Abstract
In earlier studies we used molecular methods to identify the major bacterial consortia associated with advanced dentin caries. These consortia are dominated by bacteria from the families Lactobacillaceae, Streptococcaceae, Veillonellaceae (formerly Acidaminococcaceae), Eubacteriaceae, and Lachnospiraceae from the phylum Firmicutes; Coriobacteriaceae, Bifidobacteriaceae, and Propionibacteriaceae from the phylum Actinobacteria; and Prevotellaceae from the phylum Bacteroidetes, as well as fusobacteria. The phases of infection of vital pulp tissue by dentin microorganisms remain obscure. In the present study, fluorescence in situ hybridization was performed on sections of tissue embedded in resin. Probes for 16S rRNA corresponding to the major taxa of bacteria in carious dentin were used to provide information on the characteristics of pulp infection. Lactobacilli were prominent in 7 of 8 pulps determined to be at a limited stage of infection. Established infection (6 pulps) showed a more complex profile, with lactobacilli persisting in all of the lesions and with invasion of the necrotic regions of tissue by Bacteroidetes, fusobacteria, Lachnospiraceae, and Coriobacteriaceae in particular. Advanced infections (7 pulps) were characterized by mixed anaerobic species, with a strong representation by Coriobacteriaceae and Lachnospiraceae. Lactobacilli were not represented at this stage. Typically, groups of organisms were spatially isolated within the pulp tissue. Analysis indicated that lactobacilli could invade vital pulp tissue to achieve a very high biomass that was not associated with a detectable local inflammatory infiltrate. The findings establish that invasion of the dental pulp can be associated with a pronounced selection from the complex microbial populations within carious dentin, suggesting specific pathogenicity.
Caries progression is a protracted process involving a sequence commencing with decalcification of the enamel by organic acids produced by fermentative Gram-positive bacteria, including lactobacilli and mutans streptococci (22). Failure of the enamel barrier exposes the underlying dentin to colonization by microorganisms carried in saliva, and typically the established dentin flora is complex and variable between carious teeth (4, 16). Recent data indicate that representation of obligate anaerobes in carious dentin is greater than previously appreciated (4, 8, 14, 15). Subsequent invasion and destruction of dental pulp provide the potential for extension of infection to adjacent bone and soft tissues.
Bacteria migrate down dentinal tubules that have been damaged by organic acids produced by components of the flora. Odontoblasts that line the pulp and extend long processes into dentin were recently demonstrated to express Toll-like receptors that mediate the response of cultured odontoblasts to bacterial products (23). Odontoblasts respond to microbial insult by both deposition of new tubular dentin and deposition of mineralized intratubular plugs. More aggressive bacterial advance is considered to be associated with death of odontoblasts, facilitating access of bacteria to the pulp. Proliferation of bacteria in the superficial aspect of pulp tissue disorganizes the odontoblast layer and kills cells either by direct toxic action or by excessive acute inflammatory response. Our previous analysis using molecular techniques indicated nine major bacterial taxa that predominate in carious dentin (4). In adults, dentinal caries typically progresses slowly, with estimates of >2 years before detectable infection of pulp. Bacterial invasion of the pulp resulting in pulp necrosis is likely to be quite rapid by comparison. Accordingly, there are significant logistical challenges in studying the sequence of pulp infection, particularly as clinical signs and symptoms provide limited diagnosis. In this investigation, findings for 21 infected pulps are presented to provide an overview of the role of bacterial taxa in the sequential invasion of the dental pulp.
MATERIALS AND METHODS
All chemicals were from Sigma unless otherwise specified.
Preparation of teeth.
Molar teeth were extracted from consenting adult patients according to a protocol approved by the Human Ethics Committee, Sydney West Area Health Service (protocol no. X07-0172). These teeth had large cavitated carious lesions, originating as occlusal caries, and were associated with painful responses but had clinical signs of vitality. Teeth were collected, bisected sagittally, and placed in freshly prepared 4% paraformaldehyde overnight at 4°C with the pulps in situ. They were then washed twice in sterile phosphate-buffered saline (PBS) and decalcified in sterile 10% EDTA-PBS for 4 weeks with daily changes. All buffers and water for the handling of tissues were prepared from 0.22-μm filtered 18-MΩ water. In some cases, the pulps fixed in situ in the split half of the tooth were removed under a binocular microscope. Both teeth and isolated pulps were dehydrated through graded alcohols and embedded in 13-mm gelatin capsules in Immuno-Bed resin (Polysciences, Inc.) (teeth) and JB-4 plastic block holders and stubs (Polysciences, Inc.) (pulps). The embedded teeth were ground down to facilitate fit with the microtome knife and mounted on JB-4 plastic stubs using araldite. Two-micrometer sections were cut using a Reichert-Jung SuperCut 2050 microtome, floated on water, and picked up on SuperFrost Plus Menzel-Gläser glass slides for subsequent staining and fluorescence in situ hybridization (FISH). The sections were stored desiccated at −20°C. For bright-field microscopy, the sections were stained with toluidine blue and periodic acid-Schiff reagent.
FISH pretreatment.
The sections were incubated with 0.2 M HCl for 10 min, washed with 18-MΩ water, treated with 1% Triton X-100 for 90 s, and washed with 18-MΩ water. The sections were then digested at 37°C with 2 mg proteinase K ml−1, 2 mg lysozyme ml−1, and 1,000 U mutanolysin ml−1 in 10 mM phosphate buffer, pH 6.7, for 40 min to facilitate penetration of the probe into the bacteria (17). The digestion was stopped with a 2-mg glycine ml−1 wash in 50 mM Tris buffer, pH 7.2. The slides were then washed in 18-MΩ water and allowed to dry. Each section was framed using a 25-μl Gene Frame (ABgene), and 25 μl of hybridization buffer containing the fluorescent probe at a concentration determined to be optimal for specific detection of each of the targeted bacterial taxa was added.
