Abstract
The aim of the current study was to develop a novel diagnostic test for detecting porcine reproductive and respiratory syndrome virus (PRRSV) using phage display technology. The N gene of PRRSV isolate HH08 was cloned following reverse transcription-PCR. Sequence comparison indicated that the N gene shared 96.4% homology to that of North American PRRSV (isolate VR2332) and 35.5% with that of European PRRSV (isolate LV), indicating that the PRRSV isolate was related to the North American PRRSV genotype. The bacterially expressed N protein was used as a target in a biopanning process using a phage display random peptide library. Seven phages expressing different peptides had a specific binding activity with the N protein. The putative binding motifs were identified by DNA sequencing. More importantly, the selected phages harboring specific peptides that recognize the N protein of PRRSV were able to efficiently distinguish PRRSV from other viruses in enzyme-linked immunosorbent assays.
Porcine reproductive and respiratory syndrome (PRRS) is one of the most important infectious diseases in the pig industry. The causative agent of the disease, porcine reproductive and respiratory syndrome virus (PRRSV), was first described in the late 1980s (33). Nowadays, PRRSV has been recognized globally as an economically important pathogen of domesticated swine (6, 11, 29, 31, 43). The unparalleled 2006 PRRS outbreaks engulfed China, and adult pigs were not exempt, indicating that some virus variants may have evolved to exhibit different pathogenicities (14, 45). PRRSV belongs to the Arteriviridae family, order Nidoviridales (2, 4, 5, 35). Two main PRRSV genotypes exist, the North American type (NA type) and the European type (EU type), which share approximately 60% identity at the nucleotide level (34). Since the first Chinese PRRSV was isolated in 1996, many isolates from different geographical locations in China have been identified (1). Most PRRSV isolates found in mainland China were identified as the NA type. Characterization of newly emerging isolates benefits the understanding of PRRSV genetic diversity.
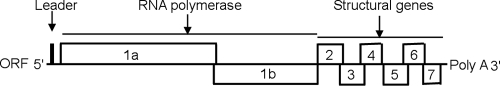
PRRSV is an enveloped virus with an approximately 14.5-kb positive-strand RNA genome that encodes a replicase polyprotein (encoded by open reading frames [ORFs] 1a and 1b) and six structural proteins (encoded by ORFs 2 to 7). The products of ORFs 2 to 4 are minor membrane-associated glycoproteins (GP2, GP3, and GP4, respectively). The products of ORFs 5 to 7 are major structural proteins: the GP5, M, and N proteins, respectively (35). A schematic diagram of the PRRSV genome is shown in Fig. 1. The viral GP5 and M proteins are incorporated in virions mainly as a disulfide-linked heterodimer or as a disulfide-linked multimer with an approximate molecular mass of 40 or 87 kDa, respectively (28, 32). They play important roles in virus neutralization by inducing cellular and humoral responses (12). The N protein of PRRSV is a basic protein with a molecular mass of 15 kDa. It is the most abundant viral protein in infected cells and constitutes about 20 to 40% of the protein content of the virion (43). Initial work indicated that pig antibodies against PRRSV are directed mainly to the N protein and to a lesser extent to the M protein and are nonneutralizing (38, 39). Since the N protein seems to be immunodominant, recombinant PRRSV N proteins are extensively used as an antigen in indirect and competition enzyme-linked immunosorbent assays (ELISAs) for detecting serum antibodies to the N protein. PRRSV sequence variability may affect the reactivity of monoclonal antibodies (MAbs) directed against the highly conserved N protein (6, 46, 50). Development of novel diagnostic tests is useful in detecting PRRSV.
FIG. 1.
Schematic diagram of the PRRSV genome. The entire genome of PRRSV is provided. The leader sequence, poly(A) tail, RNA polymerase, and structural gene regions are indicated. The framed parts from the the N terminus (5′) to the C terminus (3′) are the identified open reading frames.
Phage display peptide library technology is a powerful molecular tool. It involves the expression of random peptides or proteins on the surface of a filamentous bacteriophage (3, 8, 15, 19, 24). One significant advantage of the technology is the physical link of the phenotype to the genetic information contained within the phage particle, allowing specific screening based on binding affinity for a given target molecule by an in vivo selection process called panning. The panning procedure is shown in Fig. 2, and it contains several basic steps: (i) the exposure and binding of a phage library to an immobilized target, (ii) removal of phages that did not bind by washing, (iii) elution of specific phages binding to the target, and (iv) amplification of the phages in bacteria. The technology has been applied successfully in antibody engineering (3, 15), peptide and protein drug discovery and manufacture (19), vaccine development (21, 24), and identification of ligands (10, 22, 48).
FIG. 2.
