Abstract
Early and adequate antimicrobial therapy has been shown to improve the clinical outcome in bloodstream infections (BSI). To provide rapid pathogen identification for targeted treatment, we applied matrix-assisted laser desorption-ionization time of flight (MALDI-TOF) mass spectrometry fingerprinting to bacteria directly recovered from blood culture bottles. A total of 304 aerobic and anaerobic blood cultures, reported positive by a Bactec 9240 system, were subjected in parallel to differential centrifugation with subsequent mass spectrometry fingerprinting and reference identification using established microbiological methods. A representative spectrum of bloodstream pathogens was recovered from 277 samples that grew a single bacterial isolate. Species identification by direct mass spectrometry fingerprinting matched reference identification in 95% of these samples and worked equally well for aerobic and anaerobic culture bottles. Application of commonly used score cutoffs to classify the fingerprinting results led to an identification rate of 87%. Mismatching mostly resulted from insufficient bacterial numbers and preferentially occurred with Gram-positive samples. The respective spectra showed low concordance to database references and were effectively rejected by score thresholds. Spiking experiments and examination of the respective study samples even suggested applicability of the method to mixed cultures. With turnaround times around 100 min, the approach allowed for reliable pathogen identification at the day of blood culture positivity, providing treatment-relevant information within the critical phase of septic illness.
Despite ongoing progress in diagnosis and treatment, morbidity and mortality of bloodstream infections (BSI) have remained high (2, 11, 27). In the face of emerging resistance and a diverse spectrum of causative organisms, microbiological findings are considered crucial for optimal management of these infections (40). Current diagnosis relies on pathogen cultivation in blood culture vessels continuously monitored by semiautomated incubators (32). Gram's stain allows for preliminary identification as soon as growth of microorganisms is detected, while definite identification and susceptibility testing by phenotypic and genotypic methods are routinely performed with solid-medium subcultures (46). Thus, final results often arrive more than 72 h from sampling (17), with a minor impact on antimicrobial usage (30). Since the outcome for septic patients has been shown to depend on the adequacy of early antimicrobial chemotherapy, vital treatment decisions currently often rely on empirical knowledge and vague pathogen characterization by light microscopy (18, 30). Thus, more-rapid identification of the causative organism would be highly desirable to facilitate targeted treatment in the critical phase of septic illness (10).
In an effort to eliminate the time-consuming subcultivation step from conventional blood culture processing, fluorescence in situ hybridization (FISH), direct inoculation of biochemical typing systems, and several nucleic acid amplification-based approaches have been successfully applied for direct pathogen identification from positive blood culture bottles (3, 16, 34-35). But so far, complex sample handling, insufficient species coverage, or high assay costs have hampered widespread usage of either method in routine blood culture processing. Offering short turnaround times and broad species coverage, MALDI-TOF mass spectrometry fingerprinting based on the detection of highly abundant microbial proteins released from cells after simple chemical treatment appears to be able to overcome these drawbacks. The general applicability of the technique for differentiation of pathogenic microorganisms from agar cultures has been demonstrated in a number of recent studies (1, 4, 9, 12, 29), and dedicated fingerprinting systems are currently established in a growing number of clinical microbiology laboratories. In the present study, we established an easy-to-perform protocol to apply a commercially available mass spectrometry typing system for the identification of bacteria from positive blood culture bottles and evaluated the method's performance in a prospective survey with clinical samples.
MATERIALS AND METHODS
Chemicals and media.
The α-cyano-4-hydroxycinnamic acid (HCCA) matrix substance for MALDI-TOF measurements was obtained from Bruker (Bruker Daltonics, Billerica, MA); other chemicals of high purity (high-performance liquid chromatography [HPLC] grade) were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO). Ready-to-use agar media were obtained from Oxoid (Thermo Fisher Scientific, Waltham, MA).
Samples and study design.
The clinical evaluation part of this study was conducted during working hours from October 2008 to December 2008 at the University Medical Center Hamburg-Eppendorf, a tertiary care hospital in northern Germany. During the 9-week study period, Bactec Plus Aerobic/F and Anaerobic/F culture bottles, reported positive for microbial growth by a Bactec 9240 automated incubation system (BD, Franklin Lakes, NJ), were subjected in parallel to direct identification by mass spectrometry fingerprinting and conventional subculturing for reference identification. All samples from which a single bacterial species was recovered by conventional processing (monobacterial samples) were included in a comparative analysis of identification results.
Conventional blood culture processing and reference identification from agar subcultures.
Aspirated blood culture fluid was Gram stained for preliminary identification and inoculated on Columbia blood agar. Chocolate agar and Sabouraud's agar were additionally used if demanded by staining results. All solid media were incubated at 37°C in 5% CO2 for up to 5 days. Additional anaerobic culture on Schaedler agar was performed with anaerobic bottles. Reference identification for all microorganisms grown on agar subcultures was established by at least two independent typing methods. Biochemical identification was attempted for all isolates using GP, GN, NH, and ANC identification cards in a Vitek2 system (VT2-R05.01, AES.R05.01; bioMérieux, Marcy l'Etoile, France) as indicated by the manufacturer. For alpha-hemolytic streptococci, the API rapid ID32 Strep gallery (bioMérieux, Marcy l'Etoile, France) was used according to the manufacturer's instructions. Additional serotyping was routinely performed for beta-hemolytic streptococci (streptococcal grouping kit; Oxoid, Basingstoke, United Kingdom) and salmonellae (salmonella typing sera; Dade Behring, Marburg, Germany). In addition to biochemical typing, Columbia blood agar cultures from all isolates were subjected to mass spectrometry fingerprinting with a MALDI-Biotyper system (Bruker Daltonics, Billerica, MA). Samples were prepared in duplicate by direct deposition (23) and measured with default parameter settings for spectrum acquisition and processing. Only results with concordance scores above the manufacturer's proposed cutoff for reliable species-level identification (2.0) were retained. If no corresponding species-level identification could be obtained from biochemical typing and mass spectrometry fingerprinting, full-length 16S rRNA gene sequences were acquired (37) and compared to annotated sequences from the Ribosomal Database Project (release 10, update 12 [http://rdp.cme.msu.edu/index.jsp]) for identification (7). The definitive species-level identification from conventional blood culture processing was regarded as a sample's reference identification (ID).