FISH method.
The FISH method used was adapted from that of Hugenholtz et al. (9), using 25% formamide stringency, except for the Veillonellaceae probe, which was reacted in 40% formamide to preserve specific detection. The slides were incubated overnight at 46°C using a Hybaid OmniSlide system (Hybaid) and then washed at 48°C using the corresponding NaCl concentration and the Hybaid wash module. After a cold 18-MΩ water wash, the slides were dried and coverslipped using ProLong Gold antifade reagent (Molecular Probes). The slides were viewed using an Olympus BX60 fluorescent microscope and images captured using a Leica DFC500 camera.
Acridine orange.
The slides were treated with 1% Triton X-100 for 90 s, rinsed with water, and stained with 0.01% acridine orange in 20 mM Tris buffer, pH 7.2, for 8 min, rinsed with water, and coverslipped using water. Images were captured without delay.
Bacterial cultures.
Lactobacillus acidophilus (ATCC 4356) and Streptococcus mutans (LT11) were obtained from the culture collection of the Institute of Dental Research, Sydney, Australia. Pseudoramibacter alactolyticus (JCM 6480) (from the family Eubacteriaceae); Propionibacterium acnes (JCM 6425) (from the family Propionibacteriaceae); Selenomonas infelix (JCM 8545) (from the family Veillonellaceae); Olsenella profusa (JCM 14553), Atopobium rimae (JCM 10299), and Atopobium parvulum (JCM 10300) (from the family Coriobacteriaceae); and Prevotella oralis (JCM 6330), Prevotella veroralis (JCM 6290), Prevotella multiformis (JCM 12541), Prevotella salivae (JCM 12084), Prevotella shahii (JCM 12083), and Prevotella buccae (JCM 12245) (from the family Prevotellaceae) were obtained from the Japanese Culture Collection, Riken BioResource Center, Ibaraki, Japan. Another species from the family Prevotellaceae, Prevotella denticola (ATCC 35308), was a gift from David Dymock, University of Bristol, Bristol, United Kingdom. Prevotella melaninogenica (ATCC 25845) and Fusobacterium nucleatum (ATCC 25586) were obtained from the American Type Culture Collection (ATCC) as described previously (14). Veillonella dispar (ATCC 17748) was also obtained from the ATCC. Lachnospira multipara (DSMZ 3073) (from the family Lachnospiraceae) was obtained from DSMZ, Braunschweig, Germany. Stenotrophomonas maltophilia, a clinical isolate from asymptomatic infiltrative keratitis, and an Acinetobacter sp., a clinical isolate from contact lens-induced acute red eye, were kindly provided by Mark Willcox from the Centre for Eye Research, Sydney, Australia.
Design and validation of probes.
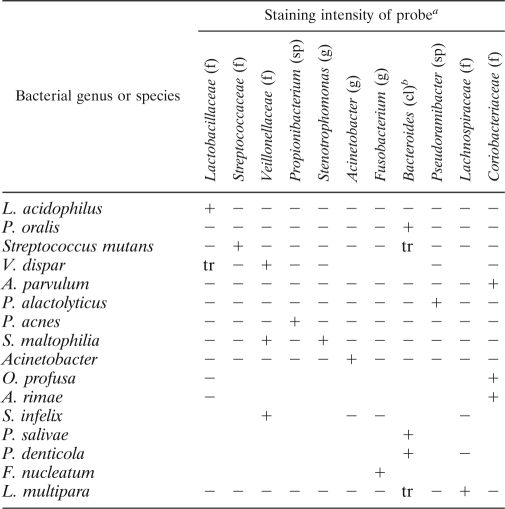
Oligonucleotide probes with 5′-end modifications were custom synthesized by Invitrogen. For localization of all bacteria in tissue sections by in situ hybridization, a universal bacterial probe (18) was labeled at the 5′ end with fluorescein isothiocyanate. For dual-staining purposes, another universal bacterial probe, EUB338 (1), labeled at the 5′ end with Alexa 594, was also used. The probes used for detection of particular bacterial taxa are detailed in Table 1. Each probe was validated against other taxa to eliminate false-positive results due to cross-reactivity. This was achieved by standardized fixation and resin embedding of the representative organisms cultured to exponential phase in batch culture. The probes were coded and applied to sections containing the bacteria. Staining intensities were recorded, and the decoded information was prepared (Table 2).
TABLE 1.
DNA sequence of FISH probes
| In situ probe | 5′-end modification/color | DNA sequence (5′ → 3′) |
|---|---|---|
| Universal | FLO/green, Alexa 594/red | CGTATTACCGCGGCTGCTGGCAC |
| EUB 338 | Alexa 488/green, Alexa 594/red | GCTGCCTCCCGTAGGAGT |
| Lactobacillaceae | Alexa 488/green | GTCCATTGTGGAAGATTCCC |
| Bacteroides | Alexa 594/red | ATACGCGTTACKCACCCGTG |
| Streptococcaceae | Alexa 594/red | TAGCCGTCCCTTTCTGGT |
| Veillonellaceae | Alexa 532/orange | TCATCCTCTCAGACCGGCTAC |
| Stenotrophomonas | Alexa 488/green | GCTGGATTTCTTTCCCAACAA |
| Acinetobacter | Alexa 488/green | TCCTCCTCGCTTAAAGTGCTT |
| Coriobacteriaceae | Alexa 488/green, Marina Blue/blue | CCGGTCGGTCTCTCAACC |
| Pseudoramibacter alactolyticus | Alexa 488/green | ACTCCCGATTTCTCGAGGC |
| Propionibacterium | Alexa 488/green | ACTCGCGCTTCGTCATG |
| Fusobacterium | Alexa 488/green | CTAATGGGACGCAAAGCTCTC |
| Lachnospiraceae | Alexa 594/red, Alexa 488/green | TTCCCTGCTGATAGAGCTTTACATAC |
| Gammaproteobacteria | Alexa 488/green | GCCTTCCCACATCGTTT |
TABLE 2.