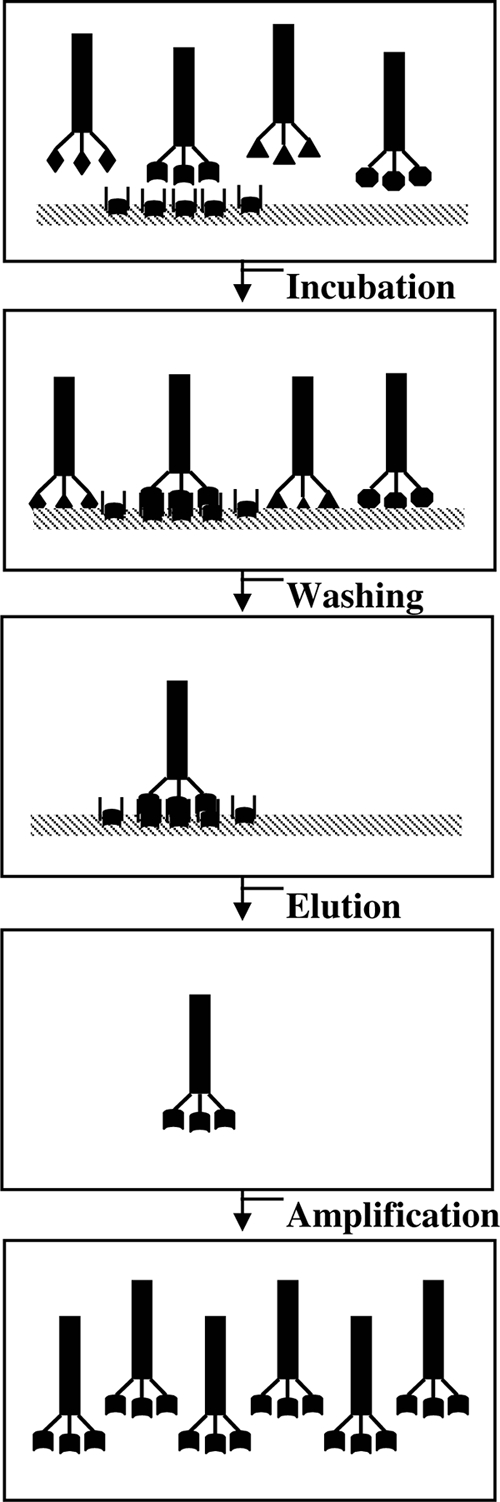
Schematic diagram of panning procedure. The panning steps of phage display technology are summarized. The “ ” symbol indicates the specific peptides on the ends of the phages; “
” symbol indicates the specific peptides on the ends of the phages; “ ” indicates target proteins; “▧” indicates immobilized support for a target. Other symbols on the ends of phages indicate unspecific exogenous peptides.
” indicates target proteins; “▧” indicates immobilized support for a target. Other symbols on the ends of phages indicate unspecific exogenous peptides.
In this study, we cloned the N gene of a Chinese PRRSV isolate that is phylogenetically related to the NA-type viruses. The bacterially expressed N protein was used as a target in a panning assay using a phage display random library. The identified phages harboring the specific peptides reacted with the PRRSV N protein. The sensitive and specific phage-based ELISA owns a broad perspective in distinguishing PRRSV from other pathogens.
MATERIALS AND METHODS
Cells and viruses.
PRRSV isolates CH-1a and HH08 were propagated in Marc-145 cells (a monkey kidney cell line) maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% newborn bovine serum (Excell Biology, Shanghai, China) at 37°C and 5% CO2 in air.
Cloning and sequence analysis of PRRSV N gene.
Total RNA was isolated from purified PRRSV isolate HH08 using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. The sense primer P1 5′-CCCCGAATTCATGCCAAATAACAACG-3′ and antisense primer P2 5′-CCCCAAGCTTGTCATGCTGAGGGTGAT-3′ were used in subsequent reverse transcription (RT)-PCR performed using an RT-PCR kit (TaKaRa, Japan). The primers contained the EcoRI and HindIII enzyme sites, respectively. The PCR profile included 95°C for 5 min, 30 cycles of 94°C for 1 min, 56.2°C for 30 s, and 72°C for 1 min, and a final extension of 72°C 10 min. The N gene was directly cloned into the TA cloning vector pMD18-T (TaKaRa, Japan), and three recombinant plasmids were sequenced. A representative PRRSV NA-type isolate, VR2332, and an EU-type isolate, LV, as well as several isolates from different geographic locations in China, were included in the sequence comparison, homologous identity, and phylogenetic tree analysis. These bioinformatic analyses were performed using the DNASTAR software program as described previously (41).
Expression and purification of the N protein.
The full-length N gene was inserted into the EcoRI and HindIII sites of a prokaryotic expression vector, pET-30a (Novagen), and a recombinant plasmid, designated pET-PRRSV-N, was purified using a commercially available kit (Nanjing Keygen Biotech. Co., Ltd., China). The authenticity of the insert was confirmed by sequencing. After pET-PRRSV-N was transformed into BL21(DE3)pLysS host cells, the expression of the histidine (His) tag-fused N protein was induced by isopropyl-β-d-thiogalactoside (IPTG) and analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (25). The protein of interest was designated His-N and purified by a nickel affinity chromatography column (Qiagen, Germany) according to the manufacturer's instructions.