Direct identification by mass spectrometry fingerprinting.
Microorganisms were recovered from blood culture fluid by stepwise centrifugation (44) and prepared for mass spectrometry fingerprinting by formic acid extraction (1). To this end, 6 ml culture fluid was drawn from positive culture bottles and centrifuged at 140 × g for 10 min in a benchtop centrifuge to sediment blood cells. Four ml supernatant was recovered and mixed with 2 ml distilled water to lyse residual erythrocytes. Bacteria were pelleted by centrifugation at 1,000 × g for 5 min, washed with 1 ml distilled water, and suspended in 300 μl distilled water. The suspension was transferred to a 1.5-ml reaction tube (Eppendorf, Hamburg, Germany), mixed with 900 μl ethanol, and centrifuged for 2 min at 13,000 × g in a microcentrifuge. Supernatant was discarded, and residual ethanol was removed after repeated centrifugation. The pellet was resuspended in 50 μl 70% formic acid and mixed with 50 μl acetonitrile. After a final centrifugation, 1-μl aliquots of the supernatant were spotted in duplicate on 96 spot ground steel targets (Bruker Daltonics, Billerica, MA) and air dried at room temperature. After evaporation of volatile components, the sample spots were overlain with 2 μl matrix solution (saturated solution of α-cyano-4-hydroxy cinnamic acid in 50% acetonitrile with 2.5% trifluoroacetic acid) and air dried at room temperature. Mass spectra were generated with a Microflex LT mass spectrometer operated by the MALDI-Biotyper automation control (Bruker Daltonics, Billerica, MA). Three hundred shots per sample spot were acquired using the recommended instrument settings for bacterial identification (linear positive mode, 20-Hz laser frequency, 20-kV acceleration voltage, 18.5-kV IS2 voltage, 250- ns extraction delay, and 2,000 to 20,000 m/z range). Automated spectrum processing (smoothing, baseline subtraction, and peak picking) and species identification were done with the MALDI-Biotyper 2.0 application. The software compares acquired sample spectra to reference spectra in the provided database and compiles a list of best matching database records. The degree of spectral concordance is expressed by a logarithmic identification score (38). In this study, the species identity of the highest-scoring database match from duplicate measurements was considered a sample's direct ID.
Data analysis.
Statistical analysis included results from all blood cultures that grew a single bacterial isolate. For each sample, the matching result from direct mass spectrometry fingerprinting (direct ID) was compared to reference identification from conventional processing (reference ID) and classified in the nominal correctness of identification variable (1 = agreement of direct ID and reference ID; 0 = discordant results). In addition, the data set assembled for statistical analysis also comprised the identification score and various sample characteristics: culture condition (1 = anaerobic bottle; 0 = aerobic bottle), culture age (days between sample receipt and processing of the positive culture bottle), date of measurement (study day), Gram reactivity (1 = Gram positive; 0 = Gram negative), and type of the recovered microorganism (1 = Staphylococcus aureus; 2 = coagulase-negative staphylococci; 3 = Streptococcaceae; 4 = Enterococcaceae; 5 = Enterobacteriaceae; 6 = Pseudomonadales/Xanthomonadales [nonfermenter]; 7 = other; the particular taxonomic level for grouping was chosen to retain group sizes of at least 10 samples).
For clinical use, matching results from mass spectrometry fingerprinting are commonly classified via identification score cutoffs to prevent the reporting of unspecific results. A manufacturer-proposed threshold of 2.0 was initially used to indicate reliable species-level identification. To investigate the impact of lower thresholds on test performance and to facilitate comparison with recently published results, species-level cutoffs between 1.3 and 2.0 were retrospectively applied to our study data and compared with respect to the number of rejected correct and incorrect direct IDs and the resulting rate of correctly identified (identification score above cutoff and correct direct ID), misidentified (identification score above cutoff and incorrect direct ID), and unidentified (identification score below cutoff) samples.
The influence of sample characteristics on identification results was assessed in a mixed-model approach accounting for cluster effects due to the inclusion of multiple aerobic and anaerobic culture bottles per patient (leading to pseudoreplicate observations on isogenic bacteria) (33). Using the MIXED procedure from the SPSS statistics software (version 16; SPSS Inc., Chicago), identification scores from direct mass spectrometry fingerprinting were modeled as a function of sample characteristics (culture condition, culture age, date of measurement, Gram reactivity, and type of organism) as fixed effects and replication as a random effect. Replication was addressed through a patient strain variable, which identified isolates of common patient source, species identity, and susceptibility pattern. The basic model was refined by introduction of interaction between factors and by stepwise elimination of insignificant predictors (P > 0.05). Fits of the derived candidate models were evaluated by comparison of information criteria (Aikake information criterion [AIC] and Bayesian information criterion [BIC]) obtained by maximum-likelihood estimation. Parameter values of the final model were determined by restricted maximum-likelihood estimation.
Spiking experiments.
