Abstract
A total of 412 methicillin-resistant Staphylococcus aureus (MRSA) strains isolated between October 2006 and June 2009, representing a mixed hospital- and community-associated patient population from Mumbai, India, were evaluated. MRSA was characterized by multiplex PCR amplification of the Panton-Valentine leukocidin (PVL) gene and the mecA gene, staphylococcal cassette chromosome mec (SCCmec) typing, and multilocus sequence typing (MLST). PCR results were compared with patient risk factors (CDC guidelines) and antimicrobial susceptibility profiles. A total of 395 MRSA strains were mecA positive, and 224 were PVL gene positive. A total of 97 mecA-positive strains were SCCmec III (25%), 136 were SCCmec IV (34%), and 162 were SCCmec V (41%). All SCCmec III strains were multidrug resistant, and all patients had risk factors. Of the SCCmec IV and V strains, 73% were multidrug susceptible and 72% of the associated patients had no risk factors. The multidrug susceptibility and absence of patient risk factors in 72% of cases with SCCmec IV and SCCmec V MRSA demonstrate the presence of community-associated MRSA (CA-MRSA) in Mumbai. Twenty-one percent of these patients had risk factors, signifying CA-MRSA infiltration into hospitals. MLST showed clonal expansion of multidrug-susceptible sequence type (ST) 22 (SCCmec IV) and ST 772 (SCCmec V), both of which feature in Asian studies and may be slowly replacing the multidrug-resistant ST 239 (SCCmec III) in hospitals. The PVL gene-positive methicillin-sensitive S. aureus (MSSA) strains were ST 30 and were postulated to be related to the penicillin-resistant S. aureus phage type 80/81, notorious for its virulence in the 1950s.
Staphylococcus aureus causes disease ranging from mild skin infection to sepsis, pneumonia, and toxic shock syndrome (32). In the preantibiotic era, mortality from S. aureus disease was high and introduction of penicillin had a dramatic impact, which was short-lived due to the emergence of penicillinase-producing S. aureus (2, 4, 20). Methicillin was introduced in 1959, and methicillin-resistant Staphylococcus aureus (MRSA) strains rapidly emerged and became a major problem in hospitals in the 1960s. These nosocomial MRSA strains were multidrug resistant (MDR), many being susceptible only to glycopeptides (1, 2, 4, 32). The past 10 years have seen reports of reduced susceptibility (vancomycin-intermediate S. aureus [VISA]) and resistance (vancomycin-resistant S. aureus [VRSA]) to vancomycin (29).
The genome of methicillin-resistant staphylococci contains a 21- to 67-kb heterologous mobile genetic element, termed staphylococcal cassette chromosome mec (SCCmec), harboring the mecA gene and other resistance determinants. Methicillin resistance is mediated by production of an altered penicillin-binding protein, PBP-2a, encoded by the mecA gene (1, 2, 5, 9, 15). The tendency of most strains to be heterogeneously resistant to methicillin, oxacillin, and the cephalosporins accounts for many problems that clinical microbiology laboratories encounter with MRSA detection by phenotypic susceptibility methods (15). SCCmec characterization relies on variations of the mec gene complex, cassette chromosome recombinase (ccr) complex, and junkyard regions. Five SCCmec types (I to V) have been confirmed in S. aureus (13, 14, 21, 26). SCCmec VI and VII types have also been discovered, but they are rarer (27).
MRSA strains were previously known for being associated with nosocomial infections (HA-MRSA). However, MRSA strains now cause community-acquired infections (CA-MRSA), especially skin and soft tissue infections (SSTI) and necrotizing pneumonia (2). Although diversity in genomes and antibiograms exists, most CA-MRSA strains carry Panton-Valentine leukocidin (PVL) genes and possess a small mobile SCCmec type IV, V, or VI, which is more easily transferable than the larger SCCmec types I, II, and III in HA-MRSA (1, 2). PVL toxin is a bicomponent leukocidin encoded by the lukS-PV and lukF-PV genes, which reside on a prophage. PVL toxin causes leukocyte destruction, tissue necrosis, and increased disease severity (1, 2, 18). CA-MRSA poses a significant threat to public health. HA-MRSA and CA-MRSA differ on the basis of SCCmec types and antimicrobial susceptibilities. Patients with HA-MRSA infections have risk factors like history of exposure to an intensive care unit (ICU), hospital stay, antimicrobial exposure, surgery, and invasive procedures. CA-MRSA infections, on the other hand, are indicated among people living in crowded areas, sharing personal items and gym equipment, as well as intravenous drug users, sports participants, and homosexuals (5, 31).