Specificity of bacterial probes used for FISH
cl, class; f, family; g, genus; sp, species. Symbols: +, positive; −, negative; tr, trace.
The Bacteroides probe could also detect other Prevotella species from the family Prevotellaceae (i.e., Prevotella veroralis, Prevotella multiformis, Prevotella shahii, Prevotella buccae, and Prevotella melaninogenica).
Assessment of pulp infection.
Isolated pulps or bisected tooth halves containing the pulps were embedded in Immuno-Bed resin (Polysciences, Inc.), and 2-μm sections were prepared using a Reichert-Jung SuperCut 2050 microtome. Brackets of sections were retained at intervals, with bracket reference slides stained with toluidine blue (0.05%) to determine morphology and evidence of microbial invasion. The brackets containing sections with the most advanced infection were selected for detailed analysis, and representative infection was determined by assessment of the sections at various depths throughout the brackets. Given the protracted nature of caries progression and the relatively imprecise clinical diagnostic criteria currently available, it was not possible to impute a temporal aspect to the assessment. Stages of infection were arbitrarily designated as limited (less than one-third of the surface area of the pulp was infected), established (between one-third and two-thirds of the surface area of the pulp was infected), and advanced (greater than two-thirds of the surface area of the pulp was infected, but some remaining intact tissue was indicative of remnants of vital pulp). Twenty-one teeth met these criteria (10 from females, mean age 34 years, and 11 from males, mean age 43 years). Seven additional teeth that met the clinical criteria either shattered on splitting or demonstrated frankly necrotic pulps.
DNA extraction.
DNA was extracted from 3 to 5 sections (2-μm thickness each) of resin-embedded lesions 1 and 9 (see Table 3). Samples were incubated with cell lysis buffer as described previously (17). After addition of SDS (1%), DNA was extracted using the QIAamp DNA minikit (Qiagen) per the manufacturer's instructions, with a final elution volume of 200 μl.
TABLE 3.
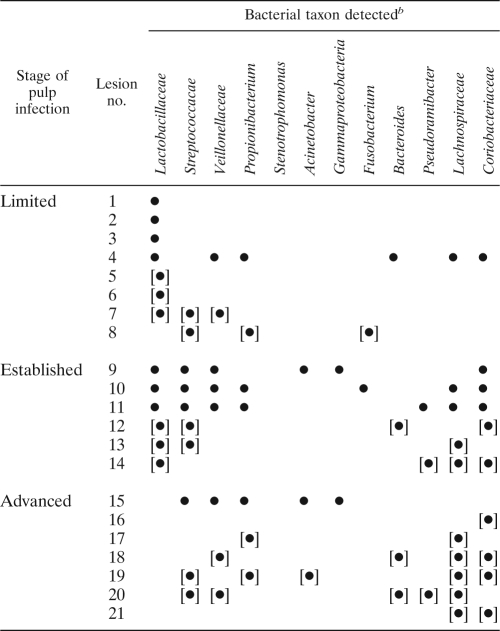
Bacterial taxa detected in stages of pulp infectiona
Braces indicate lesions of pulpal tissue only.
A bullet indicates detection.
PCR.
Lactobacillus genus-specific primers or Lactobacillus species-specific primers were used to amplify the signal in DNA isolated from dental pulp, as described previously (2).
DNA sequencing.
PCR amplicons were purified using a Mo Bio PCR cleanup kit (GeneWorks) and sequenced at the Westmead Genomics Facility (Sydney, Australia) or the Australian Genome Research Facility (Brisbane, Australia) using the appropriate forward primer. Sequence identity was established using the Basic Local Alignment Search Tool (BLAST) to search nucleotide databases accessed through NCBI (http://www.ncbi.nlm.nih.gov/).
Anti-Lactobacillus rhamnosus (formerly classified Lactobacillus casei subsp. rhamnosus) polysaccharide antibody staining.
Rabbit anti-hexosamine (anti-H)-type wall and capsular polysaccharide antibodies and rabbit anti-rhamnose (anti-R)-containing wall and capsular polysaccharide antibodies were used. The reactivities of these sera have been previously described (25). Anti-H serum reacts exclusively with the H polysaccharide, while anti-R serum reacts predominantly with the R polysaccharide and shows only weak reactivity with the H polysaccharide. Relative to equivalent concentrations of a rabbit immunoglobulin control (Dako), dilutions of the antisera were applied to 2-μm resin sections that were previously prepared for immunohistochemistry by pepsin digestion and blocking in 20% horse serum. Dilutions of each antiserum (in 10% fetal bovine serum in PBS, pH 7.4) were applied for 1 h at 37°C, and binding was detected using goat anti-rabbit immunoglobulin conjugated with Alexa Fluor 488 (Invitrogen). Dilutions of primary antisera that produced maximum specific staining were selected and the results captured using a Leica DFC500 12-megapixel camera. Further manipulation of the images using Leica Application Suite software was standardized to maintain relationships.
Statistical analysis.
Fisher's exact test, a variant of the chi-square test appropriate for small samples, was used to compare the distributions of bacterial genera in grades of infection. Also, a Mann-Whitney U test was performed to analyze the complexity of the microflora in a limited pulp infection compared with that in a more extensive infection. A P value of <0.05 was considered significant.
RESULTS
Validation of probes.
