Abstract
The GenoType MTBDRsl test rapidly detects resistance to ethambutol, fluoroquinolones, and second-line aminoglycosides (amikacin and kanamycin) and cyclic peptide (capreomycin) in Mycobacterium tuberculosis. A set of 41 multidrug-resistant (MDR) M. tuberculosis strains, 8 extensively drug-resistant (XDR) M. tuberculosis strains, and 3 non-MDR M. tuberculosis strains were tested by the MTBDRsl test and by DNA sequencing of the resistance-determining regions in gyrA and gyrB (fluoroquinolones [FQ]), rpsL (streptomycin), rrs and tlyA (aminoglycosides and/or cyclic peptide), and embB (ethambutol). The sensitivity and specificity of the MTBDRsl test were as follows: 87% and 96%, respectively, for fluoroquinolones; 100% for both for amikacin; 77% and 100%, respectively, for kanamycin, 80% and 98%, respectively, for capreomycin; and 57% and 92%, respectively, for ethambutol. Analysis of the discrepant results indicated that three FQ-resistant strains (including one XDR strain) with mutations in gyrB were missed by the MTBDRsl test and that one FQ-susceptible strain, identified as resistant by the MTBDRsl test, had a double mutation (T80A-A90G) in GyrA that did not confer resistance to FQ. Five strains (including two XDR strains) without mutations in rrs were monoresistant to aminoglycosides or cyclic peptide and were missed by the MTBDRsl test. Finally, 12/28 ethambutol-resistant strains had no mutation at codon 306 in embB, while 2/24 ethambutol-susceptible strains had such a mutation. In conclusion, the MTBDRsl test efficiently detects the most common mutations involved in resistance to fluoroquinolones, aminoglycosides/cyclic peptide, and ethambutol and accurately assesses susceptibility to amikacin. However, due to mutations not included in the test (particularly in gyrB) or resistance mechanisms not yet characterized (particularly those related to ethambutol resistance and to monoresistance to aminoglycosides or cyclic peptide), the wild-type results yielded by the MTBDRsl test should be confirmed by drug susceptibility testing.
Every year, there are more than 8 million new cases of tuberculosis (TB) worldwide, and 400,000 of these are multidrug resistant (MDR), defined as resistance to both isoniazid and rifampin, the two main anti-TB drugs (31). For the past 20 years, treatment of MDR TB was based on the association of ethambutol (ETB) with fluoroquinolones (FQ) and aminoglycosides/cyclic peptide (A-CP), i.e., the aminoglycosides kanamycin (KAN) and amikacin (AMK) and the cyclic peptide capreomycin (CAP), which are second-line drugs, or alternatively, streptomycin, which is a first-line drug (32). As a consequence of inadequate use of second-line treatments, extensively drug-resistant (XDR) tuberculosis, defined as MDR with resistance to FQ and at least one of the three injectable second-line drugs (amikacin, kanamycin, and capreomycin) has emerged. XDR strains currently represent a worrisome threat to global health, since the proportion of overall successful treatment outcome for this form of the disease is only 50% (17, 31).
Resistance to antituberculous drugs is related to mutations affecting the function and/or expression of chromosome-encoded targets. The main mechanism of acquired resistance to fluoroquinolones in Mycobacterium tuberculosis is an alteration of the DNA gyrase (consisting of two subunits, GyrA and GyrB, encoded by the gyrA and gyrB genes) (2), which is associated with decreased drug activity levels within the cell. Most mutations conferring bacterial resistance to fluoroquinolones occur in a short segment termed the quinolone resistance-determining region (QRDR) in the gyrA gene (mutations mostly affecting residues A90 and D94 and, more rarely, G88 and S91), and less frequently in gyrB (mutations mostly affecting D472 and N510) (2, 7, 18, 20, 33).
