Abstract
An echovirus 3 (Echo3) strain (strain LR31G7) was isolated from a sewage treatment plant in Greece in 2005. Full-genome molecular, phylogenetic, and SimPlot analyses were conducted in order to reveal the evolutionary pathways of the isolate. Nucleotide and phylogenetic analyses of part of the VP1 genomic region revealed that the isolated strain correlates with Echo3 strains isolated during the same year in France and Japan, implying that the same virus circulated in Europe and Asia. LR31G7 was found to be a recombinant that shares the 3′ part of its genome with an Echo25 strain isolated from asymptomatic infants in Norway in 2003. Nucleotide and SimPlot analyses of the VP1-2A junction, where the recombination was located, revealed the exact recombination breakpoint (nucleotides 3357 to 3364). Moreover, there is evidence that recombination events had occurred in 3B-3D region in the evolutionary history of the isolate. Our study indicates that recombination events play major roles in enterovirus evolution and that the circulation of multirecombinant strains with unknown properties could be potentially dangerous for public health.
Enteroviruses are among the most common viruses infecting humans. Although most infections are mild or asymptomatic, they can cause severe and potentially fatal diseases, such as acute hemorrhagic conjunctivitis, aseptic meningoencephalitis, and acute flaccid paralysis (39). They are small, nonenveloped viruses with a positive-strand RNA of approximately 7,500 nucleotides (nt). The enterovirus genome consists of three major regions: the 5′ untranslated region (5′ UTR), the 3′ untranslated region (3′ UTR), and the open reading frame (ORF). The ORF is translated into a long polyprotein which contains the four structural viral proteins (VP1 to VP4) and the seven nonstructural viral proteins (2A to 2C and 3A to 3D). Human enteroviruses (HEVs) include immunologically and genetically distinct types and are classified into five species, HEV species A (HEV-A) to HEV-D and polioviruses, which show a high level of molecular relatedness to HEV-C (7).
An enterovirus population does not exist as a single genotype but exists as a group of correlated sequences named quasispecies. Quasispecies arise as a result of a high mutation rate due to the lack of proofreading activity of the viral RNA polymerase and short generation times (19, 23). Even though high mutation rates result in nonviable viruses, it can lead to a swarm of mutations potentially beneficial to the population level, in which, under certain circumstances and after a long silent period, a dominant sequence that is adapted to new environments and the challenges that it encounters during infection may emerge (44).
Recent publications also state that enteroviruses within species exist not as delimited lineages but as a pool of independently evolving genome fragments that frequently recombine to give rise to new virus variants (30, 33, 45). There is a reservoir of a limited number of capsid sequences and a variety of functional regions that recombine and give rise to new viral genotypes exhibiting modified pathogenic properties (29). Recombination has been extensively described in polioviruses (2, 11, 14, 21, 22, 41, 42, 43), but much less attention has been devoted to recombination in nonpoliovirus enteroviruses. Recent studies based on parts of the enterovirus genome revealed that intertypic recombination is a frequent phenomenon in circulating nonpoliovirus enteroviruses (6, 24, 28, 45). Although sequences of all the prototype strains of enteroviruses are available, only a restricted number of full-genome studies of modern enterovirus strains have been reported so far (10, 29).
In the study described here, we present a full-genome analysis of an echovirus 3 (Echo3) recombinant strain (strain LR31G7) isolated in 2005 from the sewage treatment plant of the city of Larissa, Thessaly, Greece. Comparison of the full-genome sequence of isolate LR31G7 with that of prototype Echo3 strain Morrisey as well as with that of the only available fully sequenced Echo3 isolate (isolate PicoBankDM1E3) was conducted. Finally, phylogenetic analysis of the VP1 region was conducted by comparing the sequence of the VP1 region of isolate LR31G7 with the sequences of the VP1 regions of the E3 prototype strain and other E3 isolates.
MATERIALS AND METHODS
Enterovirus isolation from sewage.
An enterovirus strain (strain LR31G7) was isolated from the sewage treatment plant of the city of Larissa, Thessaly, Greece, in October 2005. The environmental sample was concentrated by the two-phase separation method recommended by WHO (48), and the virus was isolated from Rd cells. In order to avoid viral mixtures, 10-fold serial dilutions were prepared and inoculated into 96-well plates. The last dilution in which a cytopathic effect was observed was inoculated into a 25-cm2 flask containing Rd cells. The presence of the virus was confirmed by PCR with primer pair UG52-UC53 (17), following RNA extraction and reverse transcription.
Primary characterization of the isolate.