Probes were inclusive of diverse species within the taxa and were demonstrated to be specific under the stringency conditions employed (Table 2). Cross-reactivity of the Veillonellaceae probe with Pseudoramibacter alactolyticus was prevented by increasing the formamide concentration to 40%. Family-specific probes were demonstrated to produce positive staining for multiple species tested within the particular family. For instance, the probe for Coriobacteriaceae detected Atopobium rimae, Atopobium parvulum, and Olsenella profusa. Similarly, the Veillonellaceae probe detected Veillonella dispar and Selenomonas infelix. The Bacteroides probe could also detect other Prevotella species from the family Prevotellaceae, i.e., P. veroralis, P. multiformis, P. shahii, P. buccae, and P. melaninogenica in addition to P. oralis, P. denticola, and P. salivae.
Patterns of infection.
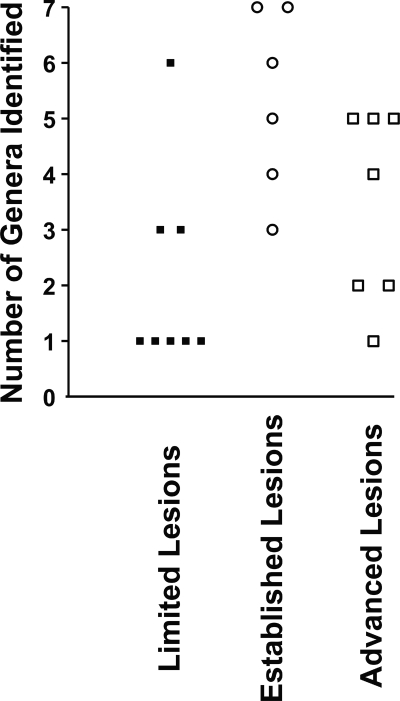
Unambiguous detection of morphologically distinct bacteria in FISH analysis was the basis for inclusion. Comparison of taxa within stages of infection of the pulp was based on this detection without reference to the abundance or distribution of the bacteria. The findings for the 21 pulps studied are summarized in Fig. 1. Fewer taxa were detected in limited lesions than in either established lesions considered alone or established lesions considered together with advanced lesions (P = 0.01, Mann-Whitney U test). Limited lesions had only one taxon present more frequently than established lesions (P = 0.031, Fisher's exact test) or established and advanced lesions considered together (P = 0.014). However, between limited and advanced lesions considered alone, there was no statistically significant difference in the numbers of samples where more than one taxon was detected (P = 0.119, Fisher's exact test).
FIG. 1.
Plot showing the number of genera identified in each sample tested according to lesion type. More taxa were identified in subjects with established and advanced lesions than in those with limited lesions (P = 0.01).
Microbial associations with lesions differing in extent.
Details of patterns of infection are displayed in Table 3. Lactobacilli were prominent in limited stages of pulp infection but poorly represented in advanced infection (see Fig. 2 and Fig. 3). Lactobacilli were present in 7/8 limited pulp infections and 6/6 established lesions but absent in advanced pulp lesions (P < 0.001, Fisher's exact test). Streptococci were not polarized in this way and were typically scattered throughout phases of infection. Bacteroides organisms were present in some limited, established, and advanced infections (Fig. 3h). Lachnospiraceae were more prevalent in established and advanced lesions considered together than in limited lesions (P = 0.024, Fisher's exact test) and also in advanced lesions relative to limited lesions (P = 0.041, Fisher's exact test). Coriobacteriaceae were more prevalent in established than in limited lesions (P = 0.026, Fisher's exact test) as well as in established and advanced lesions combined (P = 0.024, Fisher's exact test). However, there was no statistically significant difference in the incidence of Coriobacteriaceae between established and advanced lesions (Table 3 and Fig. 3g).
FIG. 2.
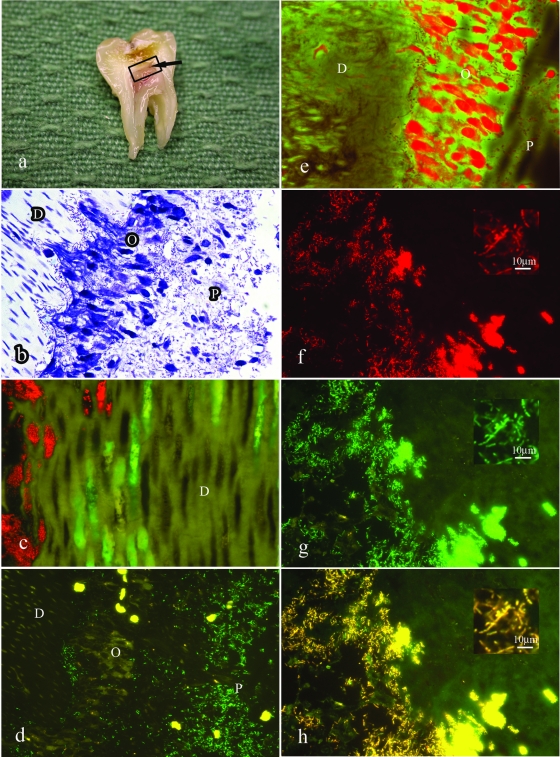
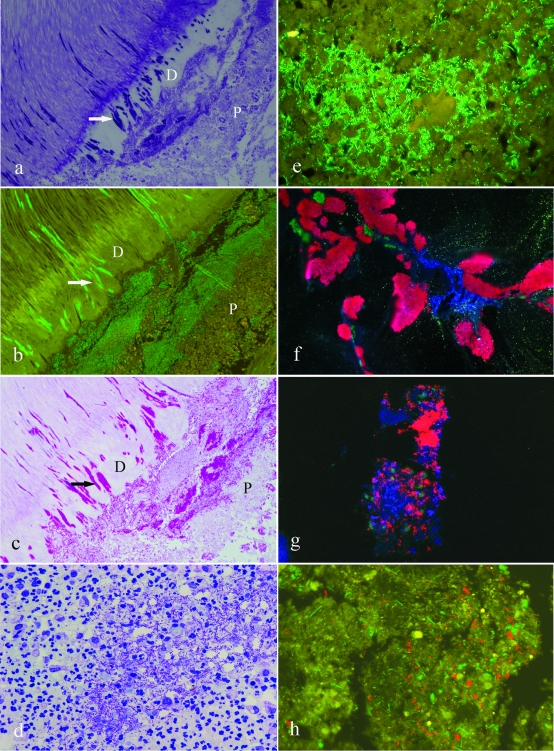
FISH analysis of lesion 1 from Table 3, showing predominance of lactobacilli in limited stages of pulp infection. (a) A split section of a tooth showing an extensive carious lesion and inflammation of the dental pulp. The arrow indicates the possible site of perforation of infection through the dentin layer. (b) Toluidine blue-stained section of the tooth showing the spread of infection along dentinal tubules (D) and invasion of the odontoblast layer (O). Note the lack of inflammatory infiltrate in the adjacent pulp tissue (P). (c) Dual-stained FISH probes showing streptococci (red) and lactobacilli (green) invading along separate tubules. (d) FISH probe for lactobacilli (green) showing invasion of the odontoblast layer in the lesion shown in panel b. Note the yellow autofluorescence of the erythrocytes in the blood vessels adjacent to the odontoblast layer. (e) The odontoblast layer (from panel d) stains intensely with acridine orange, indicating a high RNA content and the viability of these cells. (f) Higher magnification of panel e showing the universal FISH probe. (g) Field identical to that in panel f showing the FISH probe for lactobacilli. (h) Superimposition of images from panels f and g showing no detectable red-only organisms; i.e., all bacteria detected by the universal probe were also reactive for the probe for lactobacilli.