CA-MRSA cases are being reported all over the world, yet there have not been many studies regarding the actual incidence of CA-MRSA infections in India (5, 8). This study comprises MRSA strains isolated in the laboratory, including simultaneous documentation of clinical histories, risk factors, and antimicrobial susceptibility patterns. PCR amplification of PVL genes, SCCmec typing, and multilocus sequence typing (MLST) enabled characterization of MRSA (4, 6, 16, 18, 23, 24, 32). This gave insight into SCCmec types isolated in the laboratory and the frequency of PVL+ MRSA and CA-MRSA.
MATERIALS AND METHODS
Bacterial reference strains and clinical isolates.
PVL+ S. aureus and SCCmec reference strains were kindly provided by Jerome Etienne, INSERM, Lyon, France, and Dilip Mathai and Anand Manoharan, CMC, Vellore, India. The local MRSA strains were collected in the microbiology laboratory. The hospital, being a tertiary care center, also receives numerous requests for culture from other hospitals, nursing homes, and general practitioners from all over Mumbai, India. Thus, the MRSA in this study represents a mixed patient population from hospitals and communities in the city.
Isolation and confirmation of MRSA by phenotypic methods.
MRSA strains were isolated from clinical samples (nasal and axillary screening swabs, blood, pus, soft tissue, wound swabs, respiratory samples, and body fluids) from October 2006 to June 2009. Staphylococci were identified morphologically and biochemically by standard laboratory procedures. S. aureus was identified by tube coagulase and latex agglutination test for coagulase and protein A (Bio-Rad), along with mannitol fermentation on mannitol salt agar (Himedia). Methicillin resistance was confirmed using a cefoxitin disk on Mueller-Hinton agar (Oxoid) and observation of growth on oxacillin screen agar (Himedia) according to CLSI guidelines. Antimicrobial susceptibility testing was performed by disk diffusion on Mueller-Hinton agar (Oxoid) according to CLSI guidelines.
Documentation of patient risk factors and antimicrobial susceptibility patterns.
A standard form was filled out for each patient whose sample grew MRSA, with details like the name, age, sex, phone number, and hospital admission specifics. Clinical diagnosis along with risk factors (Centers for Disease Control and Prevention [CDC] guidelines) for acquiring MRSA (previous ICU or hospital stay, antimicrobial exposure, surgery or invasive procedures, etc.) was documented, along with antimicrobial susceptibility patterns (by disk diffusion according to CLSI guidelines) (5).
DNA extraction.
Rapid DNA extraction was performed from bacterial colonies as previously described (32).
Molecular characterization by PCR amplification and DNA sequencing.
Five PCR assays carried out using an MJ Research PTC-200 thermocycler were used for characterization of the MRSA. The first four were multiplex PCRs, and the last was a multilocus sequence typing (MLST) assay, comprised of PCRs for seven housekeeping genes in S. aureus along with DNA sequencing methods.
MRSA-PVL PCR-1.
The PCR mixture contained four sets of primers: PVL (toxin gene), mecA (methicillin resistance), nuc (specific for S. aureus), and SS756 (IC) (Staphylococcus species) (18, 32). PCR was performed in a 25-μl reaction volume with 2.5 μl DNA, 1× Taq buffer, 100 μmol deoxynucleoside triphosphate (dNTP) mixture (Fermentas), 5 mM MgCl2, 12.5 pmol SS756 (IC), 7.5 pmol each PVL, mecA, and nuc primers (MWG Biotech, India), and 1 unit Hotstar Taq DNA polymerase (Qiagen). Amplification was performed with denaturation at 95°C for 15 min, followed by 10 cycles of 95°C for 45 s, 67°C for 45 s, and 72°C for 1.5 minutes; 25 cycles of 95°C for 45 s, 57°C for 45 s, and 72°C for 1.5 minutes; and extension at 72°C for 10 min.