Enterovirus RNA was extracted from the inoculated Rd cells by the method described by Casas et al. (9) and stored at −20°C. The isolated RNA was reverse transcribed into cDNA. Five microliters of extracted RNA was incubated with 1 μl of random d(N6) primers (New England Biolabs), 1 μl of 40 mM deoxynucleoside triphosphates (dNTPs), and 5 μl of RNase- and DNase-free distilled water for 5 min at 65°C. After the reaction mixture was cooled on ice, 8 μl of the reaction mixture containing 4 μl Moloney murine leukemia virus (MMLV) reaction buffer (Invitrogen, Life Technologies, Paisley, United Kingdom), 200 U MMLV reverse transcriptase, 2 μl of 0.1 mM dithiothreitol, 20 U RNase inhibitor (Promega Corporation, Madison, WI), and 0.5 μl RNase- and DNase-free distilled water was added. cDNA synthesis was completed by incubation at 25°C for 10 min, 37°C for 50 min, and finally, 70°C for 15 min for inactivation of the MMLV reverse transcriptase. For the characterization of isolate LR31G7, VP1 amplification by reverse transcription (RT)-PCR with primer pair 292-222 was performed as described previously (37).
Genome amplification.
Table 1 provides details about the primer pairs that were used in order to obtain sequence information from all genomic regions of isolate LR31G7. Ten of these primers were used for the first time in the present study. They were designed with the aid of Primer3 software, obtained online from the Whitehead Institute (http://www.genome.wi.mit.edu/genomesoftware/other/). For the design of primer pairs CHR3-CHR4 and CHR5-CHR6, the sequences of all HEV-B prototype strains were used. The rest of the primer pairs were designed on the basis of the sequences of virus isolate LR31G7. All primers were synthesized by Metabion (Martinsried, Germany).
TABLE 1.
Primers used in the present study
| Primer | Position | Polarity | Sequence (5′-3′) | Reference or source |
|---|---|---|---|---|
| 72437 | 001-020 | Sense | TTAAAACAGCTCTGGGGTTG | Mulders et al. (32) |
| 216616 | 545-565 | Antisense | GAAACACGGACACCCAAAGTA | Blomqvist et al. (3) |
| 0340F | 310-333 | Sense | TAGATCAGGCYGATGAGTCACCGC | Lukashev et al. (29) |
| 1200R | 1177-1196 | Antisense | GGGAATTTCCACCACCACCC | |
| BL870 | 870-890 | Sense | CGACAGGATTTCACACAGGA | Present study |
| BR3231 | 3231-3211 | Antisense | GCTTTTCACATACGGGCTAA | |
| BL1369 | 1369-1389 | Sense | GAGGTTGTCGCAGCTTCTCT | Present study |
| BR2580 | 2580-2560 | Antisense | GGGAACCACTTGTGAGGTGT | |
| 292 | 2612-2627 | Sense | MIGCIGYIGARACNGG | Oberste et al. (37) |
| 222 | 2969-2951 | Antisense | CICCIGGIGGIAYRWACAT | |
| EUG3a | 2946-2965 | Sense | TGGCAAACTTCCWCCAACCC | Caro et al. (8) |
| EUG3b | 2946-2965 | Sense | TGGCAAACATCTTCMAATCC | |
| EUG3c | 2946-2965 | Sense | TGGCAGACTTCAACHAACCC | |
| EUC2 | 4413-4433 | Antisense | TTTGCACTTGAACTGTATGTA | |
| EUC2a | 4428-4448 | Antisense | GGTTCAATACGGCATTTG | |
| EUC2b | 4428-4448 | Antisense | GGTTCAATACGGTGTTTGCT | |
| CHR1 | 4284-4308 | Sense | CNTCHCARAGTGAYCARGARCARYT | Kottaridi et al. (24) |
| CHR2 | 5084-5081 | Antisense | GTAYACYGGTGGWCCYTGRAAKA | |
| BL4709 | 4709-4729 | Sense | TACATCCCCATTTGTGTTGG | Present study |
| BR6063 | 6063-6043 | Antisense | TGCTGGCTCCTTGTTACCTT | |
| CHR3 | 5047-5065 | Sense | CIACYCTWGARGCRCTVTT | Present study |
| CHR4 | 5860-5841 | Antisense | GACRTGAGIACHCCRCCRCA | |
| 5850F | 5837-5859 | Sense | CAGTGYGGIGGIGTICTCATGTC | Lukashev et al. (30) |
| 6500R | 6531-6506 | Antisense | AGRTTGCCAAAYGTYTGYCTCATTGC | |
| CHR5 | 6485-6507 | Sense | ATCCAGYTTGAAYGAYTCIGIRG | Present study |
| CHR6 | 7250-7227 | Antisense | GAAYTCYTCRTAYTCKTGCTCYCC |
The viral RNA was reverse transcribed into cDNA as described above for all primer pairs, with the exception of EUC2, EUG3a, EUG3b, and EUG3c, for which RT with antisense primers EUC2a and EUC2b was carried out, as has been described previously (8).