FIG. 3.
Progressive infection of the dental pulp. (a) Toluidine blue stain of lesion 9 (Table 3) showing extensive accumulation of bacteria and necrosis of the superficial region of the pulp tissue. Note the tight packing of bacteria within distorted tubules within the reactionary noncalcified dentin layer (D) (arrow) and the extrusion of the bacteria within these tubules into the pulp chamber (P). (b) FISH probe for lactobacilli in lesion 9 (Table 3). Image overlays of identical fields probed for lactobacilli and with the universal probe indicate that all of the bacteria detected were reactive with the probe specific for lactobacilli (data not shown). (c) Periodic acid-Schiff stain of the field shown in panel b. The lactobacilli are intensely reactive, suggesting the presence of a capsular polysaccharide. (d) Further extension of the infection of the pulp. Toluidine blue stain of lesion 6 (Table 3) showing the view from the middle region of the pulp tissue. Extensive infection is evident in association with inflammatory infiltrate. (e) FISH staining for lactobacilli in the region demonstrated in panel d. Lactobacilli were the only organisms detected in this lesion. (f) FISH stain of lesion 10 (Table 3) showing an enface image of the surface of carious dentin, revealing multiple bacterial forms: streptococci (red), Coriobacteriaceae (blue), and lactobacilli (green) in a zone of tissue breakdown. (g) Triple stain of the established pulpal lesion of the specimen described for panel f. FISH probes for lactobacilli (green), streptococci (red), and Coriobacteriaceae (blue) show sparse representation of lactobacilli in this established infection lesion. (h) Dual stain with FISH probes for Lachnospiraceae (green) and Bacteroides (red) for advanced pulpal infection in specimen 18 (Table 3). The organisms are dispersed throughout the degenerate tissue, showing no evidence of coassociation or clumping.
In limited lesions, the presence of high densities of lactobacilli was associated with a highly variable tissue response. For instance, in lesion 1 (Table 3), there was no detectable local inflammatory infiltrate (Fig. 2b), with the lactobacilli dispersing freely in the pulp tissue. In lesion 6 (Table 3), the lactobacilli were associated with an intense inflammatory reaction leading to a localized abscess (Fig. 3d). However, the organisms were predominantly extracellular (Fig. 3d) and were apparently metabolically active, as indicated by an intense signal for 16S rRNA (Fig. 3e). No statistically significant relationships between the taxa comprising Streptococcus, Veillonellaceae, Propionibacterium, Stenotrophomonas, Acinetobacter, Gammaproteobacteria, Fusobacterium, Bacteroidetes, and Pseudoramibacter were apparent for any of the lesion types identified.
Associations between taxa.
Polymicrobial infections typical of established and advanced stages were characterized either by bacterial taxa showing a diffuse and scattered presence throughout the affected tissue or by individual groups present in tightly concentrated masses. Evidence of coassociation was minimal, except for intricate mass associations of Bacteroidetes and Coriobacteriaceae. Although Lachnospiraceae and Coriobacteriaceae were often coincident with each other, this was not statistically significant (Fisher's exact test), and there were no other clear associations between taxa in the samples studied. This was confirmed by the in situ analysis, where there was evidence for spatial separation of the infecting organisms either in dense colonies of homologous organisms (Fig. 3g) or of scattered bacteria representing separate taxa (Fig. 3h).
Characteristics of limited infection.