Spiked samples were used to investigate the influence of the bacterial concentration on the accuracy of direct mass spectrometry fingerprinting. Suspensions of the test strains Staphylococcus aureus Newman and Escherichia coli DH5α were prepared in phosphate-buffered saline (PBS) from fresh Columbia blood agar cultures and adjusted to defined bacterial concentrations by means of optical density at 600 nm (OD600) measurements and appropriate calibration curves. Five hundred μl of bacterial suspension was mixed with 5.5 ml sterile culture fluid from preincubated Bactec Plus Aerobic/F bottles to obtain final concentrations between 106 and 109 CFU/ml, as expected in positive blood culture bottles. At least four samples were prepared for each bacterial concentration and strain and subjected to mass spectrometry fingerprinting as described above. Mean identification scores for different experimental conditions were compared by one-way analysis of variance (ANOVA) with Bonferroni posttests (significance level, 0.05). The observed influence of bacterial concentration on identification scores was described by equation 1, which was empirically chosen to fit the study data. The parameters Sm (maximum score − background), K (constant), and bg (background) were estimated for each experimental condition by nonlinear regression on data from the respective spiking experiments.
 |
(1) |
RESULTS
Analytical sensitivity and operational characteristics.
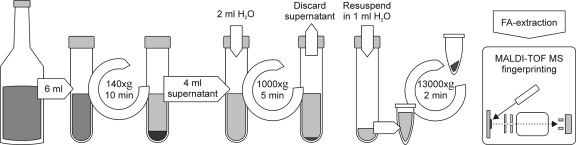
We established a simple stepwise centrifugation protocol for the purification of microorganisms from positive blood culture bottles (Fig. 1). Good separation of bacteria from blood cells and cellular debris could be demonstrated by light microscopy and by aerobic plate count. Recovery rates from routine blood cultures varied between 10 and 70% for bacterial samples but were found to be insufficient (<0.1%) for the purification of yeast.
FIG. 1.
Bacteria were recovered from blood culture fluids by differential centrifugation. Washed pellets were resuspended in 300 μl H2O, inactivated with ethanol, and prepared for subsequent mass spectrometry fingerprinting by formic acid extraction (see the text for details).
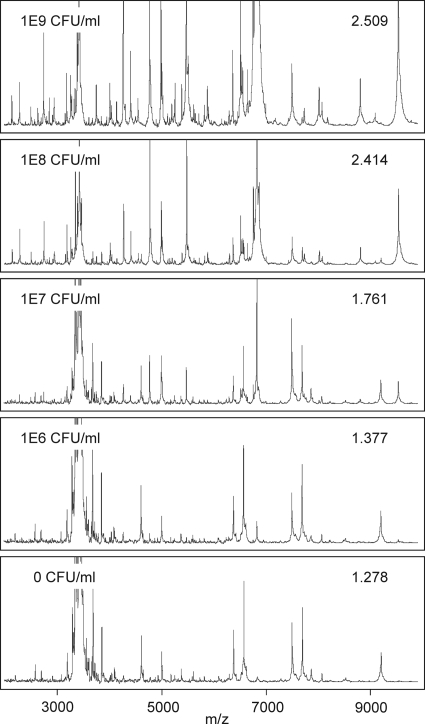
In spiking experiments, mass spectra generated from blood culture samples with a high bacterial load (>108 CFU/ml) closely resembled those from pure agar cultures. At lower inocula, nonbacterial background peaks became more prominent and influenced spectrum matching (Fig. 2). Identification scores from samples spiked with 107 CFU/ml were significantly lower than those from agar cultures, but the best database match still consistently represented the correct species identification. At 106 CFU/ml, spectra became indistinguishable from those of sterile blood cultures and yielded low-scoring incorrect direct IDs due to arbitrary matching of nonbacterial peaks (score < 1.5). No significant differences (P > 0.05) in mean identification scores were observed between the test strains Staphylococcus aureus Newman and Escherichia coli DH5α.
FIG. 2.
Mass spectra from blood culture samples spiked with different concentrations of S. aureus Newman. Lower inocula resulted in decreasing numbers of detected bacterial peaks and reduced concordance scores for the best-matching reference spectrum from the Biotyper database (inscribed numbers). At 106 CFU/ml, spectra resembled those from sterile cultures, and species identification failed.
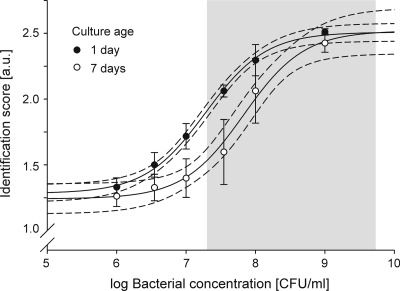
In concordance with other investigators (43), we observed bacterial concentrations at the time of blood culture positivity between 2 × 107 and 7 × 109 CFU/ml (median, 5 × 108 CFU/ml) in a random selection of 30 samples. Nonlinear regression on data from the spiking experiments with 1-day-old cultures (global fit on data for S. aureus and E. coli, R2 = 0.96) predicted satisfactory identification scores (1.9 to 2.5) within this concentration range (Fig. 3).
FIG. 3.
Effect of bacterial concentration and culture age on identification scores from direct mass spectrometry fingerprinting. Blood culture samples preincubated for 1 (filled circles) or 7 days (open circles) were spiked with different concentrations of S. aureus Newman and E. coli DH5α and subjected to mass spectrometry fingerprinting. Symbols and error bars indicate mean identification scores (in arbitrary units [a.u.]) and standard deviations from at least four independent sample preparations. Solid and dotted lines represent best fits from nonlinear regression with 95% CIs. The shaded area marks the range of bacterial concentrations at the time of culture positivity as detected in a random selection of 30 study samples. A clear association between bacterial concentration and identification scores was observed. Although advanced culture age led to reduced identification scores, the majority of clinical samples are predicted to yield sufficient scores for secure species-level identification.