The PCR mixture for each tube comprised 3 μl cDNA from isolate LR31G7, 1 μl of each primer at a concentration of 25 pmol/μl, 5 μl 10× PCR buffer, 5 μl 10 mM dNTPs, 2.5 U Paq5000 DNA polymerase (Stratagene), and double-distilled nuclease-free water up to a final volume of 50 μl/tube. For primer pairs CHR3-CHR4 and CHR5-CHR6, 40 cycles of denaturation (95°C for 30 s), annealing (45°C for 1 min), and extension (72°C for 1 min) were used. The reactions with primer pairs BL870-BR3231, BL1369-BR2580, and BL4709-BR6063 were performed for 35 cycles. Each cycle consisted of 30 s of denaturation at 95°C, 30 s of annealing at 58°C (for primer pair BL870-BR3231) or at 55°C (for primer pairs BL1369-BR2580 and BL4709-BR6063), and 2 min (for primer pair BL870-BR3231) or 1.5 min (for primer pairs BL1369-BR2580 and BL4709-BR6063) of extension at 72°C. For the other primer pairs, the PCR conditions were the same as those described in the original papers (Table 1). The PCR products were cleaned up with a PCR gel extraction kit (Qiagen GmbH, Hilden, Germany). All PCR products were sequenced at Macrogen Inc. (Seoul, South Korea) by using the primers described in Table 1.
Sequencing and sequence analysis.
The sequences were initially examined for their closest homologous sequence by using BLAST software, obtained online from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple-sequence alignments were generated by the use of ClustalW software, obtained online from the website of the European Bioinformatics Institute (htxtp://www.ebi.ac.uk/clustalw/). Phylogenetic and molecular analyses were conducted with MEGA (version 4) software. The reliability of the trees was determined by bootstrap analysis with 100 pseudoreplicate data sets. Finally, plots of nucleotide similarity between the aligned sequences were created with the aid of SimPlot software (version 3.1); the window sizes were 200 nt for full-genome analysis and 100 nt for analysis of the recombination junction (26). Partial sequences of the following Echo3 strains were used for phylogenetic analysis of isolate LR31G7 (the GenBank accession numbers are given in parentheses): CF1801081-05 (AM236930), CF1820181-05 (AM236931), Fukuoka City2005-70 (AB234341), Fukuoka City2005-97 (AB234342), 94CF858 (AJ241446), BE02-3627 (AY342794), W178-128/99 (AY208114), 810/85 (AF295490), 2392-82 (AF295453), 108-97 (AB268162), and 222-99 (AB268165). For the recombination junction analysis, the partial sequence of Echo25 strain NO-519 (GenBank accession number DQ317208) was used.
Nucleotide sequence accession number.
The almost complete genomic sequence of isolate LR31G7 (from nt 1 to nt 7189) has been deposited in the GenBank library under accession number FJ766334.
RESULTS
An enterovirus strain was isolated from the sewage treatment plant of the city of Larissa, Greece, in October 2005. Sequencing of the VP1 genomic region revealed that strain RL31G7 belongs to the Echo3 serotype, as it shows 79% (>75%) nucleotide identity and 96% (>88%) amino acid identity with the same region of the Echo3 reference strain (34). An analysis of almost the full genome (7189 nt) of the isolate was conducted, as the only completed sequenced Echo3 strains are reference strain Morrisey and isolate PicoBankDM1E3, which was associated with the induction of diabetes and which was isolated from a stool sample in Finland in 1998 (47).
Table 2 compares the levels of nucleotide and amino acid sequence identities of strain LR31G7 with those of prototype strain Morrisey and isolate PicoBankDM1E3, the only Echo3 strain that has been completely sequenced. As shown in Table 2, the sequence of isolate LR31G7 differs significantly from that of the prototype strain in all regions sequenced. The region of isolate LR31G7 with the highest degree of identity with the sequence of the prototype strain is the 5′ UTR (86%), followed by regions P2 (81%), P1 (80%), and P3 (79%). The gene with the highest degree of identity was 3B (83%), and the one with the lowest was 2B (76%). Comparison of the nucleotide and amino acid sequences of LR31G7 with those of isolate PicoBankDM1E3 revealed that there is a high degree of identity in the 5′ part of the genome (5′ UTR-VP1; 93 to 90%), but the level of identity falls from 82 to 79% in the 3′ part of the genome (2A to 3D).
TABLE 2.
