 0.047 | 0.380 | 0.500 | 0.023 | 0.047 | 0.5 | 0.032 | 0.500 | 0.250 | 1.500 | 0.125 |
| D. natronolimnaea isolate CA165 | 0.50 | <0.016 | 0.250 | 0.750 | 0.012 | 0.250 | 0.19 | 0.032 | 0.190 | 0.023 | 0.500 | <0.016 |
| D. timorensis isolate CA155 | 0.125 | 0.016 | 0.750 | 0.750 | 0.032 | 0.120 | 0.25 | 0.064 | 0.032 | 0.064 | 0.023 | 0.500 |
| R. equi ATCC 6939 | 2.00 | 0.016 | 0.380 | 0.094 | 0.047 | <0.016 | 0.064 | 0.012 | 0.190 | 0.064 | 0.125 | 0.750 |
| D. maris ATCC 616106 | 0.064 | <0.016 | 0.023 | 0.032 | 0.006 | <0.016 | 0.016 | 0.750 | 0.125 | 0.500 | 0.190 | 0.250 |
The MICs were determined by the Etest method. The medium used was Mueller-Hinton agar supplemented with sheep blood. AMK, amikacin; AMC, amoxicillin-clavulanic acid; AMP, ampicillin; CRO, ceftriaxone; CIP, ciprofloxacin; CLR, clarithromycin; COT, cotrimoxazole; IPM, imipenem; LZD, linezolid; MIN, minocycline; TOB, tobramycin; VAN, vancomycin. R. equi ATCC 6939 and D. maris ATCC 616106 were included as reference strains.
Our findings show that discrimination between R. equi and Dietzia species is problematic in the clinical microbiology laboratory and that in the absence of clear-cut markers, 16S rRNA gene sequencing is required for accurate identification of isolates of these species. Species-level identification based on the full 16S rRNA gene sequence is considered to be the “gold standard” for actinomycetes (17), but results must be treated with caution for recently established and grossly undercharacterized genera, such as Dietzia (13). In our case, eight isolates erroneously identified as R. equi could be recognized as members of the genus Dietzia and seven of them could be identified at the species level.
Dietzia bacteria appear to be widely distributed in the environment, and reports of isolates from clinical material are increasing. D. maris was detected in skin samples from healthy subjects in a recent characterization of the skin microbiota using a 16S rRNA gene-based comprehensive analysis (8), suggesting that it may be a normal human commensal. Our finding of species of Dietzia, initially wrongly identified as R. equi, in human clinical specimens highlights the potential importance of members of this newly defined bacterial genus as opportunistic human pathogens and the need for accurate methods to ensure their correct identification in the clinical setting.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences from the eight Dietzia isolates were deposited in GenBank under accession numbers GU247959 and GQ870422 to GQ870428.
Acknowledgments
This work was funded by R.E.I.P.I. (Red Española de Investigación en Patología Infecciosa). Lilian Pilares is the recipient of a fellowship from the Agencia Española para la Cooperación Internacional (A.E.C.I.). Work in the J.A.V.-B. laboratory is funded by the Horserace Betting Levy Board and The Wellcome Trust (United Kingdom).
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Azanza, M. P., R. V. Azanza, V. M. Vargas, and C. T. Hedreyda. 2006. Bacterial endosymbionts of Pyrodinium bahamense var. compressum. Microb. Ecol. 52:756-764. [DOI] [PubMed] [Google Scholar]
- 3.Bemer-Melchior, P., A. Haloun, P. Riegel, and H. B. Drugeon. 1999. Bacteremia due to Dietzia maris in an immunocompromised patient. Clin. Infect. Dis. 29:1338-1340. [DOI] [PubMed] [Google Scholar]
- 4.Bizet, C., C. Barreau, C. Harmant, M. Nowakowski, and A. Pietfroid. 1997. Identification of Rhodococcus, Gordona and Dietzia species using carbon source utilization tests (“Biotype-100” strips). Res. Microbiol. 148:799-809. [DOI] [PubMed] [Google Scholar]
- 5.Broseta, A., C. Gómez, F. Chaves, and F. Sanz. 2007. Dietzia maris bacteremia: use of molecular techniques for accurate identification of Actinomycetes. Clin. Microbiol. Newsl. 29:19-20. [Google Scholar]
- 6.Clarridge, J. E., III. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CLSI/NCCLS. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. CLSI/NCCLS document M24-A. CLSI/NCCLS, Wayne, PA. [PubMed]