Prolonged preincubation of blood culture fluid before spiking (7 days instead of 1) led to significantly reduced identification scores at lower bacterial concentrations (107 and 108 CFU/ml) and increased sample-to-sample variation. Consequently, the number of bacteria needed for reliable species identification was two to six times higher than that for 1-day-old cultures (Fig. 3).
Overall turnaround time for direct identification varied between 75 (1 sample) and 140 min (12 samples), with 10 to 70 min of hands-on time.
Analysis of monomicrobial samples.
Within the 9-week study period, 304 blood cultures (156 aerobic and 148 anaerobic bottles from 145 patients), reported positive by the automated incubation system on average 1.22 days after sample entry, were analyzed by direct mass spectrometry fingerprinting and conventional processing (individual identification results and sample characteristics are provided in file S1 in the supplemental material). Three samples that showed no sign of microbial growth by conventional processing and eight samples that grew yeast were excluded from further analysis. Sixteen samples (5.3%) that grew more than one bacterial isolate by conventional processing were selected for separate evaluation, since the fingerprinting software did not support automated analysis of mixed samples. From the remaining 277 monobacterial cultures (137 aerobic and 140 anaerobic bottles from 135 patients), a total of 147 distinct patient strains were recovered (Table 1). Species distribution of these strains was in good agreement with larger surveys from similar populations (47).
TABLE 1.
Species distribution and identification results for monobacterial samples
| Species | No. of patient strains | No. of samples | No. of samples with correct direct IDa |
|---|---|---|---|
| Staphylococcus aureus | 22 | 60 | 54 |
| Coagulase-negative staphylococci | 48 | 77 | 74 |
| Staphylococcus capitis | 2 | 2 | 2 |
| Staphylococcus epidermidis | 35 | 61 | 60 |
| Staphylococcus haemolyticus | 4 | 5 | 4 |
| Staphylococcus hominis | 5 | 5 | 5 |
| Staphylococcus lugdunensis | 1 | 2 | 2 |
| Staphylococcus pettenkoferi | 1 | 1 | 0 |
| Staphylococcus saccharolyticus | 1 | 1 | 1 |
| Streptococcaceae | 9 | 15 | 13 |
| Streptococcus agalactiae | 2 | 4 | 4 |
| Streptococcus anginosus | 1 | 2 | 2 |
| Streptococcus gallolyticus | 2 | 3 | 3 |
| Streptococcus pneumoniae | 1 | 1 | 1 |
| Streptococcus pyogenes | 2 | 3 | 3 |
| Streptococcus thermophilus | 1 | 2 | 0 |
| Enterococcaceae | 16 | 25 | 24 |
| Enterococcus faecalis | 5 | 13 | 13 |
| Enterococcus faecium | 11 | 12 | 11 |
| Enterobacteriaceae | 40 | 78 | 77 |
| Citrobacter freundii | 2 | 3 | 3 |
| Enterobacter cloacae | 5 | 6 | 6 |
| Proteus mirabilis | 1 | 2 | 2 |
| Klebsiella oxytoca | 1 | 1 | 1 |
| Klebsiella pneumoniae | 4 | 8 | 8 |
| Escherichia coli | 21 | 42 | 42 |
| Salmonella enterica | 5 | 14 | 14 |
| Serratia marcescens | 1 | 2 | 1 |
| Nonfermenters | 6 | 15 | 15 |
| Acinetobacter baumannii | 1 | 1 | 1 |
| Acinetobacter ursungii | 1 | 1 | 1 |
| Pseudomonas aeruginosa | 2 | 5 | 5 |
| Stenotrophomonas maltophilia | 2 | 8 | 8 |
| Other species | 6 | 7 | 5 |
| Corynebacterium striatum | 2 | 3 | 3 |
| Eubacterium brachy | 1 | 1 | 0 |
| Micrococcus luteus | 1 | 1 | 1 |
| Propionibacterium acnes | 1 | 1 | 0 |
| Rothia mucilaginosa | 1 | 1 | 1 |
| Total | 147 | 277 | 262 |
Agreement of direct ID and reference ID.
Identification results from direct mass spectrometry fingerprinting showed good accuracy compared to reference identification from conventional processing. The direct ID matched the reference ID in 262 of 277 monobacterial samples (95%), corresponding to 138 of 147 (94%) patient strains that could be correctly identified at the day of their first occurrence in a positive blood culture. In 261 of these samples, species-level agreement was achieved, while the best matching reference strain for a single Acinetobacter baumannii isolate was referred to only by genus (Acinetobacter sp.) in the Biotyper database. Identification scores ranged from 1.134 to 2.598 (median, 2.228) and were tightly correlated with the correctness of direct identification (point-biserial correlation, 0.583; P < 0.001). Incorrect direct IDs were ranked among the lowest-scoring samples and could be addressed by identification score cutoffs. Threshold values of 2.0 and 1.7, recommended by the manufacturer to establish species- and genus-level identification, effectively rejected 100% of incorrect direct IDs and led to an identification rate (correct direct ID and score above threshold) of 87%. The proportion of species-level IDs could be markedly increased from 75% to 81% by adopting the 1.9 cutoff recently proposed for the analysis of agar cultures by direct sample deposition (41). Retrospective evaluation suggested performance benefits from even more permissive cutoffs. Under the requirement of perfect specificity, considerable reduction of the number of falsely rejected direct IDs raised identification rates up to 92% (Table 2). A supportive test for the homogeneity of the list of best-matching reference spectra, which indicates unspecific matching of nonbacterial background peaks and could thus minimize the risk of gross misidentifications through lowered cutoffs, was developed with the help of spiking experiments and successfully applied to low-scoring identification results (score < 2.0) from our study (see file S2 in the supplemental material).