SCCmec-typing PCR-2.
The PCR mixture contained nine sets of primers: CIF, ccrC, RIF, mecV, DCS, ccrß, KDP, mecIII, and mecR1 (targeting constituents of SCCmec), as well as mecA-149 (IC) (methicillin resistance) (24). PCR was performed in a 25-μl reaction volume with 2.5 μl DNA, 1× Taq buffer, 100 μmol dNTP mixture (Fermentas), 4 mM MgCl2, 12.5 pmol each CIF, DCS, ccrß, KDP, mecIII, mecR1, and mecA-149 (IC), 2.5 pmol each RIF and mecV, 20 pmol ccrC primers (MWG Biotech, India), and 0.75 units Hotstar Taq DNA polymerase (Qiagen). Amplification was performed with denaturation at 95°C for 15 min; 30 cycles of 95°C for 25 s, 56°C for 25 s, and 72°C for 1 min; and extension at 72°C for 4 min.
SCCmec IV-subtyping PCR-3.
The PCR mixture contained six sets of primers: IVa, IVb, IVc, IVd, IVg, and IVh (SCCmec IV subtypes), as well as ccrß-210 (IC) (SCCmec IV strains) (23). PCR was performed in a 25-μl reaction volume with 2.5 μl DNA, 1× Taq buffer, 100 μmol dNTP mixture (Fermentas), 4 mM MgCl2, 5 pmol each ccrß-210, IVa, and IVb, 10 pmol each IVc and IVd, 20 pmol each IVg and IVh (MWG Biotech, India), and 0.5 units Hotstar Taq DNA polymerase (Qiagen). Amplification was performed with denaturation at 95°C for 15 min; 35 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 1.5 minutes; and extension at 72°C for 5 min.
ccr-typing PCR-4.
The PCR mixture contained primers ccrABα1-F, ccrABα2-F, ccrABα3-F, ccrABβ2-R, ccrABα4-F, ccrABα4-R, ccrCΥ-F, and ccrCΥ-R (ccr types) (16). PCR was performed in a 25-μl reaction volume with 2.5 μl DNA, 1× Taq buffer, 100 μmol dNTP mixture (Fermentas), 5 mM MgCl2, 30 pmol each ccrABα3-F and ccrABβ2-R, 25 pmol each ccrABα4-F and ccrABα4-R, 0.25 pmol ccrABα1-F, 0.125 pmol ccrABα2-F, 0.75 pmol each ccrCΥ-F and ccrCΥ-R primers (MWG Biotech, India), and 2 units Hotstar Taq DNA polymerase (Qiagen). Amplification was performed with denaturation at 95°C for 15 min, followed by 10 cycles of 95°C for 2 min, 60°C for 1 min, and 72°C for 2 min; 20 cycles of 95°C for 2 min, 50°C for 1 min, and 72°C for 2 min; and extension at 72°C for 2 min.
MRSA-MLST PCR-5.
Each PCR mixture contained a set of primers (arc, aro, glp, gmk, pta, tpi, and yqi) (6). PCRs were performed in a 25-μl reaction volume with 2.5 μl DNA, 1× Taq buffer, 100 μmol dNTP mixture (Fermentas), 4 mM MgCl2, 12.5 pmol primers (MWG Biotech, India), and 0.5 unit Hotstar Taq DNA polymerase (Qiagen). Amplification was performed with denaturation at 95°C for 15 min, followed by 30 cycles of 95°C for 1 min, 51°C for 1 min, and 72°C for 1 min and extension at 72°C for 5 min.
Electrophoresis of PCR amplicons.
The amplicons from PCR-1, -2, -3, and -5 were separated on 4% agarose gels in 1× TAE buffer (Sigma) using a 50-bp reference ladder (Fermentas), and those of PCR-4 were separated on 2% agarose gels in 1× TAE buffer (Sigma) using a 100-bp reference ladder (Fermentas). Gels were documented under a UV transilluminator. Amplicons of PCR-5 were purified using a gel extraction spin column PCR cleanup kit (Chromous Biotech) and sequenced by the ABI 3130 genetic analyzer (6). All PCRs were validated using appropriate positive and negative controls and by DNA sequencing.