- 8.Dekio, I., M. Sakamoto, H. Hayashi, M. Amagai, M. Suematsu, and Y. Benno. 2007. Characterization of skin microbiota in patients with atopic dermatitis and in normal subjects using 16S rRNA gene-based comprehensive analysis. J. Med. Microbiol. 56:1675-1683. [DOI] [PubMed] [Google Scholar]
- 9.Funke, G., F. N. Renaud, J. Freney, and P. Riegel. 1997. Multicenter evaluation of the updated and extended API (RAPID) Coryne database 2.0. J. Clin. Microbiol. 35:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Ladrón, N., M. Fernández, J. Agüero, B. González Zörn, J. A. Vázquez-Boland, and J. Navas. 2003. Rapid identification of Rhodococcus equi by a PCR assay targeting the choE gene. J. Clin. Microbiol. 41:3241-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, A. L., R. J. Koerner, S. Natarajan, J. D. Perry, and M. Goodfellow. 2008. Dietzia papillomatosis sp. nov., a novel actinomycete isolated from the skin of an immunocompetent patient with confluent and reticulated papillomatosis. Int. J. Syst. Evol. Microbiol. 58:68-72. [DOI] [PubMed] [Google Scholar]
- 13.Koerner, R. J., M. Goodfellow, and A. L. Jones. 2009. The genus Dietzia: a new home for some known and emerging opportunistic pathogens. FEMS Immunol. Med. Microbiol. 55:296-305. [DOI] [PubMed] [Google Scholar]
- 14.Navas, J., B. González-Zörn, N. Ladrón, P. Garrido, and J. A. Vázquez-Boland. 2001. Identification and mutagenesis by allelic exchange of choE, encoding a cholesterol oxidase from the intracellular pathogen Rhodococcus equi. J. Bacteriol. 183:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nesterenko, O. A., T. M. Nogina, E. I. Kasumova, E. I. Kvasnikow, and S. G. Batrakov. 1982. Rhodococcus luteus nom. nov. and Rhodococcus maris nom. nov. Int. J. Syst. Bacteriol. 32:1-14. [Google Scholar]
- 16.Ocampo-Sosa, A. A., D. A. Lewis, J. Navas, F. Quigley, R. Callejo, M. Scortti, D. P. Leadon, U. Fogarty, and J. A. Vázquez-Boland. 2007. Molecular epidemiology of Rhodococcus equi based on traA, vapA, and vapB virulence plasmid markers. J. Infect. Dis. 196:763-769. [DOI] [PubMed] [Google Scholar]
- 17.Patel, J. B., R. J. Wallace, B. A. Brown-Elliott, T. Taylor, C. Imperatrice, D. G. B. Leonard, R. W. Wilson, L. Mann, K. C. Jost, and I. Nachamkin. 2004. Sequence-based identification of aerobic actinomycetes. J. Clin. Microbiol. 42:2530-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pidoux, O., J. N. Argenson, V. Jacomo, and M. Drancourt. 2001. Molecular identification of a Dietzia maris hip prosthesis infection isolate. J. Clin. Microbiol. 39:2634-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes, G., J. L. Navarro, C. Gamallo, and M. C. de las Cuevas. 2006. Type A aortic dissection associated with Dietzia maris. Interact. Cardiovasc. Thorac. Surg. 5:666-668. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-Lázaro, D., D. A. Lewis, A. A. Ocampo-Sosa, U. Fogarty, L. Makrai, J. Navas, M. Scortti, M. Hernández, and J. A. Vázquez-Boland. 2006. Internally controlled real-time PCR method for quantitative species-specific detection and vapA genotyping of Rhodococcus equi. Appl. Environ. Microbiol. 72:4256-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 22.Takai, S., Y. Sasaki, T. Ikeda, Y. Uchida, S. Tsubaki, and T. Sekizaki. 1994. Virulence of Rhodococcus equi from patients with and without AIDS. J. Clin. Microbiol. 32:457-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Von Bargen, K., and A. Haas. 2009. Molecular and infection biology of the horse pathogen Rhodococcus equi. FEMS Microbiol. Rev. 33:870-891. [DOI] [PubMed] [Google Scholar]
- 24.Yamamura, H., P. Lisdiyanti, R. Ridwan, S. Ratnakomala, R. Sarawati, Y. Lestari, E. Triana, G. Kartina, Y. Widyastuti, and K. Ando. 2009. Dietzia timorensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 60:451-454. [DOI] [PubMed] [Google Scholar]
- 25.Yassin, A. F., H. Hupfer, and K. P. Schaal. 2006. Dietzia cinnamea sp. nov., a novel species isolated from a perianal swab of a patient with a bone marrow transplant. Int. J. Syst. Evol. Microbiol. 56:641-645. [DOI] [PubMed] [Google Scholar]