Abstract
A single dose of nevirapine (sdNVP) to prevent mother-to-child transmission of HIV-1 increases the risk of failure of subsequent NVP-containing antiretroviral therapy (ART), especially when initiated within 6 months of sdNVP administration, emphasizing the importance of understanding the decay of nevirapine-resistant mutants. Nevirapine-resistant HIV-1 genotypes (with the mutations K103N, Y181C, and/or G190A) from 21 women were evaluated 10 days and 6 weeks after sdNVP administration and at the initiation of ART. Resistance was assayed by consensus sequencing and by a more sensitive assay (oligonucleotide ligation assay [OLA]) using plasma-derived HIV-1 RNA and cell-associated HIV-1 DNA. OLA detected nevirapine resistance in more specimens than consensus sequencing did (63% versus 33%, P < 0.01). When resistance was detected only by OLA (n = 45), the median mutant concentration was 18%, compared to 61% when detected by both sequencing and OLA (n = 51) (P < 0.0001). The proportion of women whose nevirapine resistance was detected by OLA 10 days after sdNVP administration was higher when we tested their HIV-1 RNA (95%) than when we tested their HIV-1 DNA (88%), whereas at 6 weeks after sdNVP therapy, the proportion was greater with DNA (85%) than with RNA (67%) and remained higher with DNA (33%) than with RNA (11%) at the initiation of antiretroviral treatment (median, 45 weeks after sdNVP therapy). Fourteen women started NVP-ART more than 6 months after sdNVP therapy; resistance was detected by OLA in 14% of the women but only in their DNA. HIV-1 resistance to NVP following sdNVP therapy persists longer in cellular DNA than in plasma RNA, as determined by a sensitive assay using sufficient copies of virus, suggesting that DNA may be superior to RNA for detecting resistance at the initiation of ART.
A single dose of nevirapine (sdNVP) reduces mother-to-child transmission (MTCT) of HIV-1 by 47% to 58% (12, 17) but is associated with the selection of mutant viruses resistant to nonnucleoside reverse transcriptase inhibitors (NNRTI) (7, 15) and may jeopardize the effectiveness of subsequent nevirapine (NVP)-based antiretroviral treatment (ART) (5, 15, 21). However, the adverse impact of sdNVP therapy on NVP-ART efficacy seems to decrease with longer time intervals between the administration of sdNVP and the initiation of NVP-ART (ART start) (21, 29).
In 21 to 32% of sdNVP-treated mothers, consensus sequencing of HIV-1 RNA detects NNRTI mutations, which decay to undetectable levels over 1 year (7, 8, 15). More sensitive assays detect NNRTI resistance mutations in 51 to 87% of postpartum sdNVP-exposed mothers (9, 22), and these mutations remain detectable in 8 to 33% of women after a year (5, 8, 10). Comparative analyses of the decay of NVP resistance in HIV-1 DNA and RNA in blood using sensitive methods found a lower proportion of women with resistance in their HIV-1 DNA (5, 22), although the input of HIV-1 DNA was not directly determined in these studies, precluding estimations of the sensitivity limits of the assays.
We investigated the presence of three mutations (K103N, G190A, and Y181C) in HIV-1 commonly associated with sdNVP therapy that confer high-level resistance to NNRTI (14). In a cohort of women identified as having one of these NNRTI mutations, the HIV-1 RNA and DNA were examined both by consensus sequencing and by a more sensitive method, oligonucleotide ligation assay (OLA). The prevalence and concentration of these mutations were determined at three time points: 10 days and 6 weeks after sdNVP therapy and immediately before the initiation of NVP-ART. Investigating the ability to detect NVP resistance over time in HIV-1 RNA and DNA should help in the understanding of persistent HIV drug resistance and guide specimen selection for HIV-1 drug resistance testing. Such knowledge might help avoid the use of ineffective ART regimens and further selection of HIV-1 drug resistance and should help lay the groundwork for studies that seek to determine if there are specific concentrations of NVP-resistant mutants that are predictive of the virologic outcome if subsequent ART includes NNRTI.
MATERIALS AND METHODS
Study design, study population, and inclusion criteria.