Nucleotide and amino acid sequence identities between LR31G7, Echo3 prototype strain Morrisey, and strain PicoBankDM1E3
| Genomic region | Morrisey |
PicoBankDM1E3 |
||
|---|---|---|---|---|
| % nucleotide identity | % amino acid identity | % nucleotide identity | % amino acid identity | |
| 5′ UTR | 86 | 93 | ||
| P1 | 80 | 90 | ||
| VP4 | 80 | 98 | 92 | 100 |
| VP2 | 79 | 97 | 90 | 99 |
| VP3 | 82 | 97 | 91 | 99 |
| VP1 | 79 | 96 | 90 | 98 |
| P2 | 81 | 81 | ||
| 2A | 82 | 94 | 82 | 94 |
| 2B | 76 | 94 | 79 | 98 |
| 2C | 81 | 98 | 82 | 97 |
| P3 | 79 | 81 | ||
| 3A | 79 | 97 | 80 | 97 |
| 3B | 83 | 95 | 80 | 95 |
| 3C | 78 | 93 | 81 | 97 |
| 3Da | 78 | 96 | 80 | 97 |
The 1,251 nt of 3D genomic region.
In order to investigate the relationship of the sequence of strain LR31G7 to the entire sequences of the prototype strain and other strains of enterovirus species B, phylogenetic trees for each genomic region were constructed (data not shown). In phylogenetic trees for the 5′ UTR and capsid regions, LR31G7 clusters with isolate PicoBankDM1E3. In the phylogenetic trees for the structural genes, there is an absolute correlation between the serotype and the genotype, as the two Echo3 isolates (isolates LR31G7 and PicoBankDM1E3) cluster together with Echo3 prototype strain Morrisey.
As was expected, the serotype-genotype correlation that was observed in the capsid-coding region was interrupted in trees for nonstructural sequences. Analysis of the 2A region showed that isolate LR31G7 still clustered together with isolate PicoBankDM1E3, but this phylogenetic relationship between these two Echo3 isolates stopped in the 2B-coding region. It was also observed that in the 2C, 3A, 3C, and 3D phylogenetic trees, LR31G7, along with modern enterovirus isolates, belongs to a cluster separate from the prototype strain cluster. Even though there is a different classification for each genomic region in 2B, 2C, and 3A, phylogenetic trees for 3B, 3C, and 3D revealed that LR31G7 consistently clusters with coxsackie A virus 9 (CAV9) strains (strains Cuba35of93, Cuba47of93, Cuba23of00, Cuba267of90) isolated in Cuba from 1990 to 2000. Between the LR31G7 and the CAV9 isolates there are 86%, 85%, and 85 to 87% nucleotide sequence identities in the 3B, 3C, and 3D regions, respectively.
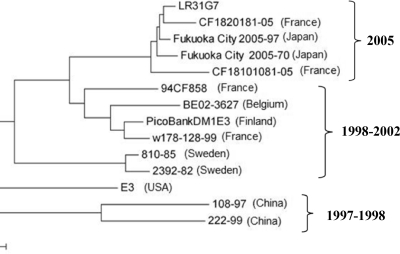
In order to correlate the sequence of isolate LR31G7 with other short sequences available from Echo3 isolates, a 269-nt region of VP1 (from nt 2692 to nt 2960) was chosen for use for the construction of a phylogenetic tree, as it occurs within most of the available sequences. As shown in Fig. 1, isolate LR31G7 clustered together with echoviruses 3 recently isolated in France (strains CF1801081-05 and CF1820181-05) and Japan (strains Fukuoka City2005-70 and Fukuoka City2005-97) in 2005. The nucleotide sequence identity of this part of VP1 between the LR31G7 and the Echo3 isolates of the 2005 cluster was 95 to 98%. The isolates of the 1998 to 2002 cluster and the 1997 to 1998 cluster present between them 86 to 91% and 75 to 78% nucleotide sequence identities, respectively.
FIG. 1.
Phylogenetic tree of 269 nt (from nt 2692 to nt 2960) of a region of VP1 of strain LR31G7 and other Echo3 strains.