RESULTS
Strain collection.
A total of 412 MRSA strains were cultured from October 2006 to June 2009. Of these, 61% were from the IPD and 39% from the OPD. Of the IPD samples, 98 came from the ICU and 154 came from the wards. Isolates were from pus/tissue (n = 186), wound swabs (n = 66), blood (n = 51), screening swabs (n = 37), respiratory tract (n = 28), fluids (n = 21), and miscellaneous sources (n = 6) (Table 1).
TABLE 1.
Frequency of the PVL gene in MRSA among each SCCmec type isolateda
| MRSA source | No. of strains | No. of strains of PVL phenotype |
|||||
|---|---|---|---|---|---|---|---|
| SCCmec III (n = 97) |
SCCmec IV (n = 136) |
SCCmec V (n = 162) |
|||||
| PVL+ | PVL− | PVL+ | PVL− | PVL+ | PVL− | ||
| Pus/tissue | 186 | 24 | 72 | 7 | 60 | 23 | |
| Wound swabs | 66 | 17 | 19 | 18 | 12 | ||
| Blood | 51 | 18 | 16 | 3 | 4 | 10 | |
| Screening swabs | 37 | 16 | 6 | 1 | 9 | 5 | |
| Respiratory | 28 | 12 | 5 | 7 | 4 | ||
| Fluids | 21 | 6 | 4 | 2 | 3 | 6 | |
| Miscellaneous | 6 | 4 | 1 | 1 | |||
| Total | 395 | 97 | 123 | 13 | 101 | 61 | |
SCCmec, staphylococcal cassette chromosome mec; PVL, Panton-Valentine leukocidin gene.
Multiplex PCR results.
A total of 412 cultured MRSA strains were subjected to PCR-1 for nuc, mecA, and PVL genes. Of these, 395 proved to be MRSA genotypically (presence of mecA and nuc) and 224 were PVL gene positive. The 395 MRSA strains were subjected to PCRs 2, 3, and 4. In PCR-2, 97 strains were SCCmec III, 136 were SCCmec IV, and 162 were SCCmec V. In PCR-3, 135 SCCmec IV strains were SCCmec IVc. One strain was SCCmec IVa. Results of PCR-4 correlated with those of PCR-2. A total of 97 strains were ccrAB3, 136 were ccrAB2, and 162 were the ccrC type. Of 412 MRSA strains, 17 proved to be methicillin-resistant coagulase-negative staphylococci (MRCNS) and PVL+ methicillin-susceptible S. aureus (MSSA) (Table 1).
Antimicrobial susceptibility patterns.
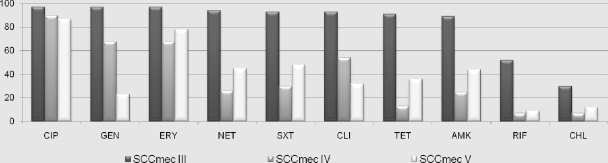
Of the SCCmec III strains, 38% were MDR and the rest were susceptible only to chloramphenicol, rifampin, vancomycin, and linezolid. Of the SCCmec IV strains, 83% were susceptible to many antimicrobial classes and the rest were susceptible to three classes, none being MDR. Of the SCCmec V strains, 64% were susceptible to many antimicrobial classes, 24% were susceptible to three classes, and 12% were MDR (Fig. 1).
FIG. 1.
Antimicrobial resistance patterns of MRSA from the laboratory. CIP, ciprofloxacin; GEN, gentamicin; ERY, erythromycin; SXT, trimethoprim-sulfamethoxazole; NET, netilmicin; AMK, amikacin; TET, tetracycline; CLI, clindamycin; RIF, rifampin; CHL, chloramphenicol (resistance data represented as percentages among SCCmec types III, IV, and V).
Patient independent risk factors.
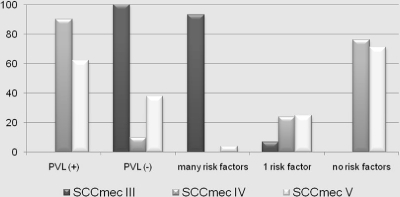
All patients with SCCmec III strains had risk factors for MRSA infections. Of the patients with SCCmec IV strains, 24% had risk factors and 76% had no documented risk factors. Of the patients with SCCmec V strains, 32% had risk factors and 68% had no documented risk factors (Table 2; Fig. 2).