Biopanning and enrichment analysis.
Phage display was performed according to the manufacturer's instructions (New England Biolabs) with modifications. For the first round of panning, 96-well plates were coated with the protein of interest at a concentration of 10 μg/well in 0.1 M NaHCO3 (pH 8.6) buffer overnight at 4°C. Next, the plates were blocked for 1 h at 4°C with blocking buffer (5% nonfat dry milk and 0.05% Tween 20 in phosphate-buffered saline [PBS]), followed by six washes with TBST (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% [vol/vol] Tween 20). The phage library diluted in TBST was added to the plate at a final concentration of 2 × 1011 (100 μl/well) and gently rocked at room temperature for 30 min. Unbound phages were removed by washing 10 times with TBST. Subsequently, bound phages were eluted by adding 100 μl elution buffer (0.2 M glycine-HCl [pH 2.2]) for 30 min with gentle rocking at room temperature. The eluate containing the bound phage neutralized with 15 μl 1 M Tris-HCl (pH 9.1) was collected. The titer of the phages was determined, and the phages were amplified in Escherichia coli ER2738 and purified by polyethylene glycol precipitation. The second, third, and fourth rounds of panning were repeated under similar panning conditions in addition to the increased concentration of Tween 20 (0.5% [vol/vol]) in TBST. In each round of panning, the titer of the phages in washing buffer (here referred to as Wash) and that in the elution buffer (here referred to as Output) were determined, and their ratio was analyzed to evaluate the enrichment efficiency.
Binding analysis of individual phage using ELISA.
Ten phage clones were subjected to ELISA analysis. Briefly, ELISA plates were coated with the N protein diluted in 0.1 M NaHCO3 (pH 8.6) (10 μg/well) overnight at 4°C. The mixed phages from the phage display library were used as a control. The following day, the plates were blocked with 1% bovine serum albumin (BSA) in TBS buffer (TBSB) for 2 h at room temperature. The plates were washed six times with TBST and then incubated with selected individual phage at a concentration of 1.5 × 1011 in 0.1 M NaHCO3 (pH 8.6) for 1 h at 37°C. After six washes with TBST, the M13 polyclonal antibody (diluted 1:1,000 in TBSB; Abcam) was added to the wells for another 1 h at 37°C. After six washes with TBST, the wells were incubated with the horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody (diluted 1:1,000 in TBSB; Sigma). The color was developed using o-phenylenediamine (OPD), and the optical density (OD) value was read using an ELISA reader at a wavelength of 405 nm. Ten positive phage clones were amplified and precipitated with polyethylene glycol-NaCl. Each phage clone DNA was purified using a plasmid extraction kit (Qiagen, Germany). The purified DNA template and the primers +130M13 (5′-TCACCTCGAAAGCAAGCTGA-3′) and −28M13 (5′-CCCTCATAGTTAGCGTAACG-3′) were used to PCR amplify the gene encoding the exogenous peptides of M13. DNA sequencing was carried out by the Borun Shanghai Company. The deduced amino acid sequences were analyzed using the DNASTAR software program.
Specificities of phages harboring specific peptides that recognize the N protein.
To analyze the specificities of the selected phages, the phages were used as diagnostic reagents to detect a panel of viruses. These viruses included PRRSV isolate HH08, PRRSV strain CH-1a, transmissible gastroenteritis coronavirus (TGEV), porcine epidemic diarrhea coronavirus (PEDV), pseudorabies virus (PRV), infectious bronchitis coronavirus (IBV), and a trivalent live vaccine against swine fever, swine erysipelas, and Pasteurella multocida. The viruses were treated with lysis buffer (0.1% SDS, 10 mmol/liter Tris [pH 7.4], and 1 mmol/liter EDTA) and transferred to ELISA plates at a concentration of 1 μg/well overnight at 4°C. Subsequent ELISA steps were carried out as described above.
Nucleotide sequence accession number.
The sequence of the N gene of PRRSV isolate HH08 was submitted to the GenBank database and was allocated accession number GQ184823.
RESULTS
Sequence analysis of N region of PRRSV isolate HH08.