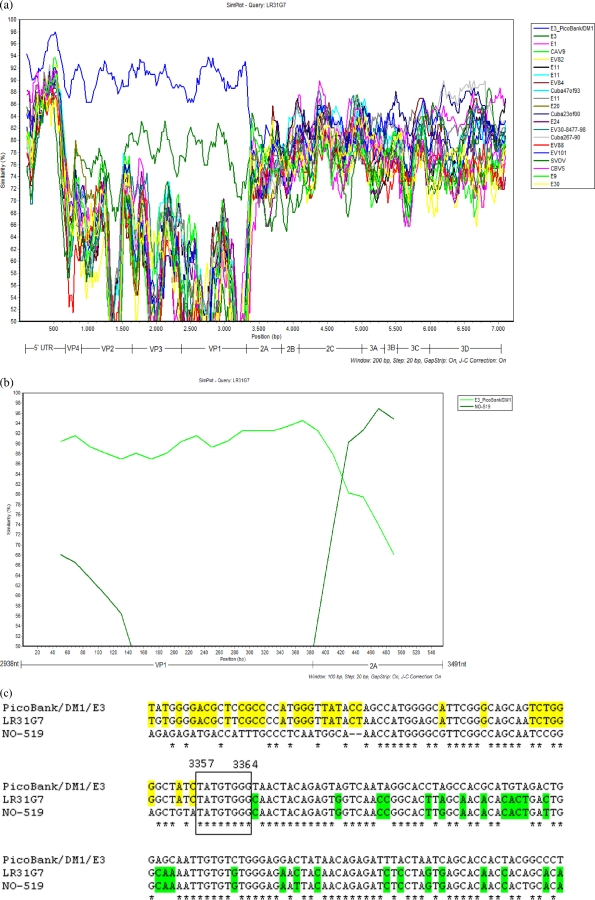
The results of SimPlot analysis of the full-genome sequence of isolate LR31G7 is presented in Fig. 2a. In the 5′ UTR and the capsid-coding region, the strain with the highest degree of identity to the sequence of LR31G7 was PicoBankDM1E3. Passing to the 2A-coding region, there was an abrupt change in the degree of identity, which fell to 81%. It is obvious that a recombination event had occurred that gave naissance to a strain that in the 5′ half of the genome is Echo3 and in which the rest of the genome is derived from another enterovirus B. Full-genome SimPlot analysis could not help with the detection of the donor of the functional genomic regions of isolate LR31G7, as the sequences of only a small number of completely sequenced enterovirus strains are available in GenBank. For this reason, we studied in detail the VP1-2A region, where the recombination was located. The 20 nt of the 3′ end of VP1 and the 20 nt of the 5′ end of the 2A region had the highest degrees of identity (92%) with the same sequences of isolate PicoBankDM1E3, but 100 nt downstream from that point, the strain that was the most closely related to LR31G7 was an Echo25 strain, strain NO-519, which was isolated in Norway in 2003, with which LR31G7 had 96% nucleotide identity. The results of SimPlot analysis of the sequences of the VP1-2A regions of isolates LR31G7, PicoBankDM1E3, and NO-519 are presented in Fig. 2b, in which the recombination between the last two strains is obvious. The high degree of nucleotide sequence identity between LR31G7 and isolates PicoBankDM1E3 and NO-519 allowed the identification of the exact location of the recombination. As shown in Fig. 2c, the recombination junction was located from nt 3357 to nt 3364.
FIG. 2.
(a) SimPlot analysis of full genome of LR31G7. E, echovirus; EV, enterovirus; SVDV, swine vesicular disease virus; CBV5, serotype 5 coxsackievirus group B. (b) SimPlot analysis of recombination junction sequences between strain PicoBankDM1E3 and strain NO-519. (c) Sequence of recombination junction of LR31G7. Identities between LR31G7 and PicoBankDM1E3 are highlighted in yellow, while identities between LR31G7 and NO-519 are highlighted in green.
DISCUSSION
In the present study, the full-genome sequence of an enterovirus isolate (isolate LR31G7) was analyzed. The strain was isolated from the sewage treatment plant of the city of Larissa, Thessaly, Greece, in October 2005. Primary characterization of the isolate by sequencing of part of the VP1 capsid region revealed that LR31G7 belongs to the Echo3 serotype. To our knowledge, no clinical Echo3 isolate identical to LR31G7 has been reported in Greece. Previous studies in Greece revealed that the most frequently isolated serotypes were Echo6, Echo13, and Echo30 in an outbreak of aseptic meningitis in 2001 (46) and Echo6 and Echo15, implicated in aseptic meningitis and encephalitis cases in 2005 to 2007 (15, 40).
Comparison of the nucleotide sequence of LR31G7 with the nucleotide sequences of prototype strain Morrisey and the only full sequenced Echo3 strain, strain PicoBankDM1E3, showed that the sequence of the Greek isolate differs from the sequences of strains Morrisey and PicoBankDM1E3 by 17% to 24% and 7% to 21%, respectively, in different genomic regions. The Greek strain is more closely related to isolate PicoBankDM1E3 than to the prototype Echo3 strain.
Phylogenetic comparison of part of the VP1 region of strain LR31G7 with the VP1 regions of other Echo3 isolates revealed that LR31G7 is related to Echo3 strains isolated in the same year (2005) in Asia and Europe, implying a temporal and not a topological correlation of these viruses. The close phylogenetic relationship of these viruses, along with the high degree of nucleotide sequence identity (95 to 98%) between them, leads to the conclusion that the same virus circulated in Greece, France, and Japan in 2005. At present, the rapid transmission of viruses from one region to another allows the viruses to circulate worldwide and not in the narrow topological boundaries of a specific population. From the information in the phylogenetic tree in Fig. 1, we can also hypothesize that isolate LR31G7 circulated in Europe for almost 7 years before it passed on to Asia. Nevertheless, it is difficult to come to a sound conclusion about the epidemiology of LR31G7, as Echo3 has been little studied and few Echo3 sequences are deposited in GenBank.