TABLE 2.
Patient breakdown and risk factors for HA-MRSA among the SCCmec types
| Patient status or risk factor | No. of strains per group |
|||
|---|---|---|---|---|
| SCCmec III | SCCmec IV | SCCmec V | Total | |
| Patient location | ||||
| Inpatient status | 97 | 54 | 91 | 242 |
| Outpatient status | 82 | 71 | 153 | |
| Risk factors for HA-MRSA | ||||
| Time from admission to culture >48 h | 72 | 16 | 10 | 98 |
| Previous hospitalization | 78 | 23 | 35 | 136 |
| Interhospital transfer | 23 | 6 | 7 | 36 |
| ICU stay | 87 | 3 | 8 | 98 |
| Hospital stay | 95 | 14 | 23 | 132 |
| Surgery | 63 | 12 | 17 | 92 |
| Catheterization | 96 | 14 | 23 | 133 |
| Intubation | 84 | 3 | 7 | 94 |
| Other invasive procedures | 78 | 20 | 37 | 135 |
| Immunocompromised status, chronic illness | 95 | 25 | 35 | 155 |
| Antimicrobial exposure | 75 | 29 | 32 | 136 |
| Patients with other risk factors | 97 | 32 | 52 | 181 |
| Patients without risk factors | 104 | 110 | 214 | |
FIG. 2.
Characteristics of the MRSA strains in the study: PVL gene positivity and patient risk factors.
Multilocus sequence typing.
MLST was performed for 13 representative SCCmec III MRSA strains and 42 representative SCCmec IV and SCCmec V MRSA strains, with similar antimicrobial susceptibility patterns and PVL positivity. For comparative purposes, MLST was also performed for 11 PVL+ MSSA strains. Each PCR amplicon was sequenced and analyzed (4, 5). All 13 SCCmec III MRSA strains were sequence type (ST) 239, with the allelic profile 2-3-1-1-4-4-3. All 29 SCCmec IV MRSA strains and 13 SCCmec V MRSA strains were PVL gene positive. All 29 SCCmec IV strains were ST 22, with the allelic profile 7-6-1-5-8-8-6, and all 13 SCCmec V strains were ST 772, with the allelic profile 1-1-1-1-22-1-1. All 11 PVL+ MSSA strains were ST 30, with the allelic profile 2-2-2-2-6-3-2 (Table 3).
TABLE 3.
Characterization of HA-MRSA, CA-MRSA, and PVL+ MSSA by MLSTa
| SA type (n) | SCCmec (n) | PVL status (n) | MLST profileb | ST:CCc | Epidemic clone |
|---|---|---|---|---|---|
| MRSA (55) | III (13) | − (13) | 2-3-1-1-4-4-3 | 239:8 | Brazilian (SLV of ST 8) |
| IV (29) | + (28) | 7-6-1-5-8-8-6 | 22:22 | United Kingdom (EMRSA-15) | |
| V (13) | + (13) | 1-1-1-1-22-1-1 | 772:1 | Asia (SLV of ST 1) | |
| MSSA (11) | NAd | + (11) | 2-2-2-2-6-3-2 | 30:30 | Southwest Pacific (USA 1100) |
MLST, multilocus sequence type.
MLST profile: arc-aro-glp-gmk-pta-tpi-yqi.
ST, sequence type; CC, clonal complex.
NA, not applicable.