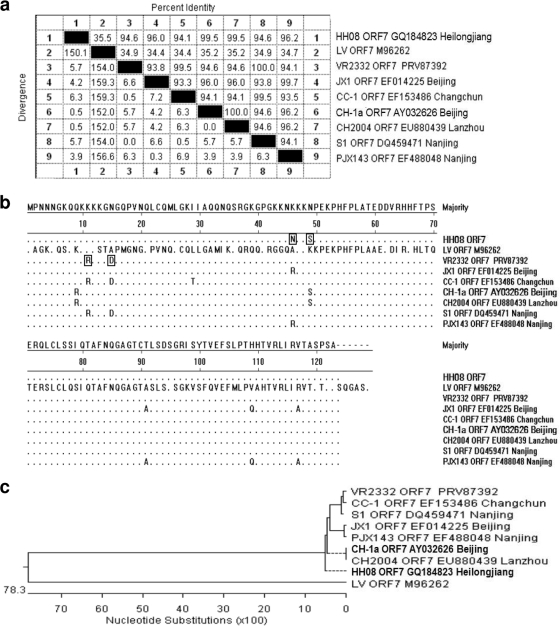
The N gene of PRRSV isolate HH08 was cloned. Sequencing results showed that the N gene consists of 372 nucleotides. Sequence comparison indicated that the N gene shared very high homology to that of NA-type PRRSVs. There was only 35.5% homologous identity with the representative EU-type isolate, LV (Fig. 3a). The N region of PRRSV HH08 was compared with those of several PRRSV isolates from different provinces in China. The sequence alignment results indicated the isolate shares significantly higher homologous identity with NA-type PRRSVs than the representative EU-type virus (LV). Amino acid comparison indicated that four sites were changed, Arg11Lys, Asp15Asn, Lys46Asn, and Asn49Ser, between the NA prototype strain VR2332 and the HH08 isolate (Fig. 3b). Whether these mutations are related to the pathogenicity of PRRSV HH08 remains unclear. The topology of the phylogenetic tree indicated that PRRSV isolate HH08 was located in a subclade of the NA-type PRRSVs (Fig. 3c). Sequence comparison and phylogenetic tree analysis indicate that most Chinese PRRSV isolates are of the NA type, resulting in the endemic prevalence in China.
FIG. 3.
Phylogenetic relationships among the PRRSV isolates. Homologous identity among the PRRSVs is shown in panel a. The isolate names and isolate places are provided; sequence alignment at the amino acid level in the N region of the PRRSVs is shown in panel b. The majority sequence shown at top is the consensus sequence, and four amino acid changes between the NA prototype strain VR2332 and the HH08 isolate are boxed. Using the MEGALIGN program in DNASTAR with the Jotun Hein method, a constructed phylogenetic tree is shown in panel c. The isolate names, GenBank accession numbers, and isolation places are indicated.
Expression of N protein in E. coli.
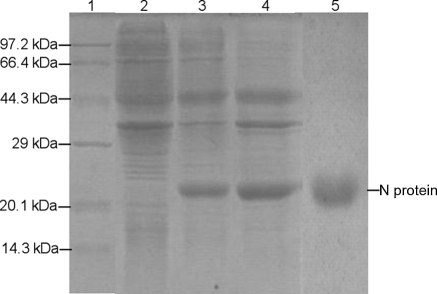
Using conventional molecular techniques, the PRRSV N protein was expressed in E. coli. SDS-PAGE analysis indicated that the protein was expressed in the form of both soluble and inclusion bodies (Fig. 4). Maximal expression of the protein was achieved at 37°C at 5 h post-IPTG (1 mmol/liter) induction. The molecular mass of the N protein is approximately 21 kDa, as expected.
FIG. 4.
Expression and purification of the PRRSV N protein. Protein expression was induced in the recombinant bacteria harboring the PRRSV N gene. SDS-PAGE results are provided. Lane 1, protein molecular mass marker; lane 2, control bacteria bearing the empty vector; lane 3, soluble N protein; lane 4, inclusion body N protein; lane 5, purified N protein.
Biopanning to the bacterially expressed N protein.
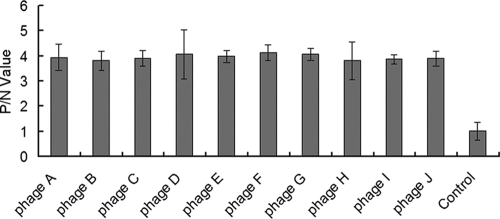
Four rounds of biopanning were performed using the bacterially expressed N protein as a target. The enrichment efficacies of the phages in each round were compared. As shown in Table 1, the Output/Input value was increased gradually. In contrast, Wash/Output decreased with increased biopanning times. The binding activities of 10 selected phages were assayed using ELISA. The results showed that they had a specific binding activity for the PRRSV N protein (Fig. 5).
TABLE 1.
Efficacy of panning to PRRSV N proteina
| Buffer or ratio | Titration value for round |
|||
|---|---|---|---|---|
| First | Second | Third | Fourth | |
| Input | 1.5 × 1011 | 1.5 × 1011 | 1.5 × 1011 | 1.5 × 1011 |
| Output | 3.1 × 105 | 4.6 × 106 | 3.5 × 107 | 6.8 × 109 |
| Wash | 4.5 × 107 | 4.6 × 106 | 6.5 × 106 | 7.1 × 105 |
| Wash/output | 1.4 × 102 | 1 | 0.1 | 0.9 × 10−4 |
| Output/input | 2 × 10−6 | 4 × 10−5 | 3.3 × 10−4 | 4.6 × 10−2 |
The titration values for phages in input buffer (Input), elution buffer (Output), and last washing buffer (Wash) and ratios of Wash/Output and Output/Input values in each round of panning are provided.