TABLE 2.
Evaluation of test performance for different identification score cutoffs
| Cutoff | No. (fraction) of: |
Estimated analytical sensitivityd [CFU/ml] | ||||
|---|---|---|---|---|---|---|
| Correct direct IDs rejected | Incorrect direct IDs rejected | Identified samplesa | Misidentified samplesb | Unidentified samplesc | ||
| 1.3 | 0 (0.00) | 4 (0.27) | 262 (0.95) | 11 (0.04) | 4 (0.01) | ND |
| 1.4 | 0 (0.00) | 12 (0.80) | 262 (0.95) | 3 (0.01) | 12 (0.04) | ND |
| 1.5 | 6 (0.02) | 15 (1.00) | 256 (0.92) | 0 (0.00) | 21 (0.08) | 4.1 × 106 |
| 1.6 | 13 (0.05) | 249 (0.90) | 28 (0.10) | 6.6 × 106 | ||
| 1.7 | 22 (0.08) | 240 (0.87) | 37 (0.13) | 9.8 × 106 | ||
| 1.8 | 31 (0.12) | 231 (0.83) | 46 (0.17) | 1.4 × 107 | ||
| 1.9 | 39 (0.15) | 223 (0.81) | 54 (0.19) | 1.9 × 107 | ||
| 2.0 | 55 (0.21) | 207 (0.75) | 70 (0.25) | 2.7 × 107 | ||
Identification score ≥ cutoff; correct direct ID.
Identification score ≥ cutoff; incorrect direct ID.
Identification score < cutoff.
Bacterial concentration needed to reach cutoff as calculated from spiking experiments (see Fig. 3).
All misidentified samples (8 aerobic and 7 anaerobic bottles from 13 patients) yielded scores below 1.5 (range, 1.266 to 1.429), and the best-matching reference spectrum often represented species uncommon to blood cultures (e.g., Azoarcus anaerobius (four times), Paecilomyces lilacinus (two times), Streptomyces griseus (two times), and Thauera linaloolentis). Mass spectra from 14 of these samples mainly comprised peaks attributable to blood culture components indicative of insufficient bacterial concentrations. Twelve of the 15 isolates recovered from misidentified samples were correctly identified by mass spectrometry fingerprinting from agar subcultures. Three isolates, identified as Staphylococcus pettenkoferi and Streptococcus thermophilus (two samples from a single patient) by full-length sequencing of their 16S rRNA genes, were not amenable to mass spectrometry identification since these species were not represented in the reference spectrum database. In contrast to other misidentified samples, the direct identification spectrum of one S. thermophilus sample showed a number of supposed bacterial peaks and could be correctly identified after addition of the appropriate user-generated reference spectrum to the fingerprinting database.
Only 1 of 15 misidentified samples (7%) grew a Gram-negative isolate. Accordingly, identification scores from direct mass spectrometry fingerprinting were correlated with Gram positivity (point-biserial correlation, −0.355; P < 0.001). The more sophisticated linear mixed-effect model analysis, considering sample correlation and interactions between influencing factors, identified the type of recovered organism, culture age, and culture conditions of certain organism types as significant predictors of identification scores (Table 3; a complete list of parameter estimates is provided in file S3 in the supplemental material). According to the model, samples growing enterococci or members of the family of Enterobacteriaceae yielded significantly better identification scores, while aerobic cultures of Staphylococcus aureus were associated with markedly reduced scores (Table 4). All six misidentified S. aureus samples were from aerobic culture bottles. Culture conditions had no significant effect in the context of other species. The mean identification score for the population of patient strains was estimated to be 2.11 (95% confidence interval [CI], 2.06 to 2.17). In agreement with results from spiking experiments, the culture age was estimated to account for a decline in identification scores by 0.09 (95% CI, 0.06 to 0.12) for each day until detection of microbial growth.
TABLE 3.
Significant predictors of identification scores in the linear mixed-effect model analysis of study data from monobacterial samples
| Parameter | Estimate | SE | P value | 95% CI |
|
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Intercept | 2.051 | 0.138 | <0.001 | 1.779 | 2.323 |
| TOMa | |||||
| Enterococcaceae | 0.416 | 0.149 | 0.006 | 0.122 | 0.710 |
| Enterobacteriaceae | 0.394 | 0.139 | 0.005 | 0.121 | 0.668 |
| Culture condition of TOMb | |||||
| S. aureus from aerobic bottle | −0.315 | 0.047 | <0.001 | −0.408 | −0.222 |
| Days to culture positivity | −0.089 | 0.016 | <0.001 | −0.122 | −0.057 |
Recovery of other types of microorganisms (TOMs) did not significantly influence predicted identification scores.
Culture conditions had no significant effect with other TOMs.
TABLE 4.
Estimated mean identification scores in the linear mixed-effect model analysis of study data from monobacterial samples
| Samples | Mean score | SE | 95% CI |
|
|---|---|---|---|---|
| Lower | Upper | |||
| All samples | 2.113 | 0.026 | 2.061 | 2.166 |
| Staphylococcus aureus | 1.944 | 0.049 | 1.847 | 2.040 |
| From aerobic bottle | 1.786 | 0.054 | 1.679 | 1.894 |
| From anaerobic bottle | 2.101 | 0.054 | 1.995 | 2.207 |
| Coagulase-negative staphylococcia | 2.061 | 0.037 | 1.989 | 2.133 |
| Streptococcaceaea | 1.941 | 0.081 | 1.781 | 2.102 |
| Enterococcaceaea | 2.336 | 0.064 | 2.210 | 2.462 |
| Enterobacteriaceaea | 2.352 | 0.038 | 2.277 | 2.427 |
| Nonfermenterb | 2.125 | 0.096 | 1.935 | 2.316 |
| Other speciesa,c | 2.040 | 0.108 | 1.826 | 2.253 |
No significant differences between aerobic and anaerobic culture bottles.