Two hundred two women participating in an internal review board-approved clinical trial who received sdNVP therapy during labor plus zidovudine (ZDV) during the last trimester of pregnancy (17) and subsequently began NVP-ART within 18 months (15) were screened for this study. They were selected if longitudinal peripheral blood specimens were collected (10 days and 6 weeks after sdNVP therapy and at ART start) and NVP resistance was detected either by consensus sequencing of HIV-1 RNA collected 10 days postpartum (15) or by OLA of cellular HIV-1 DNA at 6 weeks postpartum. Subsequently, specimens at the three time points were evaluated using consensus sequencing and a sensitive point mutation assay (OLA). Laboratory work was performed at the Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai, Thailand, and interpreted by personnel who were blind to the clinical data.
Nucleic acid extraction.
RNA was extracted from plasma collected 10 days postpartum and sequenced using ViroSeq HIV-1 Genotyping System version 2.0 (Applied Biosystems, Foster City, CA), as previously reported (15). RNA was extracted from the 6-week-postpartum and ART start plasma specimens using the Amplicor HCV Prep test (Roche Molecular Systems, Inc., Branchburg, NJ). Cellular DNA was extracted from plasma-depleted peripheral blood cells (PBC) (1 to 3 ml) using QiAmp DNA Midi kits (Qiagen, Inc., Valencia, CA).
HIV-1 RNA and DNA quantification, amplification, and sequencing.
Plasma HIV-1 viral load was determined as previously described (15). When the 6-week and ART start HIV-1 RNA had not been previously sequenced, >100 viral copies (estimates based on plasma viral load) were reverse transcribed to cDNA using the GeneAmp RNA PCR core kit (Applied Biosystems). The HIV-1 DNA in 1 μg of PBC-derived DNA was quantified using real-time PCR of the HIV-1 long terminal repeat (LTR) (1) in an ABI7000 system (Applied Biosystems). HIV-1 pol was amplified by nested PCR with first-round primers (PRA and IBR1) and second-round primers (PRB and IBR2) (3). A maximum of 2.5 μg of DNA was submitted to each first-round 100-μl PCR mixtures, and 2 μl of the first-round product was used for nested second-round PCR. Up to six first-round amplifications were performed to amplify a total of 100 to 150 copies of HIV-1 DNA. When multiple first-round amplifications were performed, 2 μl of each first-round product was combined in a single second-round PCR. Amplicons of appropriate size were confirmed by visualization on a 1% agarose gel. Bidirectional sequences were generated from day 10 DNA and week 6 RNA and DNA specimens, as described previously (http://www.hivfrenchresistance.org/ANRS-procedures.pdf), using forward primer NEI35 (19) or 5seq-RT (5′-AAACAATGGCCATTGACAGAAGA-3′) and reverse primer IBR-2 (3).
Oligonucleotide ligation assay.
OLAs utilize the specificity of ligase to join allele-specific probes to distinguish single nucleotide polymorphisms. OLA has been adapted to detect single nucleotide mutations associated with HIV-1 drug resistance (11) and, with modifications, has been shown to be able to detect as little as 0.4% mutant virus (18). HIV-1 drug resistance genotypes were determined using the OLA as described previously (3) with slight variations for subtype and to optimize the optical density (OD) signal. Codon K103 reaction mixtures contained 2 μl of amplicon diluted 1:5 in nuclease-free water, 0.167 U of DNA ligase, and 0.333 pmol of each oligonucleotide. Codon Y181 reaction mixtures contained 2 μl of amplicon diluted 1:5 in nuclease-free water, 0.167 U of DNA ligase, 0.333 pmol of the wild-type oligonucleotide, and 1.333 pmol of the mutant and common oligonucleotides. Codon G190 reaction mixtures contained 2 μl of undiluted amplicon, 0.167 U of DNA ligase, 0.333 pmol of the wild-type oligonucleotide, and 0.667 pmol of the mutant and common oligonucleotides. Cloned subtype CRF01_AE viruses with K103N, Y181C, or G190A were used to generate 0%-, 2%-, 5%-, and 100%-mutant standards in wild-type backgrounds (3) and were analyzed on each plate along with the study participants' specimens.
OLA quality control and interpretation.
All specimens and controls were analyzed in duplicate. The mean optical density at 490 nm (OD490) was compared to the mean OD490s of the standards tested on the same plate. Specimens were considered valid if the ODs of duplicates differed by less than 25%. Specimens were interpreted as positive for the mutant codon if the mean OD490 of the specimen was equal to or greater than the mean OD490 of the 5%-mutant standard. Specimens with a mean OD490 below the 5% standard and with a wild-type OD490 of >0.6 were interpreted as having the wild-type codon.
Data analysis.