FIG. 5.
Analysis of binding of selected phages to the PRRSV N protein using ELISA. Ten selected phages, named phages a to j, were incubated with the PRRSV N protein in ELISA plates to test their binding activities for the protein as described in Materials and Methods. The ratio between the optical density (OD) value of tested individual phage and the OD value of the control is shown on the y axis; the individual phage and the control phage complex from the phage library are shown on the x axis.
Identification of putative motifs that are responsible for binding activity.
DNA of the selected phages was extracted, and the genes encoding the peptides expressed on the surfaces of the recombinant phages were amplified by primer-specific PCR. DNA sequencing indicated that among the 10 selected phages, 7 deduced peptide sequences (12 amino acids in length) were identified (Table 2). Four phages shared consensus sequences. In addition, a putative motif, Hx (xxx)L, was determined using DNASTAR software.
TABLE 2.
Deduced amino acid sequences of phage clonesa
| Clone ID | Sequence |
|---|---|
| Phage a | KHMHWHPPALNT |
| Phage b | HYQGVHSRYCYH |
| Phage c | KHMHWHPPALNT |
| Phage d | LTPTMFNMHGVL |
| Phage e | VSRHQSWHPHDL |
| Phage f | KHMHWHPPALNT |
| Phage g | HWPRPDDSFWRP |
| Phage h | HSTWKLLRLDME |
| Phage i | GPYFPTHSFLKS |
| Phage j | KHMHWHPPALNT |
Ten selected phages (phages a to j) were subjected to DNA extraction and PCR. The deduced amino acid sequences are shown. The sequences of phages a, c, f, and j are identical. Boldface indicates putative motifs that bind the PRRSV N protein.
Phages harboring specific peptides distinguish PRRSV from other viruses.
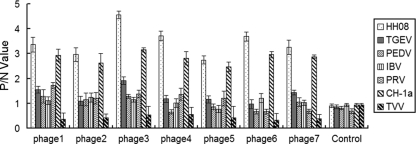
The selected phages were analyzed for their specificities in recognizing PRRSV and other porcine viral/bacterial agents. As shown in Fig. 6, these phages gave rise to a significantly higher reactivity to PRRSVs HH08 and CH-1a than to the control pathogens. In particular, the phage expressing the peptide KHMHWHPPALNT showed the highest reactivity to PRRSVs.
FIG. 6.
Phage-based ELISA for differentiating PRRSV from other pathogens. Seven phages harboring specific peptides recognizing the PRRSV N protein were identified from the 10 selected phages and were designated phages 1 to 7. Phages 1, 2, 4, 5, 6, and 7 are identical to the phages b, i, h, d, e, and g; phage 3 is identical to phages a, c, f, and j indicated in Fig. 3 and Table 2. They were incubated with the various pathogens in ELISA plates. The ratio between the OD value of individual phage and the OD value of the control is shown on the y axis. “TVV” is the abbreviation of the trivalent vaccine against swine fever, swine erysipelas, and Pasteurella multocida. The phage control is the phage complex from the phage library.
DISCUSSION
As an emerging infectious disease, PRRSV infection often leads to enormous economic loss in the pig industry. Taxonomically, the virus consists of two major subtypes, namely, the NA type and the EU type. Comparative analysis of the predicted N protein indicates that the protein is very conserved among PRRSVs within a given genotype, often displaying between 96 and 100% amino acid identity (27, 30). Pathogenic diagnosis is an effective means of monitoring PRRSV. The PRRSV N protein is very conserved and plentiful as an important viral component. Recombinant PRRSV N proteins have been used as diagnostic antigens for detecting serum antibodies against the N protein. In addition, indirect and competition ELISAs with synthetic peptides have been established to characterize the epitopes of the N protein that are recognized by a battery of monoclonal antibodies (MAbs) and by antibodies from infected pigs (39). Direct pathogen detection is very suitable for early diagnosis of PRRSV infection.
In our study, the high-level expression of the PRRSV N protein in prokaryotic cells enabled us to develop N-protein-mediated methods for detecting PRRSV. Compared with other available foreign gene expression systems, such as yeast, insect-baculovirus, mammalian cells, etc., the prokaryotic expression system owns advantages in protein yield, production cost, operation convenience, etc. (49). It is postulated that the biological properties of the bacterially expressed PRRSV N protein do not vary significantly, since it is a serine phosphoprotein (47). Many works have shown the utility of the PRRSV N protein produced in prokaryotic systems (7, 9, 28, 42). In our study, the purified protein was used to produce specific antiserum in immunized rabbits. Both the protein and antibody reacted with either PRRSV-positive serum or the N protein from other PRRSV strains in Western blotting and immunofluorescence analysis (unpublished data), indicating that the recombinant N protein may share certain antigenic properties of the “wild” N protein.