DISCUSSION
From the 412 strains culture confirmed as MRSA, 395 were mecA positive by PCR. Of these MRSA strains, 64% were found to be PVL gene positive by PCR. The highest number of MRSA strains in this study were isolated from SSTI, and 67% of these strains were PVL+. From 2006 to 2009, the isolation of MRSA from pus and tissue samples at the hospital has increased considerably compared to that from other sources. We are aware that a larger study surveying both community and hospitalized patients in a defined geographical area may be more comprehensive. However, consecutive samples were included to try and represent the situation at a tertiary care center laboratory which serves as a referral laboratory by default. Molecular characterization using multiplex PCR assays showed that 25% of the MRSA strains were SCCmec III, 34% were SCCmec IV, and 41% were SCCmec V. None of the SCCmec III MRSA strains were PVL+, but 90% of SCCmec IV and 62% of SCCmec V strains were PVL+. While all SCCmec III strains showed multidrug-resistant phenotypes, all SCCmec IV strains and around 88% of SCCmec V strains were multidrug susceptible (Fig. 1) (10). All patients with SCCmec III strain infections had multiple risk factors for MRSA infections. On the other hand, 76% of patients with SCCmec IV strains and 68% of patients with SCCmec V strains had no documented risk factors (Table 2 and Fig. 2). This particular multidrug susceptibility pattern and absence of risk factors among patients with SCCmec IV and V strains show the emergence of CA-MRSA (Fig. 1 and 2) (5). This is an alarming trend, as such a large portion of patients who had no apparent risk factors seem to have acquired these infections, hence posing a major threat to public health. Even though these “community-acquired” infections were relatively susceptible to multiple antimicrobial classes compared to HA-MRSA, 60% of patients had to be hospitalized or have surgery, as the infections were rather severe, a majority of them being caused by PVL+ MRSA (2). No SCCmec III MRSA strain in this study was PVL positive (Table 4).
TABLE 4.
Characteristics of MRSA SCCmec types in the study
| SCCmec | PVL status | Drug of resistance (% of resistant strains)a | Infection (% with infection)b |
|---|---|---|---|
| III | 100% PVL− | MDR (100) | BSI (19) |
| RTI (12) | |||
| IV, V | 75% PVL+ | ERY (74) | |
| CLI (42) | SSTI (66) | ||
| CIP (90) | BA (6) | ||
| TET (25) |
MDR, multiple drug resistance; ERY, erythromycin; CLI, clindamycin; CIP, ciprofloxacin; TET, tetracycline.
BSI, bloodstream infection; RTI, respiratory tract infection; BA, breast abscess; SSTI, other skin and soft tissue infections.
Thirteen SCCmec III MRSA strains were found to be ST 239, corresponding with the sequence of the Brazilian/Hungarian clone belonging to clonal complex (CC) 8. ST 239 strains are the endemic HA-MRSA clone in many Asian countries, although recent studies show that they are being steadily displaced by emerging CA-MRSA clones (5, 10, 12, 19, 22). ST 239, a single-locus variant (SLV) of USA 300, is thought to have evolved from ST 8 (the USA 300 clone lineage) and ST 30 (the S. aureus phage type 80/81, USA 1100 clone lineage) (5, 15, 17, 19, 28, 31). A total of 29 SCCmec IV (28 of subtype c and 1 of subtype a) MRSA strains were ST 22 (CC 22), corresponding with the sequence type of the epidemic United Kingdom EMRSA-15 strain (5, 10, 12, 22). It has now spread to other parts of the world, especially in many Asian countries. All 13 SCCmec V MRSA strains were ST 772 (CC 1), corresponding with the sequence type of other strains from India (New Delhi and Karnataka) collected by another group in a previous study, information about which has been deposited in the MLST database but is unpublished as yet. The 772 sequence type, an SLV of ST 1 (USA 400) (MW2), was first reported in Bangladesh and has also been reported in Malaysia this year (1, 25). A global epidemiological trial showed the occurrence of ST 239, ST 22, and ST 772 S. aureus in India (8). In this study, the numbers of SCCmec III MRSA strains isolated in the laboratory have been gradually decreasing from 2006 to 2009, whereas the isolation of SCCmec IV and SCCmec V MRSA has increased further with each year (10). The MLST data, along with the other parameters studied, illustrate clonal expansion of the ST 22 and ST 772 lineages among strains in Mumbai. MLST carried out for 11 PVL+ MSSA strains showed that all were ST 30 (CC 30), indicating that they are related to the USA 1100 clone from the Southwest Pacific United States (Table 3) (5, 8, 15, 28). The ST 30 PVL+ MSSA strains are postulated to be descendants of the historical penicillin-resistant epidemic S. aureus phage type 80/81 (5, 15, 28). The ST 30 clone has been implicated as the predominant clonal type in CA-MRSA isolates (SCCmec IVc) in Singapore (11). Since the present study findings show that the SCCmec IVc cassette is present among the CA-MRSA strains, it is possible that the ST 30/SCCmec IVc MRSA may emerge as an important community-acquired epidemic clone (like USA 1100) in Mumbai in the future, as it has in Singapore (11, 28).