Two specimens had orientations that provided the opportunity to examine the initial stages of invasion of bacteria from dentin into the pulp. In specimen 1 (Table 3), bacteria invading the odontoblast layer but also penetrating more deeply into the pulp tissue were detected. There was no obvious tissue response in the superficial zone rich in infecting bacteria, but there was a mononuclear leukocyte accumulation deeper into the pulp tissue. The viability of the tissue, including the odontoblast layer, was confirmed by staining with acridine orange for RNA (Fig. 2e). Only the probe for lactobacilli produced detectable staining of pulp. To analyze this further, sections were subjected to dual staining with the Alexa 488 Lactobacillus sp. probe and the Alexa 594 universal bacterial probe. The combined images indicate that all detected bacteria were lactobacilli (Fig. 2f, g, and h). This was confirmed by overlaying images obtained by probing with either the universal or lactobacilli probe, followed by photobleaching of fluorescence and reprobing of the section with the alternative FISH probe. Image overlays indicated a correspondence of staining for universal and Lactobacillus sp. probes.
In specimen lesion 9 (Table 3), the infection was more extensive, resulting in a zone of necrosis (Fig. 3a). There was evidence for invasion across a broad area of the pulp-dentin interface, with the organisms reaching a very high biomass within the affected pulp tissue. In this specimen, there was also evidence that the bacteria in the zone of invasion from carious dentin were almost exclusively lactobacilli, as indicated by comparison of specimens stained with lactobacillus probes (Fig. 3b) with those stained with universal bacterial probes (data not shown). The intensity of reactivity of 16S rRNA with the FISH probe (Fig. 3b) indicated that the organisms approaching and invading the pulp were highly metabolically active, particularly in comparison with bacteria more distally placed in the outer regions of the tubules (data not shown).
Characteristics of the invading lactobacilli.
Resin blocks containing lesions 1 and 9 were trimmed to the areas of interest, and bacterial DNA was extracted from a series of sections. Using primer sets for the major Lactobacillus spp. found in carious lesions, amplicons were detected for Lactobacillus rhamnosus primer sets only for lesion 1 and for Lactobacillus crispatus primer sets only for lesion 9 (data not shown). Identities were confirmed by sequencing (99% sequence identity). No amplicons were detected using universal bacterial 16S rRNA primer sets following extraction of resin sections containing noninfected pulp tissue.
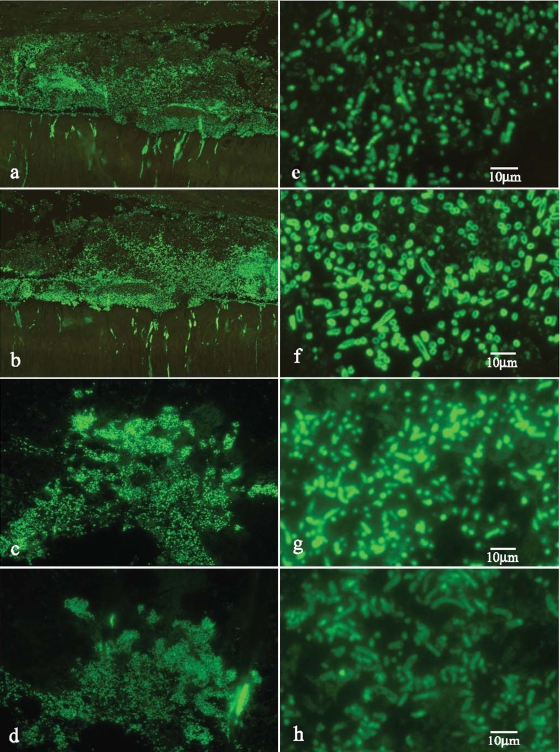
To further analyze the status of the lactobacilli invading the pulp tissue, sections were stained with periodic acid-Schiff reagent. The results shown in Fig. 3c indicate intense reactivity for the lactobacilli, suggesting the presence of capsular polysaccharide. This was confirmed by immunostaining for H and R exopolysaccharides (Fig. 4). The patterns of differential staining for the H- and R-type polysaccharides are interpreted as indicating the presence of abundant H-type polysaccharide capsular material in lesion 9 (Fig. 4f) but only trace reactivity for the antiserum directed primarily against the R-type polysaccharide (Fig. 4e). In contrast, for lesion 1 there was intense staining for the R-type polysaccharide compatible with capsular material (Fig. 4g), while there was only trace staining for the H-type polysaccharide (Fig. 4h).
FIG. 4.
Profile of a surface polysaccharide expressed by invasive lactobacilli. (a) Low-power view of a section stained with antibody to the Lactobacillus rhamnosus R-type polysaccharide in specimen 9 (Table 3), showing reactivity of lactobacilli in the region of the dentin-pulp interface. Note that the reactivity for the organisms within dentinal tubules appears to intensify at the pulpal interface. (b) Low-power view of a section stained with antibody to the Lactobacillus rhamnosus H-type polysaccharide in specimen 9 (Table 3). (c) Low-power view of a section stained with antibody to the Lactobacillus rhamnosus R-type polysaccharide in specimen 1 (Table 3). (d) Low-power view of a field similar to that in panel c stained for the H-type polysaccharide following standardized exposure. The staining is less intense than that for the R-type polysaccharide. (e) High-power view of panel a. The staining is weak and not uniform. (f) High-power view of panel b showing intense peripheral staining. The dimensions of the stained organisms suggest extensive capsular polysaccharide. (g) High-power view of panel c showing intense staining. The dimensions of the stained organisms suggest the presence of capsular polysaccharide, although the profile is not as clear as shown in panel f. (h) High-power view of panel d showing trace reactivity for only the anti-H-type polysaccharide.
DISCUSSION
The extension of infection from carious dentin into dental pulp has usually been considered to result from a failure of the dentin layer and collapse of necrotic dentin into the pulp, resulting in a macroscopic defect (19). It has been reported, however, that up to 50% of pulp infections are not associated with macroscopic defects in the pulp-dentin interface (8). The implication is that infection of pulp occurs by direct extension of invasion along the dentinal tubules, a phenomenon well known for many decades (21).