The percentage of mutant virus present in each specimen was estimated using a logarithmic trend line fitted to the ODs for 0-, 2-, 5-, and 100%-mutant standards on each plate (Excel, Microsoft, Redmond, WA), with the positive cutoff predetermined to be ≥5% mutant. Generalized estimating equation (GEE) logistic modeling was used to calculate whether the differences in the percentages of women with resistance detected by the four assay methods at ART start were statistically significant. GEE modeling adjusted the statistical comparisons to account for any dependence that may have existed among the set of four observations generated from the same blood sample using OLA and sequencing of the plasma and cellular fractions of the sample. Fisher's exact test was used to compare proportions, and a Mann-Whitney U test was used to compare distributions.
RESULTS
Study population.
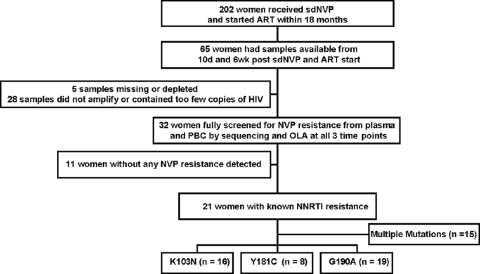
Of the 202 previously reported women who took sdNVP and started NVP-ART within the subsequent 18 months (15), 65 had had peripheral blood collected 10 days and 6 weeks after sdNVP therapy and at the initiation of NVP-ART (ART start). Of these women, 5 had plasma and PBC specimens from various time points that were missing or depleted due to previous studies, and 28 had a PBC specimen that had <100 total copies of HIV-1 DNA. The remaining 32 women were screened for NVP resistance by reviewing consensus sequences of their HIV-1 RNA from 10 days postpartum and by OLA of their HIV-1 DNA from 6 weeks postpartum; NVP resistance was not detected in 34% (11 of 32) of the women. From the 21 women with resistance, longitudinal specimens were studied to compare the dynamics of their NVP-resistant HIV-1 RNA and DNA by consensus sequencing and OLA (Fig. 1). These women were a median of 28 years old (interquartile range [IQR], 25 to 32 years) and had a median CD4 cell count of 178 cells/ml (IQR, 141 to 212 cells/ml) and a plasma HIV-1 RNA viral load of 4.70 log10 copies/ml (IQR, 4.16 to 4.94 log10 copies/ml) during pregnancy and started NVP-ART 11.6 months after sdNVP therapy (IQR, 4.72 to15.1 months). A total of 112 OLA and 115 sequencing results from these 21 women were studied. All women were determined to be infected with HIV-1 CRF01_AE, except one who was infected with a probable AE/B recombinant.
FIG. 1.
Schema for selection of the sdNVP-treated women with NVP resistance.
Decay of NVP resistance.
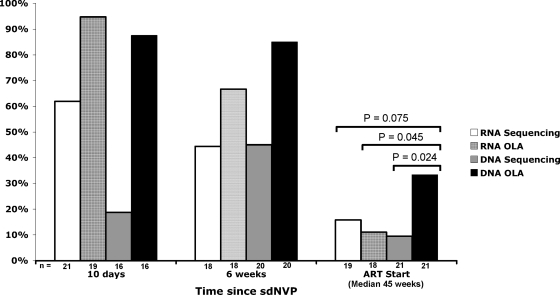
Twenty-one women were selected for this study because NVP resistance was detected in at least one of their specimens. The mutations (K103N, Y181C, and G190A) detected by consensus sequencing and OLA of HIV-1 RNA and DNA from each subject's available specimens are shown in Fig. 2. In specimens from 10 days following sdNVP therapy, HIV-1 nevirapine resistance mutations were most frequently detected in the OLA of RNA (95% of women versus 19 to 88% by the other three methods), whereas at 6 weeks following sdNVP therapy, mutations were most frequently detected by OLA of DNA (85% of women versus 44 to 66% by the other three methods). At the final time point immediately prior to the start of NVP-ART, which was a median of 45 weeks after sdNVP therapy (range, 17 to 72 weeks), nevirapine resistance was detected in 33% of women (7/21) (the highest proportion) by OLA of DNA versus 16% (3/19) by consensus sequencing of RNA (P = 0.075), 11% (2/18) by OLA of RNA (P = 0.045), and 10% (2/21) by consensus sequencing of DNA (P = 0.024) (Fig. 3). Of the seven women with mutations detected in the ART start specimen by any method, all had mutant HIV-1 detected by OLA of DNA, four had mutants that were also detected by OLA of RNA, four had mutants that were detected by consensus sequencing of DNA, and three had mutants that were detected by RNA sequencing. More than 6 months after sdNVP therapy, mutations were still detectable in two women but only by OLA of DNA, although RNA samples from two other participants were unavailable at ART start (Fig. 2).