Selection of specific ligands for the N protein is the first step in establishing a novel N-protein-based ELISA. The employment of combinatorial peptide libraries displayed on the surfaces of filamentous bacteriophage offers an efficient means of obtaining a diverse set of peptides that bind a target protein (23). Phage display has been used to isolate peptides binding specifically to isolated proteins, inorganic material, and complex target structures (44). Reports of identifying ligands for either viral pathogens or their receptors or antibodies using phage display technology are increasing (13, 16, 18, 20, 26, 40). In this study, using the immobilized N protein as a target, several phages harboring various peptides were identified in protein-based ELISA for the first time. DNA sequencing indicated that 4 of the 10 selected phages shared a consensus peptide. Although six other peptides did not show any consensus sequences, the amino acid residues H and L existed in the peptides with the highest frequency. Interestingly, a putative motif, HxxL/HxxxL/HxxxxL, was found in phages a, c, d, e, f, h, i, and j, and residue H was found in phages b and g, suggesting the H-containing motifs may be indispensable in binding to the PRRSV N protein.
Since 1996, the order Nidovirales has officially united the families Coronaviridae and Arteriviridae, and in phylogenetic analyses of key replicase domains, including the RNA-dependent RNA polymerase (RdRp) and helicase, different nidovirus subgroups were found to cluster, suggesting that they share an ancestor (2, 36). Besides being a predominant immunogen in pigs, the N protein is a major viral component that interacts with the host genome (37). Recently X-ray structure analysis of the N- and C-terminal domains of the N protein of infectious bronchitis coronavirus (IBV) was performed. The observed structural similarity between the N proteins of IBV and PRRSV suggests that members of the Coronaviridae and Arteriviridae families share a mechanism of filamentous nucleocapsid formation, with suitable alterations necessary to interact specifically with their respective genomes (17). To analyze the cross-reactivity of the phages, mammalian coronavirus TGEV, avian coronavirus IBV, porcine herpesvirus, PRV, a trivalent vaccine against swine fever, swine erysipelas, and Pasteurella multocida, and the first identified Chinese PRRSV isolate were included in ELISAs. Our result indicated that the phages had a significantly higher-affinity activity for PRRSVs than for other pathogens. The low reactivity of the control, the phage complex from the phage library, to the targets in the ELISA excluded any artifact. Compared with the cost and convenience in protein purification or peptide synthesis, the propagation of phage is relatively cheap and can be done on a large scale. At present, real-time RT-PCR is one of the most popular tests for detecting PRRSV; however, detection of viral particles with the phages may be an alternative method. It would be interesting to compare the sensitivities between the two diagnostic methods in the future. The current finding confirms that phage display technology is a powerful tool for identifying protein ligands; at the same time, phages recognizing the PRRSV N protein may be used as diagnostic reagents to monitor PRRSV infection clinically.
Acknowledgments
We thank Ulrich Neumann of the Clinic for Poultry, University of Veterinary Medicine Hannover, Hannover, Germany, for providing the anti-M13 antibody.
Funds from the Heilongjiang Provincial Science and Technology Department (ZJN0702-01) to G.L., the National Natural Science Foundation of China (30700591), the Heilongjiang Provincial Science and Technology Department (QC07C32) to J.Y., and the National Natural Science Foundation of China (30700590 and 30972195) to X.R. are acknowledged.
Footnotes
Published ahead of print on 17 March 2010.