In the 5′ part of the genome, the degree of nucleotide sequence identity between strains LR31G7 and PicoBankDM1E3 was extremely high compared with that in the rest of the genome. This correlation between LR31G7 and PicoBankDM1E3 in the 5′ UTR-2A region was also confirmed by phylogenetic analysis (data not shown). In the phylogenetic trees for this part of the genome, the sequences of the two Echo3 strains clustered together, while in the phylogenetic trees for the capsid-coding region (data not shown), the sequences of the two strains clustered together and were in the same cluster as the sequence of prototype strain Morrisey, confirming once more that these strains belong to serotype 3 (4, 25, 31). LR31G7 and PicoBankDM1E3 are separate in the phylogenetic trees for 2B-3D. These data, along with the findings of SimPlot analysis (Fig. 2), strongly support the hypothesis that a recombination event occurred between circulating enteroviruses.
Analysis of the VP1-2A junction (Fig. 2b and c) revealed not only the donor but, for the first time in nonpoliovirus enteroviruses, the exact point of recombination (from nt 3357 to nt 3364). In Fig. 2b and c, the exchange of parts of the genomes between Echo3 strain PicoBankDM1E3 and Echo25 strain NO-519 is obvious. Keeping in mind that genetic exchanges between enteroviruses occur when there is coinfection of the same cell, recombination between these two viruses is unlikely to have occurred, as PicoBankDM1E3 and NO-519 were isolated in 1998 and 2003, respectively. Consequently, genetic exchanges have recently occurred between a descendant of PicoBankDM1E3 and NO-519. We can hypothesize that the Echo3 strains circulated in the same year worldwide (Fig. 1) or direct ancestors of them have recombined with the Echo25 strain. Moreover, the possibility that all strains of the 2005 cluster in Fig. 1 are recombinants cannot be excluded. The confirmation of our assumption will be possible only when the genome sequences of these strains become available.
The findings of phylogenetic analysis of all sequenced regions and in agreement with the findings of previous studies (4, 25, 28, 29), LR31G7 clusters with strains of the same serotype only in the capsid-coding genes. In phylogenetic trees for the proteins encoded by 2A and 3A, a correlation of LR31G7 with different enteroviruses was observed; but the phylogenetic trees for the 3B, 3C, and 3D regions revealed that LR31G7 consistently clusters with the CAV9 strains isolated in Cuba from 1990 to 2000, demonstrating their close relationship. The continuous clustering of the sequence of this part of the genome between LR31G7 and CAV9 strains led us to the hypothesis that a recombination event might had occurred in the evolutionary history of strain LR31G7.
In accordance with the findings of previous studies of the 2C and 3D genomic regions (5, 24), as well as the 3A and 3C regions, modern enterovirus isolates along with LR31G7 form a cluster that is separate from most HEV-B prototype strains. This observation reinforces the statement that the 2C and 3D regions, as well as, probably, the 3A and 3C regions, are conserved among modern strains. Moreover, in the phylogenetic tree of the 3D region, LR31G7 and the recently isolated viruses belong to the Echo1 cluster, whereas according to the findings of other studies (38, 30, 29), isolates that grouped together with the Echo1 prototype strain mostly originated in the 1980s.
Recombination in polioviruses has been extensively studied (12, 16, 18, 21, 27, 43). In the last few years, more attention has been shifted toward nonpoliovirus enteroviruses, since it has been proven that many prototype strains, as well as Echo18 and CAV21 strains (1, 20, 35, 36) are recombinants and there is more and more evidence for genetic exchange between circulating enteroviruses (30, 35). Even though much progress has been made in studies of genetic exchange between enteroviruses, recombination junctions have been detected only in recombinant Sabin poliovirus strains (13, 22).
In nonpoliovirus enteroviruses, because of the genomic diversity and the paucity of high degrees of nucleotide sequence identity between recent enteroviruses, it is very difficult to allocate the exact point of genetic exchange between circulating enteroviruses.
The present work, in which the recombination breakpoint between circulating enterovirus is presented for the first time, constitutes an additional step to the study and understanding of the evolution of enteroviruses.
Acknowledgments
The work was supported by a research grant from the postgraduate program Applications of Molecular Biology-Genetics, Diagnostic Biomarkers (grant 3817), of the Department of Biochemistry & Biotechnology, School of Health Sciences, University of Thessaly.
Footnotes
Published ahead of print on 3 February 2010.