FIG. 2.
NVP resistance mutations detected in HIV-1 RNA and DNA from each woman by consensus sequencing (seq) and oligonucleotide ligation assay (OLA). Participants are listed by increasing time interval between sdNVP therapy and initiation of NVP-ART (ART start). Data for each specimen (RNA and DNA) at 10 days (10d) and 6 weeks following sdNVP therapy and just prior to ART start are shown for each woman, with the symbols in the key indicating the resistant codons detected. n/a, not available.
FIG. 3.
Percentages of women with NVP resistance (K103N, Y181C, or G190A) detected in their HIV-1 RNA or DNA by consensus sequencing and OLA.
Comparison of NVP resistance by OLA versus sequencing.
Totaling all RNA and DNA specimens analyzed, OLA detected nevirapine resistance in almost twice as many specimens as consensus sequencing did (63% of 115 versus 33% of 112 specimens, P < 0.01). All G190A mutations detected by sequencing were also detected by OLA. In addition, G190A was detected by OLA in 37 specimens that appeared to be wild type by consensus sequencing. K103N mutations detected by sequencing in two day 10 specimens were wild type by OLA, although OLA detected K103N in later specimens from these participants. K103N was detected by OLA in 14 specimens that were wild type by sequencing. Y181C was detected in three specimens by consensus sequencing that were wild type by OLA; in two of these cases, Y181C was detected by OLA at a level estimated to be between 2 and 5%, which was below the predetermined positive cutoff of ≥5% mutant. Y181C was detected by OLA in 5 specimens that were wild type by sequencing.
Comparison of drug resistance in plasma-derived RNA versus PBC-derived DNA.
The frequency of NVP resistance was greater in RNA than in DNA at the initial assessment 10 days following sdNVP therapy but persisted longer in DNA (Fig. 3). The most noticeable difference between RNA and DNA was that the percentage of women with NVP resistance detected in their HIV-1 RNA decreased (95 to 67% by OLA and 62 to 44% by consensus sequencing) but was stable or increased in DNA (88 to 85% by OLA and 19 to 45% by consensus sequencing) between day 10 and 6 weeks after therapy (Fig. 2). Beyond 6 weeks, the percentage of woman with resistance detected in both their HIV-1 RNA and DNA decreased, although at ART start, more women had resistance detectable in DNA than in RNA (7 versus 3, respectively), and no resistance mutations were detected in RNA that were not also detected in DNA.
Dynamics of single and multiple mutant codons.
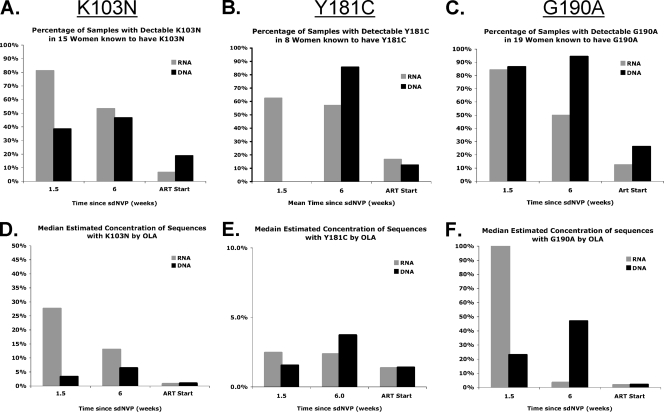
G190A was the most frequent mutation in our study population, detected in 90% (19 of 21) of women by sequencing and/or OLA; K103N was detected in 71% (15 of 21) of women, and Y181C was the least common, found in 38% (8 of 21) of women. Subtle differences were observed in the decay between the three codons, which are shown in Fig. 4. When OLA and sequencing results are combined, 76% (16 of 21) of the participants had more than one resistance mutation detected over the three time points. Among the specimens with resistance, more than one mutation was found in 60% (12 of 20) of the day 10 RNA specimens, 29% (4 of 14) of the day 10 DNA specimens, 58% (7 of 12) of the week 6 RNA specimens, 59% (10 of 17) of the week 6 DNA specimens, 33% (1 of 3) of the ART start RNA specimens, and 14% (1 of 7) of the ART start DNA specimens. Thirty-three percent (7 of 21) of participants had all three mutations detected, and in three cases, all three resistance codons were detected in the same specimen.