In India, probably due to overcrowding and poor personal hygiene, CA-MRSA has spread. Although it mainly manifests in severe soft tissue and skin infections requiring surgical drainage, it is now becoming pronounced in bacteremias affecting neonates, especially from lower economic sections, and breast abscesses in lactating mothers, becoming increasingly common in urban areas. In this study, all patients with SCCmec III MRSA had risk factors, but a significant 21% of patients with strains of the SCCmec IV and SCCmec V MRSA also had risk factors. Therefore, it can be concluded that 25% were true HA-MRSA strains and 54% were true CA-MRSA strains (5, 22, 30). Since 21% of strains had the SCCmec IV or V cassette but the patients had risk factors, the spread of these genotypes, earlier associated only with CA-MRSA, into the hospital environment is indicated (5, 30).
SCCmec characterization by multiplex PCR assays, multilocus sequence typing, antimicrobial susceptibility testing, and documentation of patient risk factors enabled successful characterization of strains isolated in the laboratory, confirming the emergence of CA-MRSA in Mumbai, with dominant epidemic clones like ST 22 (EMRSA-15) and ST 772. Since the numbers of SCCmec IV and V MRSA strains isolated in the laboratory have been increasing and the number of SCCmec III MRSA strains has been decreasing every year from 2006 to 2009, it is probable that the ST 22 and ST 772 clones may be replacing the ST 239 clone in the hospital (5, 10, 12, 22). There have been many studies conducted and reviews written in the last few years about CA-MRSA in hospital settings, blurring the fine line that differentiates it from HA-MRSA (5, 22, 30). It can also be concluded that defining CA-MRSA merely by the absence of risk factors for health care exposure in patients would underestimate the real burden of its incidence today, since many CA-MRSA clones have successfully established themselves in hospital settings (5, 30). Since it has been hypothesized that all the major MRSA clones could have evolved from one common ancestor, S. aureus phage type 80/81, the ST 30 PVL+ MSSA strains in the hospitals or communities today may one day evolve into “successful” MRSA epidemic clones themselves (28). With the recent pandemic swine origin influenza virus A (H1N1), CA-MRSA is adept enough to cause severe pneumonia with increased morbidity and mortality (3, 7). Just as the SCCmec IV ST 22 (EMRSA-15) has spread over Europe and Asia, the SCCmec V ST 772, which currently seems to be native to Asia, may emerge as a powerful global epidemic MRSA strain in the years to come.
Acknowledgments
All reference strains in this study were the very generous and kind gift of Jerome Etienne with his dedicated team at INSERM, Lyon, France, and Dilip Mathai and Anand Manoharan from Christian Medical College, Vellore.
We thank the National Health and Education Society (NHES), of the P.D. Hinduja National Hospital and Medical Research Centre, for funding this study.
We have no conflict of interest to declare.