REFERENCES
- 1.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Blomqvist, S., A. L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573-580. [DOI] [PubMed] [Google Scholar]
- 3.Blomqvist, S., A. Skytta, M. Roivainen, and T. Hovi. 1999. Rapid detection of human rhinoviruses in nasopharyngeal aspirates by a microwell reverse transcription-PCR-hybridization assay. J. Clin. Microbiol. 37:2813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolanaki, E., C. Kottaridi, P. Markoulatos, L. Margaritis, and T. Katsorchis. 2005. Nucleotide analysis and phylogenetic study of the homology boundaries of coxsackie A and B viruses. Virus Genes 31:307-320. [DOI] [PubMed] [Google Scholar]
- 5.Bolanaki, E., C. Kottaridi, P. Markoulatos, L. Margaritis, and T. Katsorchis. 2006. Evolution of 2B and 2C genomic parts of species B coxsackie viruses. Phylogenetic study and comparison with other regions. Virus Genes 32:249-259. [DOI] [PubMed] [Google Scholar]
- 6.Bolanaki, E., C. Kottaridi, P. Markoulatos, Z. Kyriakopoulou, L. Margaritis, and T. Katsorchis. 2007. Partial 3D gene sequences of coxsackie viruses reveal interspecies exchanges. Virus Genes 35:129-140. [DOI] [PubMed] [Google Scholar]
- 7.Brown, B., M. S. Oberste, K. Maher, and M. A. Pallansch. 2003. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J. Virol. 77:8973-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for “serotyping” of human enteroviruses. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 9.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 53:25-36. [DOI] [PubMed] [Google Scholar]
- 10.Chua, B. H., P. C. McMinn, S. K. Lam, and K. B. Chua. 2001. Comparison of the complete nucleotide sequences of echovirus 7 strain UMMC and the prototype (Wallace) strain demonstrates significant genetic drift over time. J. Gen. Virol. 82:2629-2639. [DOI] [PubMed] [Google Scholar]
- 11.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahourou, G., S. Guillot, O. Le Gall, and R. Crainic. 2002. Genetic recombination in wild-type poliovirus. J. Gen. Virol. 83:3103-3110. [DOI] [PubMed] [Google Scholar]
- 13.Dedepsidis, E., V. Pliaka, Z. Kyriakopoulou, C. Brakoulias, S. Levidiotou-Stefanou, A. Pratti, Z. Mamuris, and P. Markoulatos. 2008. Complete genomic characterization of an intertypic Sabin 3/Sabin 2 capsid recombinant. FEMS Immunol. Med. Microbiol. 52:343-351. [DOI] [PubMed] [Google Scholar]
- 14.Dedepsidis, E., Z. Kyriakopoulou, V. Pliaka, C. Kottaridi, E. Bolanaki, S. Levidiotou-Stefanou, D. Komiotis, and P. Markoulatos. 2007. Retrospective characterization of a vaccine-derived poliovirus type 1 isolate from sewage in Greece. Appl. Environ. Microbiol. 73:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frantzidou, F., K. Dumaidi, A. Spiliopoulou, A. Antoniadis, and A. Papa. 2007. Echovirus 15 and autumn meningitis outbreak among children, Patras, Greece, 2005. J. Clin. Virol. 40:77-79. [DOI] [PubMed] [Google Scholar]
- 16.Georgopoulou, A., and P. Markoulatos. 2001. Sabin type 2 polioviruses with intertypic vaccine/vaccine recombinant genomes. Eur. J. Clin. Microbiol. Infect. Dis. 20:792-799. [DOI] [PubMed] [Google Scholar]
- 17.Georgopoulou, A., P. Markoulatos, N. Spyrou, and N. C. Vamvakopoulos. 2000. Improved genotyping vaccine and wild-type poliovirus strains by restriction fragment length polymorphism analysis: clinical diagnostic implications. J. Clin. Microbiol. 38:4337-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland, J., and E. Domingo. 1998. Origin and evolution of viruses. Virus Genes 16:13-21. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, P. J., C. North, P. D. Minor, and G. Stanway. 1989. The complete nucleotide sequence of coxsackievirus A21. J. Gen. Virol. 70:2943-2952. [DOI] [PubMed] [Google Scholar]
- 21.Karakasiliotis, I., E. Paximadi, and P. Markoulatos. 2005. Evolution of a rare vaccine-derived multirecombinant poliovirus. J. Gen. Virol. 86:3137-3142. [DOI] [PubMed] [Google Scholar]
- 22.Karakasiliotis, I., P. Markoulatos, and T. Katsorchis. 2004. Site analysis of recombinant and mutant poliovirus isolates of Sabin origin from patients and from vaccinees. Mol. Cell. Probes 18:103-109. [DOI] [PubMed] [Google Scholar]
- 23.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kottaridi, C., E. Bolanaki, Z. Kyriakopoulou, E. Dedepsidis, A. Pratti, and P. Markoulatos. 2007. Possible recombination and gene adaptation exchanges among clinical echovirus strains: crossing the temporal and topological barriers. Diagn. Microbiol. Infect. Dis. 58:407-412. [DOI] [PubMed] [Google Scholar]