FIG. 4.
The dynamics of NVP-resistant-HIV-1 selection and decay are compared in the HIV-1 RNA and DNA of subjects known to harbor mutations at each codon. The prevalence of samples with detectable resistance (A to C) and the concentrations (D to F) of NVP-resistant variants as determined by oligonucleotide ligation assay (OLA) at each point in time are shown by codon. For all codons, the prevalence of resistance and the estimated concentrations of resistant viruses peak sooner in RNA but decay more slowly in DNA.
Estimating the concentration of NVP resistance mutations.
The concentration of mutant viruses over time was estimated using OLA by comparing the ODs of samples to a standard curve. The median concentrations of the mutants with individual resistance codons are shown in Fig. 4. Maximum mutant concentrations were observed in day 10 RNA sequences, except for the Y181C mutant concentration, which peaked in the week 6 DNA specimen. For all three codons, the concentration of mutant virus decreased in RNA but increased in DNA between day 10 and 6 weeks after sdNVP therapy. The estimated concentration of mutant virus by OLA was associated with the likelihood of resistance being detected by consensus sequencing. The median mutant concentration in specimens with resistance detected by OLA but not by sequencing (n = 45) was 18%. In contrast, when resistance was detected by both sequencing and OLA (n = 51), the median concentration of mutant virus by OLA was 61% (P < 0.0001). By ART start, the median concentration of mutant virus in RNA and DNA for all women was quite low (1 and 2%, respectively). In total, there were five specimens from four women that appeared to contain greater than 50% mutant virus by OLA, and yet resistance was not detected by sequencing. The raw sequencing chromatograms from these specimens were reviewed, and four of the five had subtle minor peaks corresponding to the resistance codon detected by OLA.
DISCUSSION
In this study, the kinetics of HIV-1 resistance following sdNVP therapy differed in RNA and DNA. The most important observation was that at ART start, testing DNA by OLA detected NNRTI resistance in the greatest percentage of women (33% versus 10 to 16% using either OLA of RNA or sequencing of DNA or RNA). These results are consistent with viral-dynamics models (24) and predicted by experimental data (25), with a rapid turnover of plasma HIV-1 RNA compared to a relatively slower turnover of most infected cells and a very slow decay of a small subset of cells with HIV-1 DNA (4), but contradict the only previous longitudinal study of NVP resistance in DNA following sdNVP therapy, which suggested that resistance decays more quickly in DNA than in RNA (22). Our study differs because we quantified the input of HIV-1 DNA copies assayed in order to ensure our ability to detect resistant mutants present in as little as 5% of the viral population. This required assaying up to 15 μg of DNA, which is a substantially greater input than that of other published protocols (22) and may explain why we detected more resistance in DNA.
Consistent with other studies (7, 8, 9, 10, 22), we observed a gradual decay of NVP resistance in the peripheral blood. Our study design did not allow us to assess the rate of decay, but resistance was detected more than 6 months following sdNVP therapy. Specifically, 2 of 14 women starting NVP-ART more than 6 months following sdNVP therapy had NVP resistance detected in DNA. These women had mutations detected at 13 and 15 months following sdNVP therapy, suggesting that mutants may not decay to clinically insignificant levels by 6 months postpartum (21) but, in some women, will persist longer (20). Furthermore, our study confirms the potential increased sensitivity of point mutation assays, with OLA able to detect resistance about twice as often as sequencing (6). Consensus sequencing is less sensitive in the detection of resistant variants present at less than a 25% to 50% concentration (27), whereas OLA in this study was designed to detect mutants when present at a concentration as low as 5% of the virus population, suggesting that perhaps half of the women with resistance detected by OLA had drug resistance concentrations of between 5% and 25%, which is similar to others' observations (28). Additional resistant variants were likely present at concentrations less than 5% of the population in additional specimens, and recent clinical studies suggest that even very low concentrations of virus with NVP resistance can be clinically significant (5, 13, 16, 23, 28).