REFERENCES
- 1.An, T. Q., Y. J. Zhou, G. Q. Liu, Z. J. Tian, J. Li, H. J. Qiu, and G. Z. Tong. 2007. Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: coexistence of two NA-subgenotypes with great diversity. Vet. Microbiol. 123:43-52. [DOI] [PubMed] [Google Scholar]
- 2.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 3.Chames, P., and D. Baty. 2000. Antibody engineering and its applications in tumor targeting and intracellular immunization. FEMS Microbiol. Lett. 189:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z. Y., and P. G. W. Plagemann. 1995. Detection of related positive strand RNA virus genomes by reverse transcription polymerase chain reaction using degenerate primers for common replicase sequences. Virus Res. 39:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conzelmann, K. K., N. Visser, P. van Woensel, and H. J. Thiel. 1993. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology 193:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dea, S., C. A. Gagnon, H. Mardassi, B. Pirzadeh, and D. Rogan. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 145:659-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dea, S., L. Wilson, D. Therrien, and E. Cornaglia. 2000. Competitive ELISA for detection of antibodies to porcine reproductive and respiratory syndrome virus using recombinant E. coli-expressed nucleocapsid protein as antigen. J. Virol. Methods 87:109-122. [DOI] [PubMed] [Google Scholar]
- 8.Devlin, J. J., L. C. Panganiban, and P. E. Devlin. 1990. Random peptide libraries: a source of specific protein binding molecules. Science 249:404. [DOI] [PubMed] [Google Scholar]
- 9.Doan, D. N., and T. Dokland. 2003. Structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Structure 11:1445-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrlich, G. K., and P. Bailon. 2001. Identification of model peptides as affinity ligands for the purification of humanized monoclonal antibodies by means of phage display. J. Biochem. Biophys. Methods 49:443-454. [DOI] [PubMed] [Google Scholar]
- 11.Faaberg, K. S., and P. G. Plagemann. 1997. ORF 3 of lactate dehydrogenase-elevating virus encodes a soluble, nonstructural, highly glycosylated, and antigenic protein. Virology 227:245-251. [DOI] [PubMed] [Google Scholar]
- 12.Faaberg, K. S., J. D. Hocker, M. M. Erdman, D. L. H. Harris, E. A. Nelson, M. M. Torremorell, and P. G. Plagemann. 2006. Neutralizing antibody responses of pigs infected with natural GP5 N-glycan mutants of porcine reproductive and respiratory syndrome virus. Viral Immunol. 19:294-304. [DOI] [PubMed] [Google Scholar]
- 13.Hall, P. R., B. Hjelle, H. Njus, C. Ye, V. Bondu-Hawkins, D. C. Brown, K. A. Kilpatrick, and R. S. Larson. 2009. Phage-display selection of cyclic peptides that inhibit Andes virus infection. J. Virol. 83:8965-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, J., Y. Wang, and K. S. Faaberg. 2006. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 122:175-182. [DOI] [PubMed] [Google Scholar]
- 15.Hayden, M. S., L. K. Gillilandt, and J. A. Ledbetter. 1997. Antibody engineering. Curr. Opin. Immunol. 9:201-212. [DOI] [PubMed] [Google Scholar]
- 16.Houimel, M., and K. Dellagi. 2009. Peptide mimotopes of rabies virus glycoprotein with immunogenic activity. Vaccine 27:4648-4655. [DOI] [PubMed] [Google Scholar]
- 17.Jayaram, H., H. Fan, B. R. Bowman, A. Ooi, J. Jayaram, E. W. Collisson, J. Lescar, and B. V. Prasad. 2006. X-Ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 80:6612-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karthe, J., K. Tessmann, J. Li, R. Machida, M. Daleman, D. Häussinger, and T. Heintges. 2008. Specific targeting of hepatitis C virus core protein by an intracellular single-chain antibody of human origin. Hepatology 48:702-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kay, B. K., A. V. Kurakin, and R. Hyde-DeRuyscher. 1998. From peptides to drugs via phage display. Drug Discov. Today 3:370-378. [Google Scholar]
- 20.Kirsch, M. I., B. Hülseweh, C. Nacke, T. Rülker, T. Schirrmann, H. J. Marschall, M. Hust, and S. Dübel. 2008. Development of human antibody fragments using antibody phage display for the detection and diagnosis of Venezuelan equine encephalitis virus (VEEV). BMC Biotechnol. 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klemm, P., and M. A. Schembril. 2000. Fimbrial surface display systems in bacteria: from vaccines to random libraries. Microbiology 146:3025-3032. [DOI] [PubMed] [Google Scholar]
- 22.Ladner, R. C., and A. C. Ley. 2001. Novel frameworks as a source of high-affinity ligands. Curr. Opin. Biotechnol. 12:406-410. [DOI] [PubMed] [Google Scholar]
- 23.Larson, R. S., D. C. Brown, C. Ye, and B. Hjelle. 2005. Peptide antagonists that inhibit Sin Nombre virus and hantaan virus entry through the beta3-integrin receptor. J. Virol. 79:7319-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesinski, G. B., and J. Westerink. 2001. Novel vaccine strategies to T-independent antigens. J. Microbiol. Methods 47:135-149. [DOI] [PubMed] [Google Scholar]
- 25.Liu, B., G. Li, X. Sui, J. Yin, H. Wang, and X. Ren. 2009. Expression and functional analysis of porcine aminopeptidase N produced in prokaryotic expression system. J. Biotechnol. 141:91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, W., Y. Chen, M. Wang, Y. Chen, Z. Zheng, H. Song, H. Chen, Y. Guan, M. H. Ng, J. Zhang, and N. Xia. 2009. Peptide mimics of a conserved H5N1 avian influenza virus neutralization site. Biochem. J. 419:133-139. [DOI] [PubMed] [Google Scholar]