- 25.Kottaridi, C., E. Bolanaki, Z. Mamuris, C. Stathopoulos, and P. Markoulatos. 2006. Molecular phylogeny of VP1, 2A, and 2B genes of echovirus isolates: epidemiological linkage and observations on genetic variation. Arch. Virol. 151:1117-1132. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 27.Kyriakopoulou, Z., C. Kottaridi, E. Dedepsidis, E. Bolanaki, S. Levidiotou-Stefanou, and P. Markoulatos. 2006. Molecular characterization of wild-type polioviruses isolated in Greece during the 1996 outbreak in Albania. J. Clin. Microbiol. 44:1150-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 29.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2005. Recombination in circulating human enterovirus B: independent evolution of structural and non-structural genome regions. J. Gen. Virol. 86:3281-3290. [DOI] [PubMed] [Google Scholar]
- 30.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2003. Recombination in circulating enteroviruses. J. Virol. 77:10423-10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirand, A., C. Archimbaud, C. Henquell, Y. Michel, M. Chambon, H. Peigue-Lafeuille, and J. L. Bailly. 2006. Prospective identification of HEV-B enteroviruses during the 2005 outbreak. J. Med. Virol. 78:1624-1634. [DOI] [PubMed] [Google Scholar]
- 32.Mulders, M. N., J. H. Reimerink, M. Stenvik, I. Alaeddinoglu, H. G. van der Avoort, T. Hovi, and M. P. Koopmans. 1999. A Sabin vaccine-derived field isolate of poliovirus type 1 displaying aberrant phenotypic and genetic features, including a deletion in antigenic site 1. J. Gen. Virol. 80:907-916. [DOI] [PubMed] [Google Scholar]
- 33.Oberste, M. S., K. Maher, and M. A. Pallansch. 2004. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 78:855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberste, M. S., S. Peñaranda, and M. A. Pallansch. 2004. RNA recombination plays a major role in genomic change during circulation of coxsackie B viruses. J. Virol. 78:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberste, M. S., S. Peñaranda, K. Maher, and M. A. Pallansch. 2004. Complete genome sequences of all members of the species human enterovirus A. J. Gen. Virol. 85:1597-1607. [DOI] [PubMed] [Google Scholar]
- 37.Oberste, M. S., W. A. Nix, K. Maher, and M. A. Pallansch. 2003. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 26:375-377. [DOI] [PubMed] [Google Scholar]
- 38.Oprisan, G., M. Combiescu, S. Guillot, V. Caro, A. Combiescu, F. Delpeyroux, and R. Crainic. 2002. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 83:2193-2200. [DOI] [PubMed] [Google Scholar]
- 39.Palacios, G., and M. S. Oberste. 2005. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11:424-433. [DOI] [PubMed] [Google Scholar]
- 40.Papa, A., L. Skoura, K. Dumaidi, A. Spiliopoulou, A. Antoniadis, and F. Frantzidou. 2009. Molecular epidemiology of echovirus 6 in Greece. Eur. J. Clin. Microbiol. Infect. Dis. 28:683-687. [DOI] [PubMed] [Google Scholar]
- 41.Paximadi, E., I. Karakasiliotis, D. Papaventsis, G. Papageorgiou, and P. Markoulatos. 2008. Recombinant Sabin environmental isolates in Greece and Cyprus. J. Appl. Microbiol. 104:1153-1162. [DOI] [PubMed] [Google Scholar]
- 42.Paximadi, E., I. Karakasiliotis, E. Bolanaki, A. Krikelis, and P. Markoulatos. 2007. Vaccine derived bi- and multi-recombinant Sabin strains. Virus Genes 35:541-548. [DOI] [PubMed] [Google Scholar]
- 43.Paximadi, E., I. Karakasiliotis, Z. Mamuris, C. Stathopoulos, V. Krikelis, and P. Markoulatos. 2006. Genomic analysis of recombinant Sabin clinical isolates. Virus Genes 32:203-210. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz-Jarabo, C. M., A. Arias, E. Baranowski, C. Escarmís, and E. Domingo. 2000. Memory in viral quasispecies. J. Virol. 74:3543-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santti, J., T. Hyypiä, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siafakas, N., P. Markoulatos, and S. Levidiotou-Stefanou. 2004. Molecular identification of enteroviruses responsible for an outbreak of aseptic meningitis; implications in clinical practice and epidemiology. Mol. Cell. Probes 18:389-398. [DOI] [PubMed] [Google Scholar]
- 47.Williams, C. H., S. Oikarinen, S. Tauriainen, K. Salminen, H. Hyöty, and G. J. Stanway. 2006. Molecular analysis of an echovirus 3 strain isolated from an individual concurrently with appearance of islet cell and IA-2 autoantibodies. Clin. Microbiol. 44:441-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.World Health Organization. 3 March 2003, posting date. Guidelines for environmental surveillance of poliovirus circulation. World Health Organization, Geneva, Switzerland. http://www.who.int/vaccines-documents/DocsPDF03/www737pdf.