Not recovered from anaerobic culture bottles.
Corynebacterium striatum, Eubacterium brachy, Micrococcus luteus, Propionibacterium acnes, and Rothia mucilaginosa.
Mixed cultures.
In accordance with previous blood culture surveys, 5.5% (16 of 297) of our study samples represented mixed cultures of two or more isolates, and the majority (14 of 16 samples) remained unrecognized by Gram stain (3, 16). As for monomicrobial samples, the highest-scoring database hit correctly identified one of the recovered isolates if identification scores exceeded 1.5 (13 of 16 samples). The median identification score for the best hit (2.164; range, 1.238 to 2.497) was also up to the results from pure cultures. Although the software used for spectrum processing and species identification did not feature automated analysis of mixed samples, a second isolate was recognized in four study samples by manual inspection of the list of best-matching reference spectra, since the acquired mass spectra yielded high concordance scores with spectra from two different species. This behavior could be reproduced with balanced mixtures (ratio, 1:2 to 2:1) of two test strains in spiked samples.
DISCUSSION
We have applied a commercially available MALDI-TOF mass spectrometry fingerprinting system for the rapid identification of bacteria purified from positive blood culture bottles. Although originally devised for use with pure agar cultures, the system successfully identified bacteria from crude sample preparations without modifications to the detection algorithm or reference database. Even without specific measures to reject unspecific identification results, accuracy compared to results with reference methods exceeded the 90% limit, which has been proposed as a minimal requirement for selecting an identification system for clinical use (31).
The unambiguous association between correctness of identification and identification scores facilitated reliable rejection of incorrect direct IDs even by permissive score cutoffs. As expected, the manufacturer-proposed threshold for species-level identification (2.0) was found to underestimate the correctness of spectrum matching in a sample population, where low concordance scores mostly reflected suboptimal sample conditions (prominent background signal and low analyte concentration) rather than a low degree of relatedness between the sample isolate and the best database match. The report of species-level identification for lower-scoring samples would increase test performance and appears to be safe unless groups with high spectral similarity between member species are involved (e.g., viridans streptococci). Modified identification algorithms, considering the presence of (nonbacterial) background peaks (45), might improve identification rates for crude samples without the specificity tradeoffs involved in lower score thresholds.
The bacterial concentration and culture age were found to substantially influence direct identification results in spiking experiments and clinical samples. Prolonged blood culture incubation increases the amount of nonbacterial sample components (e.g., cellular debris) that cannot be removed by differential centrifugation and subsequently impair spectrum quality by masking specific bacterial signals. Since most clinically relevant blood cultures are reported positive during the first days of incubation (36), this phenomenon affects only a minority of samples. Because of the similarity of spectra from misidentified samples to those from sterile blood culture bottles, we suspect suboptimal analyte concentration as the most relevant cause of unsuccessful direct identification. A combination of low bacterial numbers at the time of positivity and insufficient recovery could result in final bacterial concentrations below the assumed limit of detection. Reports from other investigators suggest that the observed negative correlation of Gram positivity and identification scores and the subaverage performance with aerobic S. aureus cultures are probably related to lower bacterial numbers in Gram-positive blood culture samples at the time of positivity (8, 43). Analytical sensitivity might be improved by processing of larger sample volumes or by affinity purification (13).
The reduced identification rate for aerobic S. aureus cultures (23 of 29 samples correctly identified) was compensated by good results from anaerobic bottles (31 of 31 samples correctly identified). Five of the six misidentified samples were recovered the same day from an accompanying anaerobic bottle, so that direct mass spectrometry fingerprinting is suitable for rapid diagnosis of this important bloodstream pathogen from paired cultures.
Compared to conventional processing with solid-medium subcultures, direct mass spectrometry fingerprinting shortens the time to result by at least 1 work day, providing accurate species-level identification within 2 h from blood culture positivity. Besides numerous studies that have documented the importance of early adequate antimicrobial therapy on patient outcome (14-15, 19, 21), a decent analysis of Staphylococcus aureus and Pseudomonas aeruginosa bacteriemias demonstrated increased 30-day mortality if more than 2 days (45 and 52 h, respectively) elapsed between administration of appropriate antibiotics and collection of index blood culture (25-26). In our study, more than 80% of identification results were available the day after sample entry in our laboratory, facilitating targeted treatment optimization within the critical phase of septic illness. These treatment adjustments can improve therapeutic efficacy, minimize adverse effects, reduce costs, and lessen the risk of resistance development (5, 22, 24). Rapid pathogen identification from positive blood culture bottles has been shown to more readily entail changes of antibiotic therapy than results from conventional blood culture processing (17, 30). Consequently, a plethora of techniques, which vary considerably in terms of turnaround time, species coverage, and usability, have already been put to the task. While agglutination assays and simple biochemical tests (e.g., tube coagulase test) are fast and inexpensive, they often lack sensitivity and are limited to a few species (28). Modified probe assays (fluorescence in situ hybridization [FISH]) equal direct mass spectrometry fingerprinting regarding time to result but require multiple species- or genus-specific assays in parallel to cover the most frequently isolated bloodstream pathogens. The set of probes evaluated by Peters and coworkers would cover 95% of our study samples, facilitating species-level identification in 60% of cases at best (35). Nucleic acid amplification techniques offer superior sensitivity and can be designed to achieve almost universal species coverage or to provide additional information on antimicrobial susceptibility (16, 34). But as for sequence-based identification directly from venipuncture blood (39), routine use in blood culture processing is impeded by complex sample handling and high assay costs. Various groups have successfully applied direct inoculation of biochemical typing systems for rapid pathogen identification from blood culture bottles. Using this approach, a clinical benefit of accelerated microbiological diagnosis was demonstrated in at least two studies (17, 44). Nevertheless, biochemical typing requires a considerable time to result, and some authors reported poor accuracy with Gram-positive samples (6, 8). Compared to the aforementioned methods, direct mass spectrometry fingerprinting offers a favorable combination of easy sample handling, broad species coverage, and short turnaround time. Without the need for additional consumables or software, it can be readily applied by laboratories already running an appropriate typing system for routine bacterial identification. And while improved identification algorithms and automated purification protocols might increase sensitivity and facilitate sample handling, accuracy results from this and two other recently published studies (20, 42) already warrant use of the technique in routine diagnosis of BSI.