Resistance to CAP and to AMK-KAN is caused by mutations at positions 1401 and 1402 and position 1484, respectively, in the rrs gene with the following expression patterns: mutants with the rrs mutation A1401G display low-level resistance to CAP and high-level resistance to AMK and KAN (21). In contrast, the rrs C1402T mutants are susceptible to AMK and have low-level resistance to KAN and high-level resistance to CAP. Finally, the rrs G1484T mutants display high-level resistance to all 3 drugs. In addition to these modifications in rrs, mutations in the tlyA gene, which encodes a putative rRNA methyltransferase, are suggested to confer resistance to CAP (22). The mutations at positions 1401, 1402, and 1484 in rrs do not confer resistance to streptomycin (STR). STR resistance is associated with point mutations (i) in the rpsL gene encoding the ribosomal protein S12, and (ii) in the S12-interacting regions of the 16S rRNA gene, rrs near positions A514 to C517 and C904 to A907 (13, 26). Over the past 3 decades, several investigations showed that the mutations involved in resistance to STR do not confer cross-resistance to AMK, KAN, or CAP (19).
Ethambutol (ETB) inhibits arabinosyltransferases encoded by the embCAB operon, which encompasses three contiguous genes, namely, embC, embA, and embB, that are involved in the biosynthesis of the cell wall components arabinogalactan and lipoarabinomannan (28). ETB resistance is reported to be most frequently associated with mutations in the embCAB operon, particularly, mutations in embB codon 306. The alterations identified in this codon are thought to represent a rapid screening method for the detection of ETB resistance in clinical isolates (14, 16, 29). However, several reports indicate that only 50% of ETB-resistant M. tuberculosis isolates carry a mutation in embB codon 306 (10, 23, 24, 29).
Since WHO advises the use of at least 4 active drugs against MDR TB, the rapid determination of drug resistance is an important prerequisite for the initiation of effective chemotherapy to successfully treat patients and prevent additional resistance traits leading to treatment failure from being acquired (32). Molecular assays were developed to quickly predict drug resistance in clinical isolates of M. tuberculosis. A DNA strip assay based on a combination of PCR and reverse hybridization testing the rpoB and katG genes (the GenoType MTBDR test and its extended version, MTBDRplus) was developed to detect resistance to rifampin (RIF) and isoniazid (INH) (i.e., MDR strains) (3, 4, 11). Very recently, a new DNA strip assay, the GenoType Mycobacterium tuberculosis drug-resistant second-line assay (MTBDRsl; Hain Lifescience), was developed to detect resistance to ethambutol, fluoroquinolones, and injectable aminoglycosides/cyclic peptides, i.e., to detect XDR strains among MDR strains. In a very recent report, Hillemann et al. presented the first evaluation of the MTBDRsl test (12). In this study, the MTBDRsl test detected mutations in 91% of strains resistant to FQ, 85% resistant to AMK, 87% resistant to CAP, and 69% of the strains resistant to ETB (12).
The goal of the present study was to evaluate the ability of the GenoType MTBDRsl DNA strip assay to detect resistance to fluoroquinolones, aminoglycosides/cyclic peptides, and ethambutol in MDR clinical isolates displaying a wide variety of molecular mechanisms of resistance and representing a broad range of additional resistance to ETB, FQ, second-line aminoglycosides and/or cyclic peptide (KAN, AMK, and CAP), and STR, including XDR strains. The molecular mechanisms accounting for drug resistance in these strains were first established by DNA sequencing of the resistance-determining regions in the following genes: gyrA and gyrB for fluoroquinolones, rpsL and rrs for streptomycin, rrs for kanamycin-amikacin-capreomycin, tlyA for capreomycin, and embB for ethambutol. The results of the MTBDRsl test were compared to the sequencing data and to the results of phenotypic drug susceptibility testing (DST).
MATERIALS AND METHODS
Strains.