In this study, OLA identified G190A as the most common mutation, which contrasts with previously published studies of patients from Africa (7) and consensus sequencing of our entire Thai cohort (15), in whom K103N was the most common mutation selected. G190A has been noted to be more common among subtype CRF01_AE viruses than among subtype B viruses (see http://hivdb.stanford.edu/cgi-bin/MutPrevBySubtypeRx.cgi) and may be more likely selected by MTCT regimens which contain zidovudine, consistent with past observations that zidovudine alters the selection of specific NVP resistance codons (2, 26). However, even in this study, K103N was more frequent than G190A (48% versus 33%) when the day 10 postpartum specimens were analyzed by consensus sequencing, suggesting that viral variants with G190A may be present at lower levels and/or persist longer once selected. A high percentage of women in this study had multiple resistance mutations: 71% (15 of 21) of women had more than one resistance mutation detected. Looking for multiple mutations increased the number of women with detectable mutants (n = 21) by 31% compared with what would have been detected (n = 16) had we assayed for K103N only. This supports the concept that studies of NVP resistance following sdNVP therapy that focus exclusively on K103N substantially underestimate resistance. Our observations suggest that further research is needed to determine which mutant codons are most informative to monitor after sdNVP therapy, which may differ if the patient is given a short course of ZDV or an NNRTI “tail.”
One important caveat of this study is that the participants are not representative of women exposed to sdNVP therapy. In order to more efficiently study the decay of NVP resistance, women were selected for this study because they were known to have NVP-resistant HIV-1. These women received a short course of ZDV beginning at 28 weeks of gestation in addition to sdNVP therapy, and all except one had infection with subtype CRF01_AE. In addition, this analysis was restricted to 49% (32 of 65) of the women in the Program for HIV Prevention and Treatment 2 study who started NVP within 18 months and had study specimens collected at 10 days and 6 weeks postpartum and at ART start. Furthermore, some women were excluded because of specimens with too little HIV-1 DNA, which suggests that we may have sampled women with relatively higher DNA viral loads in their PBC. In addition, our methodology did not allow us to differentiate between integrated proviral DNA and unintegrated cDNA; this distinction could influence the clinical impact of the mutations and warrants further study. Finally, some missing samples likely limited the power of the study, and the study is too small to accurately model the decay of each of the resistance codons.
The dynamics of selection and persistence of NVP resistance differed between RNA and DNA following sdNVP therapy. While mutants were detected more readily in RNA than in DNA within days of sdNVP therapy, the mutants remained detectable longer in DNA. Furthermore, utilization of a sensitive point mutation assay to screen for three resistance mutations markedly increased the detection of persistent drug resistance mutations compared to that of consensus sequencing, especially in DNA samples. This study suggests that when sufficient HIV-1 DNA is examined, testing of cell-associated HIV-1 DNA may be useful in assessing persistent drug resistance mutations.
Acknowledgments
We are grateful for the support of the Program for HIV Prevention and Treatment (PHPT) Study Group, as well as the Office of Permanent Secretary, Department of Health, Communicable Diseases Control, Food and Drug Administration, Health Sciences, and Provincial Hospitals Division, Thai Ministry of Public Health. This work was supported by grants from the National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health (grants 5 T32 HD07233 [T.A.W.], 5 K23 AI077357 [T.A.W.], 5 R01 AI058723 [L.M.F.], and 1 U01 AI068632 [IMPAACT Development Virology Laboratory] [L.M.F.]), the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (grant 5 R01 HD39615 [M.L., PHPT-2 study]), the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) (grant 12-08 [M.L., PHPT-2 study]), the Global Fund to fight AIDS, Tuberculosis and Malaria, Thailand (grant PR-A-N-008), and the Institut de Recherche pour le Développement (IRD), Marseilles, France.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Arvold, N. D., N. Ngo-Giang-Huong, K. McIntosh, V. Suraseranivong, B. Warachit, S. Piyaworawong, T. Changchit, M. Lallemant, and G. Jourdain. 2007. Maternal HIV-1 DNA load and mother-to-child transmission. AIDS Patient Care STDS 21:638-643. [DOI] [PubMed] [Google Scholar]