An unexpected finding of the current study was the prominence of lactobacilli in limited phases of infection of pulp tissue. This was notable in relation to the complexity of the carious dentin flora within a lesion, where up to 31 bacterial taxa were detected (4). Previous analysis has indicated the diversity of lactobacilli within carious dentin, where 18 different species and phylotypes, including novel forms, have been detected, with each lesion containing at least three species or phylotypes (2). In lesions 1 and 9 (Fig. 2 and 3), the lactobacilli were not accompanied by a local inflammatory response. This was evident for the lactobacilli detected in lesion 6 (Fig. 3), but the absence of detectable phagocytosis indicated failure to effectively opsonize the organisms. These responses suggested altered bacterial surfaces, particularly capsular material.
The R-type polysaccharide was dominant in the putative L. rhamnosus strain detected as the predominant organism in lesion 1. Cultured strains of L. rhamnosus demonstrate considerable variation in the abundance and location of the R-type polysaccharide, dependent in part on the concentrations of particular environmental sugars and on growth rates (25). For instance, culture of L. rhamnosus strain NCTC 6375 in ribose-containing media reduced expression of the R-type polysaccharide, whereas culture in glucose, galactose, or sucrose did not reduce R-type polysaccharide expression relative to H-type polysaccharide expression (25). The putative L. rhamnosus strain detected at the dentin-pulp interface in lesion 1 displayed a staining profile, interpreted as encapsulation, that either is largely characteristic of the strain or was modulated by undisclosed sugars within this tissue environment.
Periodic acid-Schiff staining for carbohydrate disclosed intense staining for the putative L. crispatus strain detected as dominant in specimen 9 (Table 3), indicating, for the advancing columns within dentin and bacteria within the pulp, a rich accumulation of polysaccharide. These bacteria displayed intense immunoreactivity with the anti-H-type (hexosamine-containing) polysaccharide raised against L. rhamnosus. In this context, strains of L. crispatus (formerly classified as L. acidophilus group A2) (3) contain cell wall-associated galactose and glucose but not rhamnose (11, 13). This could explain the failure of the detected organisms to react strongly with the anti-rhamnose-type polysaccharide antiserum (25) and the intense peripheral staining of the putative L. crispatus strain with the anti-H-type polysaccharide serum, where glucose is the immunodominant sugar (25). As there was evidence for polymicrobial infection of the pulp in lesion 9 (Table 3), it is possible that the intense focus of L. crispatus represents a dominant, active, secondary invasion rather than a primary invasion of vital pulp tissue, as detected for lesion 1. That is, the capacity of L. crispatus to function as a primary invader of pulp tissue remains unclear.
These two lactobacilli also reacted intensely with acridine orange and with probes for 16S rRNA, indicating copious RNA content. Rather than a toxic response, the response appears to be one of uncontrolled proliferation within peripheral pulp tissue. The subdued inflammatory response within the affected peripheral pulp tissue was noteworthy and could indicate a limited recognition capacity of the innate immune defenses. For instance, lipoteichoic acid derived from lactobacilli was not recognized by Toll-like receptor 4 expressed by odontoblasts (20). Bacterial capsular material has the capacity to promote nonresponsiveness by interacting with Toll-like receptor 2, which is also expressed by odontoblasts (13, 23). In contrast, the intense focus of lactobacilli evident in the deeper aspect of the pulp in lesion 6 (Table 3 and Fig. 3d) was associated with a dense inflammatory cell infiltrate that appeared to be ineffective in sequestering and destroying the organisms. This was evident by the histopathology and the strong signal obtained with the FISH probe, indicating preservation of rRNA.
The basis for this activity and for the apparent invasive behavior of bacteria provisionally identified as L. rhamnosus and L. crispatus is unknown, with scant evidence of parallels in other infectious processes (6). Extensive literature supports the capacity of probiotic bacteria, including certain strains of L. rhamnosus (5) and L. crispatus (24), to protect the intestine from colonization by pathogenic organisms (13). Other data also suggest the protective effects of lactobacilli. For instance, lactobacilli protect against a lethal reaction to ulceration of the intestinal mucosa induced by toxoplasmosis infection in experimental mice (7). Conversely, there is limited evidence for the pathogenic potential of lactobacilli, although aggregation of platelets (12) and expression of a surface plasminogen-activating enzyme (10) have been reported. In the present study, detection of rRNA was likely to be enhanced in actively metabolizing bacteria. This technology does not eliminate the possibility of other contributions, particularly by microaerophilic bacteria, including Acinetobacter spp., to the early phase of pulp infection. These organisms could be short-lived in this tissue environment and, therefore, not detected in FISH analysis. It was observed, however, that Acinetobacter spp. were detected in only two advanced infection lesions.
In summary, the final phase of caries extension, infection of the pulp, has some unusual features. Evidence from this study provides a strong indication for the role of lactobacilli, although it cannot be concluded that species of this genus are essential for the initiation of infection of vital pulp tissue. While the subsequent extension of infection was associated with development of a polymicrobial flora, analysis of the spatial distribution of the organisms did not support cooperation or dependencies between groups of bacteria. It was also noted that high concentrations of anaerobic bacteria were detected in regions of tissue that were clearly necrotic, indicating a favorable environment for growth of these bacteria rather than pathogenic action.
Acknowledgments
This study was supported by NIDCR grant R01 DE015272-07 and Australian National Health and Medical Research Council grant 512524.
We thank Kerstin Angner and Maruth Banavar for collection of teeth with carious lesions and John Gibbins for his valuable comments on the manuscript.