Forty-nine Mycobacterium tuberculosis complex clinical isolates (identified by GenoType Mycobacterium CM) isolated in France and received by the French Reference Center for Mycobacteria during the study period (2005 to 2009) were included based on their MDR (n = 41) or XDR (n = 8) status. Most strains also had resistance to FQ, aminoglycosides/cyclic peptide, or ethambutol. Overall, the 8 XDR strains displayed the following additional resistance patterns: ofloxacin (OFX), CAP, and ETB (n = 1); OFX, KAN, and ETB (n = 1); OFX, KAN, AMK, and CAP (n = 1); OFX, KAN, AMK, and ETB (n = 1); and OFX, KAN, AMK, CAP, and ETB (n = 4). The 41 MDR strains displayed the following additional resistance patterns: OFX (n = 5); OFX and ETB (n = 9); ETB (n = 8); KAN (n = 1); KAN and ETB (n = 1); KAN, AMK, and CAP (n = 1); KAN, AMK, CAP, and ETB (n = 3); and no additional resistance (n = 13). We also included 3 non-MDR strains selected on the basis of their resistance to fluoroquinolones or second-line aminoglycosides/cyclic peptide (one was susceptible to both RIF and INH and resistant to CAP, one was resistant to RIF and OFX and susceptible to INH, and one was susceptible to RIF and resistant to INH and OFX). In vitro drug susceptibility testing for OFX, STR, AMK, KAN, CAP, and ETB was performed on Lowenstein-Jensen medium following the proportions method (5), using the following concentrations: 2 mg/liter for OFX; 4 mg/liter for STR; 20 mg/liter (each) for AMK, KAN, and CAP; and 2 mg/liter for ETB (30).
DNA sequencing of drug resistance-associated genes.
Genomic DNA was isolated from bacteria grown on Lowenstein-Jensen medium. A loop of culture was suspended in water (500 μl) and heated at 95°C for 15 min. The DNA used for PCR amplification was obtained by heat shock extraction (1 min at 95°C and 1 min on ice, repeated five times). A volume of 5 μl was used in PCR amplifications using the oligonucleotide primers described below. For fluoroquinolone resistance, the quinolone resistance-determining regions of the gyrA and gyrB genes were amplified and sequenced using primers PRI8 (5′-YGGTGGRTCRTTRCCYGGCGA-3′) and PRI9 (5′-CGCCGCGTGCTSTATGCRATG-3′) for gyrA and primers gyrBa (5′-GAGTTGGTGCGGCGTAAGAGC-3′) and gyrBe (5′-CGGCCATCAAGCACGATCTTG-3′) for gyrB (8). For aminoglycosides/cyclic peptide resistance, the rrs, rpsL, and tlyA genes were amplified and sequenced using primers TB53 (5′-GATGACGGCCTTCGGGTTGT-3′) and TB54 (5′-TCTAGTCTGCCCGTATCGCC-3′) for the region from positions 513 to 516 in the rrs gene (13), primers TB55 (5′-GTAGTCCACGCCGTAAACGG-3′) and TB56 (5′-AGGCCACAAGGGAACGCCTA-3′) for the region around position 907 in the rrs gene (13), primers RRSA (5′-GGCGTTCCCTTGTGGCCTGTG-3′) (primer designed in-house) and RRS1539 (5′-GGGGCGTTTTGCTGGTGCTCC-3′) (1) for the region from positions 1401 to 1484 in the rrs gene, primers rpsl1 (5′-CCAACCATCCAGCAGCTGGTC-3′) and rpsl2 (5′-ATCCAGCGAACCGCGGATGA-3′) for the rpsL gene (primers designed in-house), and primers tlyAs (5′-GCATCGCACGTCGTCTTT-3′) and tlyAas (5′-GGTCTCGGTGGCTTCGTC-3′) for the tlyA gene (9). With regard to ETB resistance, the embB gene in the region of codon M306 was amplified using primers embBs (5′-CCGACCACGCTGAAACTG-3′) and embBas (5′-GTAATACCAGCCGAAGGGATCCT-3′) (15). After amplification, unincorporated nucleotides and primers were removed by filtration with Microcon 100 microconcentrators (Amicon Inc., Beverly, MA), and the amplicons were sequenced using the Big Dye Terminator cycle sequencing-ready kit following the manufacturer's instructions.
MTBDRsl test.