- 27.Magar, R., R. Larochelle, S. Dea, C. A. Gagnon, E. A. Nelson, J. Christopher-Hennings, and D. A. Benfield. 1995. Antigenic comparison of Canadian and US isolates of porcine reproductive and respiratory syndrome virus using monoclonal antibodies to the nucleocapsid protein. Can. J. Vet. Res. 59:232-234. [PMC free article] [PubMed] [Google Scholar]
- 28.Mardassi, H., B. Massie, and S. Dea. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221:98-112. [DOI] [PubMed] [Google Scholar]
- 29.Meng, X. J. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 74:309-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng, X. J., P. S. Paul, P. G. Halbur, and M. A. Lum. 1995. Phylogenetic analysis of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the U. S. A. and Europe. Arch. Virol. 140:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meulenberg, J. J. M., M. M. Hulst, E. J. De Meijer, P. L. J. M. Moonen, A. Petersen-den Besten, E. P. De Kluyver, G. Wensvoort, and R. J. M. Moormann. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meulenberg, J. J., A. Petersen-den Besten, E. P. De Kluyver, R. J. Moormann, W. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oleksiewicz, M. B., A. Bøtner, and P. Normann. 2002. Porcine B-cells recognize epitopes that are conserved between the structural proteins of American- and European-type porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 83:1407-1418. [DOI] [PubMed] [Google Scholar]
- 34.Oleksiewicz, M. B., A. Bøtner, P. Toft, P. Normann, and T. Storgaard. 2001. Epitope mapping porcine reproductive and respiratory syndrome virus by phage display: the nsp2 fragment of the replicase polyprotein contains a cluster of B-cell epitopes. J. Virol. 75:3277-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostrowski, M., J. A. Galeota, A. M. Jar, K. B. Platt, F. A. Osorio, and O. J. Lopez1. 2002. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J. Virol. 76:4241-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasternak, A. O., W. J. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 37.Pei, Y., D. C. Hodgins, C. Lee, J. G. Calvert, S. K. Welch, R. Jolie, M. Keith, and D. Yoo. 2008. Functional mapping of the porcine reproductive and respiratory syndrome virus capsid protein nuclear localization signal and its pathogenic association. Virus Res. 135:107-114. [DOI] [PubMed] [Google Scholar]
- 38.Plagemann, P. G. W. 2005. Epitope specificity of monoclonal antibodies to the N-protein of porcine reproductive and respiratory syndrome virus by ELISA with synthetic peptides. Vet. Immunol. Immunopathol. 104:50-68. [DOI] [PubMed] [Google Scholar]
- 39.Plagemann, P. G. W. 2006. Peptide ELISA for measuring antibodies to N-protein of porcine reproductive and respiratory syndrome virus. J. Virol. Methods 134:99-118. [DOI] [PubMed] [Google Scholar]
- 40.Rajik, M., F. Jahanshiri, A. R. Omar, A. Ideris, S. S. Hassan, and K. Yusoff. 2009. Identification and characterisation of a novel anti-viral peptide against avian influenza virus H9N2. Virol. J. 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren, X., J. Yin, D. Ma, and G. Li. 2009. Characterization and membrane gene-based phylogenetic analysis of avian infectious bronchitis virus Chinese strain HH06. Virus Genes 38:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez, M. J., J. Sarraseca, J. Garcia, A. Sanz, J. Plana-Durán, and J. Ignacio Casal. 1997. Epitope mapping of the nucleocapsid protein of European and North American isolates of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 78:2269-2278. [DOI] [PubMed] [Google Scholar]
- 43.Snijder, E. J., and J. J. M. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 44.Stratmann, J., B. Strommenger, K. Stevenson, and G. F. Gerlach. 2002. Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk. J. Clin. Microbiol. 40:4244-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian, K., X. Yu, T. Zhao, Y. Feng, Z. Cao, C. Wang, et al. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wootton, S. K., E. A. Nelson, and D. Yoo. 1998. Antigenic structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 5:773-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wootton, S. K., R. R. Rowland, and D. Yoo. 2002. Phosphorylation of the porcine reproductive and respiratory syndrome virus (PRRSV) nucleocapsid protein. J. Virol. 76:10569-10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yi, G., J. Qian, Z. Wang, and Y. Qi. 2003. A phage-displayed peptide can inhibit infection by white spot syndrome virus of shrimp. J. Gen. Virol. 84:2545-2553. [DOI] [PubMed] [Google Scholar]
- 49.Yin, J., G. Li, X. Ren, and G. Herrler. 2007. Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 127:335-347. [DOI] [PubMed] [Google Scholar]
- 50.Yoon, K. J., J. J. Zimmerman, M. J. McGinley, J. Landgraf, M. L. Frey, H. T. Hill, and K. B. Platt. 1995. Failure to consider the antigenic diversity of porcine reproductive and respiratory syndrome (PRRS) virus isolates may lead to misdiagnosis. J. Vet. Diagn. Invest. 7:386-387. [DOI] [PubMed] [Google Scholar]