Footnotes
Published ahead of print on 17 March 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Barbuddhe, S. B., T. Maier, G. Schwarz, M. Kostrzewa, H. Hof, E. Domann, T. Chakraborty, and T. Hain. 2008. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bearman, G. M. L., and R. P. Wenzel. 2005. Bacteremias: a leading cause of death. Arch. Med. Res. 36:646-659. [DOI] [PubMed] [Google Scholar]
- 3.Bruins, M. J., P. Bloembergen, G. J. H. M. Ruijs, and M. J. H. M. Wolfhagen. 2004. Identification and susceptibility testing of Enterobacteriaceae and Pseudomonas aeruginosa by direct inoculation from positive BACTEC blood culture bottles into Vitek 2. J. Clin. Microbiol. 42:7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbonnelle, E., J.-L. Beretti, S. Cottyn, G. Quesne, P. Berche, X. Nassif, and A. Ferroni. 2007. Rapid identification of staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 45:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, F. Y., J. E. Peacock, Jr., D. M. Musher, P. Triplett, B. B. MacDonald, J. M. Mylotte, A. O'Donnell, M. M. Wagener, and V. L. Yu. 2003. Staphylococcus aureus bacteremia: recurrence and the impact of antibiotic treatment in a prospective multicenter study. Medicine (Baltimore) 82:333-339. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. R., S. Y. Lee, B. H. Yang, and J. J. Lu. 2008. Rapid identification and susceptibility testing using the VITEK 2 system using culture fluids from positive BacT/ALERT blood cultures. J. Microbiol. Immunol. Infect. 41:259-264. [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Cueto, M., E. Ceballos, L. Martinez-Martinez, E. J. Perea, and A. Pascual. 2004. Use of positive blood cultures for direct identification and susceptibility testing with the vitek 2 system. J. Clin. Microbiol. 42:3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degand, N., E. Carbonnelle, B. Dauphin, J.-L. Beretti, M. L. Bourgeois, I. Sermet-Gaudelus, C. Segonds, P. Berche, X. Nassif, and A. Ferroni. 2008. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deresinski, S. 2007. Principles of antibiotic therapy in severe infections: optimizing the therapeutic approach by use of laboratory and clinical data. Clin. Infect. Dis. 45(Suppl. 3):S177-S183. [DOI] [PubMed] [Google Scholar]
- 11.Dombrovskiy, V. Y., A. A. Martin, J. Sunderram, and H. L. Paz. 2007. Rapid increase in hospitalization and mortality rates for severe sepsis in the United States: a trend analysis from 1993 to 2003. Crit. Care Med. 35:1244-1250. [DOI] [PubMed] [Google Scholar]
- 12.Grosse-Herrenthey, A., T. Maier, F. Gessler, R. Schaumann, H. Böhnel, M. Kostrzewa, and M. Krüger. 2008. Challenging the problem of clostridial identification with matrix-assisted laser desorption and ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe 14:242-249. [DOI] [PubMed] [Google Scholar]
- 13.Guo, Z., Y. Liu, S. Li, and Z. Yang. 2009. Interaction of bacteria and ion-exchange particles and its potential in separation for matrix-assisted laser desorption/ionization mass spectrometric identification of bacteria in water. Rapid Commun. Mass Spectrom. 23:3983-3993. [DOI] [PubMed] [Google Scholar]
- 14.Harbarth, S., J. Garbino, J. Pugin, J. A. Romand, D. Lew, and D. Pittet. 2003. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am. J. Med. 115:529-535. [DOI] [PubMed] [Google Scholar]
- 15.Ibrahim, E. H., G. Sherman, S. Ward, V. J. Fraser, and M. H. Kollef. 2000. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 118:146-155. [DOI] [PubMed] [Google Scholar]
- 16.Jordan, J. A., J. Jones-Laughner, and M. B. Durso. 2009. Utility of pyrosequencing in identifying bacteria directly from positive blood culture bottles. J. Clin. Microbiol. 47:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerremans, J. J., P. Verboom, T. Stijnen, L. H. van Roijen, W. Goessens, H. A. Verbrugh, and M. C. Vos. 2008. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J. Antimicrob. Chemother. 61:428-435. [DOI] [PubMed] [Google Scholar]
- 18.Kollef, M. H. 2008. Broad-spectrum antimicrobials and the treatment of serious bacterial infections: getting it right up front. Clin. Infect. Dis. 47(Suppl. 1):S3-S13. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, A., D. Roberts, K. E. Wood, B. Light, J. E. Parrillo, S. Sharma, R. Suppes, D. Feinstein, S. Zanotti, L. Taiberg, D. Gurka, and M. Cheang. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34:1589-1596. [DOI] [PubMed] [Google Scholar]
- 20.La Scola, B., and D. Raoult. 2009. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One 4:e8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leibovici, L., I. Shraga, M. Drucker, H. Konigsberger, Z. Samra, and S. D. Pitlik. 1998. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J. Intern. Med. 244:379-386. [DOI] [PubMed] [Google Scholar]
- 22.Leone, M., and C. Martin. 2008. How to break the vicious circle of antibiotic resistances? Curr. Opin. Crit. Care 14:587-592. [DOI] [PubMed] [Google Scholar]