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Afroz, S., N. Kobayashi, S. Nagashima, M. M. Alam, A. B. Hossain, M. A. Rahman, M. R. Islam, A. B. Lutfor, N. Muazzam, M. A. Khan, S. K. Paul, A. K. Shamsuzzaman, M. C. Mahmud, A. K. Musa, and M. A. Hossain. 2008. Genetic characterization of Staphylococcus aureus isolates carrying Panton-Valentine leukocidin genes in Bangladesh. Jpn. J. Infect. Dis. 61:393-396. [PubMed] [Google Scholar]
- 2.Boyle-Vavra, S., and R. S. Daum. 2007. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab. Invest. 87:3-9. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, V. C. C., Y.-K. Lau, K.-L. Lee, K.-H. Yiu, K.-H. Chan, P.-L. Ho, and K.-Y. Yuen. Fatal coinfection with swine origin influenza virus A / H1N1 and community-acquired methicillin-resistant Staphylococcus aureus. J. Infect., in press. doi: 10.1016/j.jinf.2009.08.021. [DOI] [PubMed]
- 4.de Lencastre, H., D. Oliveira, and A. Tomasz. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 10:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deurenberg, R. H., and E. E. Stobberingh. 2008. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8:747-763. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geddes, A. M. 2009. Influenza and bacterial pneumonia. Int. J. Antimicrob. Agents 34:293-294. [DOI] [PubMed] [Google Scholar]
- 8.Goering, R. V., R. M. Shawar, N. E. Scangarella, F. P. O'Hara, H. Amrine-Madsen, J. M. West, M. Dalessandro, J. A. Becker, S. L. Walsh, L. A. Miller, S. F. van Horn, E. S. Thomas, and M. E. Twynholm. 2008. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J. Clin. Microbiol. 46:2842-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, L.-Y., T.-H. Koh, K. Singh, M.-L. Kang, A. Kurup, and B.-H. Tan. 2005. Dissemination of multisusceptible methicillin-resistant Staphylococcus aureus in Singapore. J. Clin. Microbiol. 43:2923-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, L.-Y., Y.-L. Koh, N. L. Chlebicka, T.-Y. Tan, P. Krishnan, R. T. Lin, N. Tee, T. Barkham, and T.-H. Koh. 2006. Establishment of ST30 as the predominant clonal type among community-associated methicillin-resistant Staphylococcus aureus isolates in Singapore. J. Clin. Microbiol. 44:1090-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu, L. Y., N. Loomba-Chlebicka, Y. L. Koh, T. Y. Tan, P. Krishnan, R. T. Lin, N. W. Tee, D. A. Fisher, and T. H. Koh. 2007. Evolving EMRSA-15 epidemic in Singapore hospitals. J. Med. Microbiol. 56:376-379. [DOI] [PubMed] [Google Scholar]
- 13.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J. 2009. Understanding the evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Newsl. 31:17-23. [Google Scholar]
- 16.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, M., B. A. Diep, A. E. Villaruz, K. R. Braughton, X. Jiang, F. R. DeLeo, H. F. Chambers, Y. Lu, and M. Otto. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 106:5883-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 19.Liu, Y., H. Wang, N. Du, E. Shen, H. Chen, J. Niu, H. Ye, and M. Chen. 2009. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob. Agents Chemother. 53:512-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 21.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchese, A., L. Gualco, E. Maioli, and E. Debbia. 2009. Molecular analysis and susceptibility patterns of meticillin-resistant Staphylococcus aureus (MRSA) strains circulating in the community in the Ligurian area, a northern region of Italy: emergence of USA300 and EMRSA-15 clones. Int. J. Antimicrob. Agents 34:424-428. [DOI] [PubMed] [Google Scholar]
- 23.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J. Antimicrob. Chemother. 60:42-48. [DOI] [PubMed] [Google Scholar]
- 24.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neela, V., G. R. Ehsanollah, S. Zamberi, A. Van Belkum, and N. S. Mariana. 2009. Prevalence of Panton-Valentine leukocidin genes among carriage and invasive Staphylococcus aureus isolates in Malaysia. Int. J. Infect. Dis. 13:e131-e132. [DOI] [PubMed] [Google Scholar]
- 26.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira, D. C., C. Milheirico, and H. de Lencastre. 2006. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 50:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson, D. A., A. M. Kearns, A. Holmes, D. Morrison, H. Grundmann, G. Edwards, F. G. O'Brien, F. C. Tenover, L. K. McDougal, A. B. Monk, and M. C. Enright. 2005. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet 365:1256-1258. [DOI] [PubMed] [Google Scholar]
- 29.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, and W. R. Jarvis. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopeptide-Intermediate Staphylococcus aureus Working Group. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 30.Tenover, F. C. 2005. Community-associated methicillin-resistant Staphylococcus aureus: it's not just in communities anymore. Clin. Microbiol. Newsl. 28:33-35. [Google Scholar]
- 31.Tenover, F. C., and R. V. Goering. 2009. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J. Antimicrob. Chemother. 64:441-446. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, K., J. Sparling, B. L. Chow, S. Elsayed, Z. Hussain, D. L. Church, D. B. Gregson, T. Louie, and J. M. Conly. 2004. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 42:4947-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]