- 2.Balzarini, J., A. Karlsson, M.-J. Pérez-Pérez, M. J. Camarasa, W. G. Tarpley, and E. De Clercq. 1993. Treatment of human immunodeficiency virus type 1 (HIV-1)-infected cells with combinations of HIV-1-specific inhibitors results in a different resistance pattern than does treatment with single-drug therapy. J. Virol. 67:5353-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck, I. A., C. Crowell, R. Kittoe, H. Bredell, M. Machaba, C. Willamson, W. Janssens, S. Jallow, G. van der Groen, Y. Shao, M. Jacob, N. M. Samuel, I. L. de Rivera, N. Ngo-Giang-Huong, S. Cassol, G. Alemnji, and L. M. Frenkel. 2008. Optimization of the oligonucleotide ligation assay, a rapid and inexpensive test for detection of HIV-1 drug resistance mutations, for non-North American variants. J. Acquir. Immune Defic. Syndr. 48:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi, X., H. Gatanaga, S. Ida, K. Tsuchiya, S. Matsuoka-Aizawa, S. Kimura, and S. Oka. 2003. Emergence of protease inhibitor resistance-associated mutations in plasma HIV-1 precedes that in proviruses of peripheral blood mononuclear cells by more than a year. J. Acquir. Immune Defic. Syndr. 34:1-6. [DOI] [PubMed] [Google Scholar]
- 5.Coovadia, A., G. Hunt, E. J. Abrams, G. Sherman, T. Meyers, G. Barry, E. Malan, B. Marais, R. Stehlau, J. Ledwaba, S. M. Hammer, L. Morris, and L. Kuhn. 2009. Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clin. Infect. Dis. 48:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis, G. M., M. Mahalanabis, I. A. Beck, G. Pepper, A. Wright, S. Hamilton, S. Holte, W. E. Naugler, D. M. Pawluk, C. Li, and L. M. Frenkel. 2004. Comparison of oligonucleotide ligation assay and consensus sequencing for detection of drug-resistant mutants of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and plasma. J. Clin. Microbiol. 42:3670-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eshleman, S. H., L. A. Guay, A. Mwatha, S. P. Cunningham, E. R. Brown, P. Musoke, F. Mmiro, and J. B. Jackson. 2004. Comparison of nevirapine (NVP) resistance in Ugandan women 7 days versus 6-8 weeks after single-dose NVP prophylaxis: HIVNET 012. AIDS Res. Hum. Retroviruses 20:595-599. [DOI] [PubMed] [Google Scholar]
- 8.Flys, T., D. V. Nissley, C. W. Claasen, D. Jones, C. Shi, L. A. Guay, P. Musoke, F. Mmiro, J. N. Strathern, J. B. Jackson, J. R. Eshleman, and S. H. Eshleman. 2005. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J. Infect. Dis. 192:24-29. [DOI] [PubMed] [Google Scholar]
- 9.Flys, T. S., S. Chen, D. C. Jones, D. R. Hoover, J. D. Church, S. A. Fiscus, A. Mwatha, L. A. Guay, F. Mmiro, P. Musoke, N. Kumwenda, T. E. Taha, J. B. Jackson, and S. H. Eshleman. 2006. Quantitative analysis of HIV-1 variants with the K103N resistance mutation after single-dose nevirapine in women with HIV-1 subtypes A, C, and D. J. Acquir. Immune Defic. Syndr. 42:610-613. [DOI] [PubMed] [Google Scholar]
- 10.Flys, T. S., D. Donnell, A. Mwatha, C. Nakabiito, P. Musoke, F. Mmiro, J. B. Jackson, L. A. Guay, and S. H. Eshleman. 2007. Persistence of K103N-containing HIV-1 variants after single-dose nevirapine for prevention of HIV-1 mother-to-child transmission. J. Infect. Dis. 195:711-715. [DOI] [PubMed] [Google Scholar]
- 11.Frenkel, L. M., L. E. Wagner, S. M. Atwood, T. J. Cummins, and S. Dewhurst. 1995. Specific, sensitive, and rapid assay for human immunodeficiency virus type 1 pol mutations associated with resistance to zidovudine and didanosine. J. Clin. Microbiol. 33:342-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson, J. B., P. Musoke, T. Fleming, L. A. Guay, D. Bagenda, M. Allen, C. Nakabiito, J. Sherman, P. Bakaki, M. Owor, C. Ducar, M. Deseyve, A. Mwatha, L. Emel, C. Duefield, M. Mirochnick, M. G. Fowler, L. Mofenson, P. Miotti, M. Gigliotti, D. Bray, and F. Mmiro. 2003. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18-month follow-up of the HIVNET 012 randomised trial. Lancet 362:859-868. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, J. A., J. Li, X. Wei, J. Lipscomb, D. Irlbeck, C. Craig, A. Smith, D. E. Bennett, M. Monsour, P. Sandstrom, E. R. Lanier, and W. Heneine. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy. PLoS Med. 5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2008. Update of the drug resistance mutations in HIV-1. Top. HIV Med. 16:138-145. [PubMed] [Google Scholar]