Footnotes
Published ahead of print on 3 March 2010.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byun, R., M. A. Nadkarni, K.-L. Chhour, F. E. Martin, N. A. Jacques, and N. Hunter. 2004. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 42:3128-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cato, E. P., W. E. C. Moore, and J. L. Johnson. 1983. Synonymy of strains of “Lactobacillus acidophilus” group A2 (Johnson et al. 1980) with the type strain of Lactobacillus crispatus (Brygoo and Aladame 1953) Moore and Holdeman 1970. Int. J. Syst. Bacteriol. 33:426-428. [Google Scholar]
- 4.Chhour, K.-L., M. A. Nadkarni, R. Byun, F. E. Martin, N. A. Jacques, and N. Hunter. 2005. Molecular analysis of microbial diversity in advanced caries. J. Clin. Microbiol. 43:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Champs, C., N. Maroncle, D. Balestrino, C. Rich, and C. Forestier. 2003. Persistence of colonization of intestinal mucosa by a probiotic strain, Lactobacillus casei subsp. rhamnosus Lcr35, after oral consumption. J. Clin. Microbiol. 41:1270-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harty, D. W., H. J. Oakey, M. Patrikakis, E. B. Hume, and K. W. Knox. 1994. Pathogenic potential of lactobacilli. Int. J. Food Microbiol. 24:179-189. [DOI] [PubMed] [Google Scholar]
- 7.Heimesaat, M. M., S. Bereswill, A. Fischer, D. Fuchs, D. Struck, J. Niebergall, H. K. Jahn, I. R. Dunay, A. Moter, D. M. Gescher, R. R. Schumann, U. B. Gobel, and O. Liesenfeld. 2006. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J. Immunol. 177:8785-8795. [DOI] [PubMed] [Google Scholar]
- 8.Hoshino, E., N. Ando, M. Sato, and K. Kota. 1992. Bacterial invasion of non-exposed dental pulp. Int. Endod. J. 25:2-5. [DOI] [PubMed] [Google Scholar]
- 9.Hugenholtz, P., G. W. Tyson, and L. L. Blackall. 2002. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol. Biol. 179:29-42. [DOI] [PubMed] [Google Scholar]
- 10.Hurmalainen, V., S. Edelman, J. Antikainen, M. Baumann, K. Lahteenmaki, and T. K. Korhonen. 2007. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology 153:1112-1122. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. L., C. F. Phelps, C. S. Cummins, J. London, and F. Gasser. 1980. Taxonomy of the Lactobacillus acidophilus group. Int. J. Syst. Bacteriol. 30:53-68. [Google Scholar]
- 12.Kirjavainen, P. V., E. M. Tuomola, R. G. Crittenden, A. C. Ouwehand, D. W. Harty, L. F. Morris, H. Rautelin, M. J. Playne, D. C. Donohue, and S. J. Salminen. 1999. In vitro adhesion and platelet aggregation properties of bacteremia-associated lactobacilli. Infect. Immun. 67:2653-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lebeer, S., J. Vanderleyden, and S. C. J. De Keersmaecker. 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72:728-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin, F. E., M. A. Nadkarni, N. A. Jacques, and N. Hunter. 2002. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40:1698-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massey, W. L., D. M. Romberg, N. Hunter, and W. R. Hume. 1993. The association of carious dentin microflora with tissue changes in human pulpitis. Oral Microbiol. Immunol. 8:30-35. [DOI] [PubMed] [Google Scholar]
- 16.Munson, M. A., A. Banerjee, T. F. Watson, and W. G. Wade. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadkarni, M. A., F. E. Martin, N. Hunter, and N. A. Jacques. 2009. Methods for optimizing DNA extraction before quantifying oral bacterial numbers by real-time PCR. FEMS Microbiol. Lett. 296:45-51. [DOI] [PubMed] [Google Scholar]
- 18.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 19.Smulson, M. H., and S. M. Sieraski. 1989. Histopathology and diseases of the dental pulp., p. 142-145. In F. S. Weine (ed.), Endodontic therapy, 4th ed. The C. V. Mosby Company, St. Louis, MO.
- 20.Staquet, M. J., S. H. Durand, E. Colomb, A. Romeas, C. Vincent, F. Bleicher, S. Lebecque, and J. C. Farges. 2008. Different roles of odontoblasts and fibroblasts in immunity. J. Dent. Res. 87:256-261. [DOI] [PubMed] [Google Scholar]
- 21.Thoma, K. H. 1944. Oral pathology, a histological, roentgenological, and clinical study of the diseases of the teeth, jaws, and mouth, 2nd ed. The C. V. Mosby Company, St. Louis, MO.
- 22.van Houte, J. 1994. Role of micro-organisms in caries etiology. J. Dent. Res. 73:672-681. [DOI] [PubMed] [Google Scholar]
- 23.Veerayutthwilai, O., M. R. Byers, T. T. Pham, R. P. Darveau, and B. A. Dale. 2007. Differential regulation of immune responses by odontoblasts. Oral Microbiol. Immunol. 22:5-13. [DOI] [PubMed] [Google Scholar]
- 24.Voltan, S., I. Castagliuolo, M. Elli, S. Longo, P. Brun, R. D'Inca, A. Porzionato, V. Macchi, G. Palu, G. C. Sturniolo, L. Morelli, and D. Martines. 2007. Aggregating phenotype in Lactobacillus crispatus determines intestinal colonization and TLR2 and TLR4 modulation in murine colonic mucosa. Clin. Vaccine Immunol. 14:1138-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wicken, A. J., A. Ayres, L. K. Campbell, and K. W. Knox. 1983. Effect of growth conditions on production of rhamnose-containing cell wall and capsular polysaccharides by strains of Lactobacillus casei subsp. rhamnosus. J. Bacteriol. 153:84-92. [DOI] [PMC free article] [PubMed] [Google Scholar]