- 23.Liu, H., Z. Du, J. Wang, and R. Yang. 2007. Universal sample preparation method for characterization of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 73:1899-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livermore, D. M. 2005. Minimising antibiotic resistance. Lancet Infect. Dis. 5:450-459. [DOI] [PubMed] [Google Scholar]
- 25.Lodise, T. P., Jr., N. Patel, A. Kwa, J. Graves, J. P. Furuno, E. Graffunder, B. Lomaestro, and J. C. McGregor. 2007. Predictors of 30-day mortality among patients with Pseudomonas aeruginosa bloodstream infections: impact of delayed appropriate antibiotic selection. Antimicrob. Agents Chemother. 51:3510-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lodise, T. P., P. S. McKinnon, L. Swiderski, and M. J. Rybak. 2003. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin. Infect. Dis. 36:1418-1423. [DOI] [PubMed] [Google Scholar]
- 27.Martin, G. S., D. M. Mannino, S. Eaton, and M. Moss. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 348:1546-1554. [DOI] [PubMed] [Google Scholar]
- 28.McDonald, C. L., and K. Chapin. 1995. Rapid identification of Staphylococcus aureus from blood culture bottles by a classic 2-hour tube coagulase test. J. Clin. Microbiol. 33:50-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellmann, A., J. Cloud, T. Maier, U. Keckevoet, I. Ramminger, P. Iwen, J. Dunn, G. Hall, D. Wilson, P. Lasala, M. Kostrzewa, and D. Harmsen. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munson, E. L., D. J. Diekema, S. E. Beekmann, K. C. Chapin, and G. V. Doern. 2003. Detection and treatment of bloodstream infection: laboratory reporting and antimicrobial management. J. Clin. Microbiol. 41:495-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken. 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC.
- 32.Mylotte, J. M., and A. Tayara. 2000. Blood cultures: clinical aspects and controversies. Eur. J. Clin. Microbiol. Infect. Dis. 19:157-163. [DOI] [PubMed] [Google Scholar]
- 33.Paterson, S., and J. Lello. 2003. Mixed models: getting the best use of parasitological data. Trends Parasitol. 19:370-375. [DOI] [PubMed] [Google Scholar]
- 34.Paule, S. M., A. C. Pasquariello, R. B. Thomson, Jr., K. L. Kaul, and L. R. Peterson. 2005. Real-time PCR can rapidly detect methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive blood culture bottles. Am. J. Clin. Pathol. 124:404-407. [DOI] [PubMed] [Google Scholar]
- 35.Peters, R. P. H., P. H. M. Savelkoul, A. M. Simoons-Smit, S. A. Danner, C. M. J. E. Vandenbroucke-Grauls, and M. A. van Agtmael. 2006. Faster identification of pathogens in positive blood cultures by fluorescence in situ hybridization in routine practice. J. Clin. Microbiol. 44:119-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reisner, B. S., and G. L. Woods. 1999. Times to detection of bacteria and yeasts in BACTEC 9240 blood culture bottles. J. Clin. Microbiol. 37:2024-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sacchi, C. T., A. M. Whitney, L. W. Mayer, R. Morey, A. Steigerwalt, A. Boras, R. S. Weyant, and T. Popovic. 2002. Sequencing of 16S rRNA gene: a rapid tool for identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauer, S., A. Freiwald, T. Maier, M. Kube, R. Reinhardt, M. Kostrzewa, and K. Geider. 2008. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One 3:e2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrenzel, J. 2007. Clinical relevance of new diagnostic methods for bloodstream infections. Int. J. Antimicrob. Agents 30(Suppl. 1):S2-S6. [DOI] [PubMed] [Google Scholar]
- 40.Seifert, H. 2009. The clinical importance of microbiological findings in the diagnosis and management of bloodstream infections. Clin. Infect. Dis. 48(Suppl. 4):S238-S245. [DOI] [PubMed] [Google Scholar]
- 41.Seng, P., M. Drancourt, F. Gouriet, B. La Scola, P. E. Fournier, J. M. Rolain, and D. Raoult. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543-551. [DOI] [PubMed] [Google Scholar]
- 42.Stevenson, L. G., S. K. Drake, and P. R. Murray. 2010. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan, T. Y., L. S. Ng, and L. L. Kwang. 2008. Evaluation of disc susceptibility tests performed directly from positive blood cultures. J. Clin. Pathol. 61:343-346. [DOI] [PubMed] [Google Scholar]
- 44.Trenholme, G. M., R. L. Kaplan, P. H. Karakusis, T. Stine, J. Fuhrer, W. Landau, and S. Levin. 1989. Clinical impact of rapid identification and susceptibility testing of bacterial blood culture isolates. J. Clin. Microbiol. 27:1342-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahl, K. L., S. C. Wunschel, K. H. Jarman, N. B. Valentine, C. E. Petersen, M. T. Kingsley, K. A. Zartolas, and A. J. Saenz. 2002. Analysis of microbial mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74:6191-6199. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, M. L., M. Mitchell, A. J. Morris, P. R. Murray, L. G. Reimer, L. B. Reller, M. Towns, M. P. Weinstein, S. A. Wellstood, J. W. M. Dunne, R. C. Jerris, and D. L. Welch. 2007. Principles and procedures for blood cultures; approved guideline. CLSI document M47-A. Clinical and Laboratory Standards Institute, Wayne, PA.
- 47.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]