- 15.Jourdain, G., N. Ngo-Giang-Huong, S. Le Coeur, C. Bowonwatanuwong, P. Kantipong, P. Leechanachai, S. Ariyadej, P. Leenasirimakul, S. Hammer, and M. Lallemant. 2004. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N. Engl. J. Med. 351:229-240. [DOI] [PubMed] [Google Scholar]
- 16.Kuritzkes, D. R., C. M. Lalama, H. J. Ribaudo, M. Marcial, W. A. Meyer, C. Shikuma, V. A. Johnson, S. A. Fiscus, R. T. D'Aquila, B. R. Schackman, E. P. Acosta, and R. M. Gulick. 2008. Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. J. Infect. Dis. 197:867-870. [DOI] [PubMed] [Google Scholar]
- 17.Lallemant, M., G. Jourdain, S. Le Coeur, J. Y. Mary, N. Ngo-Giang-Huong, S. Koetsawang, S. Kanshana, K. McIntosh, and V. Thaineua. 2004. Single-dose perinatal nevirapine plus standard zidovudine to prevent mother-to-child transmission of HIV-1 in Thailand. N. Engl. J. Med. 351:217-228. [DOI] [PubMed] [Google Scholar]
- 18.Lalonde, M. S., R. M. Troyer, A. R. Syed, S. Bulime, K. Demers, F. Bajunirwe, and E. J. Arts. 2007. Sensitive oligonucleotide ligation assay for low-level detection of nevirapine resistance mutations in human immunodeficiency virus type 1 quasispecies. J. Clin. Microbiol. 45:2604-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larder, B. A., P. Kellam, and S. D. Kemp. 1991. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS 5:137-144. [DOI] [PubMed] [Google Scholar]
- 20.Lockman, S., and A5208/OCTANE Study Team. 2009. Lopinavir/ritonavir+tenofovir/emtricitabine is superior to nevirapine+tenofovir/emtricitabine for women with prior exposure to single-dose nevirapine: A5208 (“OCTANE”), abstr. 94LB. 16th Conf. Retrovir. Oppor. Infect., Montreal, Canada.
- 21.Lockman, S., R. L. Shapiro, L. M. Smeaton, C. Wester, I. Thior, L. Stevens, F. Chand, J. Makhema, C. Moffat, A. Asmelash, P. Ndase, P. Arimi, E. van Widenfelt, L. Mazhani, V. Novitsky, S. Lagakos, and M. Essex. 2007. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N. Engl. J. Med. 356:135-147. [DOI] [PubMed] [Google Scholar]
- 22.Loubser, S., P. Balfe, G. Sherman, S. Hammer, L. Kuhn, and L. Morris. 2006. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS 20:995-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzner, K. J., S. G. Giulieri, S. A. Knoepfel, P. Rauch, P. Burgisser, S. Yerly, H. F. Günthard, and M. Cavassini. 2009. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin. Infect. Dis. 48:239-247. [DOI] [PubMed] [Google Scholar]
- 24.Nowak, M. A., S. Bonhoeffer, G. M. Shaw, and R. M. May. 1997. Anti-viral drug treatment: dynamics of resistance in free virus and infected cell populations. J. Theor. Biol. 184:203-217. [DOI] [PubMed] [Google Scholar]
- 25.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188-191. [DOI] [PubMed] [Google Scholar]
- 26.Richman, D. D., D. Havlir, J. Corbeil, D. Looney, C. Ignacio, S. A. Spector, J. Sullivan, S. Cheeseman, K. Barringer, and D. Pauletti. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuurman, R., L. Demeter, P. Reichelderfer, J. Tijnagel, T. de Groot, and C. Boucher. 1999. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 37:2291-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simen, B. B., J. F. Simons, K. H. Hullsiek, R. M. Novak, R. D. Macarthur, J. D. Baxter, C. Huang, C. Lubeski, G. S. Turenchalk, M. S. Braverman, B. Desany, J. M. Rothberg, M. Egholm, and M. J. Kozal. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693-701. [DOI] [PubMed] [Google Scholar]
- 29.Weidle, P., J. Stringer, M. McConnell, J. Kiarie, T. Anekthananon, T. Jariyasethpong, D. Potter, W. Mutsotso, C. Borkowf, O. Bolu, and the NNRTI Response Study Team. 2008. Effectiveness of NNRTI-containing ART in women previously exposed to a single dose of nevirapine: a multi-country cohort study, abstr. 48. 15th Conf. Retrovir. Oppor. Infect., Boston, MA.