The GenoType MTBDRsl DNA strip is coated with 22 probes (see Fig. 1). Briefly, resistance to fluoroquinolones was based on the use of 3 wild-type probes covering GyrA codons 85 to 97. The presence of the most frequently observed mutations are confirmed by positive hybridization with 6 mutant probes (A90V, S91P, D94A, D94N/Y, D94G, and D94H; see Fig. 1). For aminoglycosides/cyclic peptide resistance, two wild-type probes cover nucleotides 1401 and 1402 and nucleotide 1484, and two mutant probes specifically detect the A1401G and G1484T exchanges. For ethambutol resistance, one wild-type probe covers codon 306, and the presence of the most frequently observed mutations, M306V and M306I, is confirmed by positive hybridization with two mutant probes (for M306I, only the base change from ATG to ATA is detected by MTBDRsl, while base changes to ATC or to ATT are not). The amplification reactions and the GenoType MTBDRsl test were performed as recommended by the manufacturer, using 5 μl of extracted DNA.
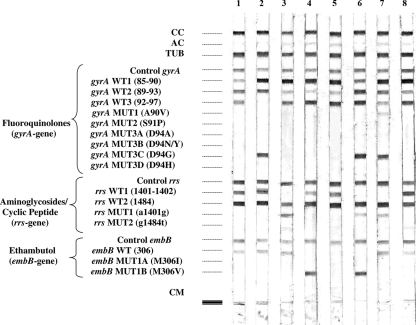
FIG. 1.
Hybridization patterns obtained with the GenoType MTBDRsl assay. The controls, targeted genes, and mutations are given to the left of the figure: CC, conjugate control; AC, amplification control (23S rRNA); TUB, M. tuberculosis complex-specific control (23S rRNA); Control gyrA, control for gyrA amplification; gyrA WT1 to WT3, gyrA wild-type probes located in regions from codons 85 to 97; gyrA MUT1 to MUT3D, gyrA mutant probes testing mutations for codons A90V, S91P, D94A, D94N/Y, D94G, and D94H; Control rrs, control for rrs amplification; rrs WT1 and WT2, rrs wild-type probes located in regions for nucleotides 1401 and 1402 and nucleotide 1484; rrs MUT1 and MUT2, rrs mutant probes testing mutations for A1401G and G1484T; Control embB, amplification control for embB; embB WT, embB wild-type probe located in codon 306; embB MUT1A and MUT1B, mutant probes testing mutations M306V and M306I (base exchange at codon 306 [ATG→ATA]); CM, colored marker. Typical hybridization patterns were obtained and are shown in the figure as follows: lane 1, H37Rv (wild type); lane 2, gyrA D94G; lane 3, rrs A1401G; lane 4, embB M306V; lane 5, embB M306I with base exchange ATG→ATC; lane 6, gyrA D94G+wt (gyrA D94G and wild type) and rrs A1401G and embB M306V; lane 7, gyrA A90V plus gyrA D94G (mixture of two mutants) and rrs A1401G; lane 8, MDR with wild-type gyrA, rrs, and embB genes showing a weak signal intensity for embB wild-type probe, and a weak false-positive hybridization signal for rrs MUT2 (G1484T) mutation probe.
Nucleotide sequence accession numbers.
The nucleotide sequences determined for the mutant genes included in the present report were deposited into the GenBank database under the following accession numbers: GU323380 to GU323383 for the GyrB mutants S458F, N510D, D472A, and T511P, respectively; GU323384 to GU323394 for the GyrA mutants D94A, D94G, D94N, D94H, G88A, G88C, T80A-A90E, T80A-A90G, T80A-A90G-D94G, D94G-A90V, and A90V, respectively; GU323395 to GU323398 for the EmbB mutants M306V, M306I (ATC), M306I (ATA), and M306I (ATT), respectively; GU323399 to GU323400 for the RpsL mutants K43R and K88R, respectively; and GU3233401 to GU3233405 for the rrs mutants A513C, C516T, A907C, A1401G, and G1484T, respectively.
RESULTS
Representative patterns obtained with the GenoType MTBDRsl test are shown in Fig. 1. The MTBDRsl DNA strip assay produced easily interpretable results, although some of the probes included in the test yielded rather weak hybridization intensities. In particular, the wild-type probe for embB often produced faint bands for strains with a wild-type embB gene (Fig. 1, lane 8), but those bands were considered positive, since their intensities were about as strong as or stronger than that of the amplification control (AC) band (Fig. 1). Conversely, very weak bands showing a hybridization intensity less than that of the amplification control band were occasionally observed with the rrs MUT2 (G1484T) mutation probe for strains confirmed by DNA sequencing to have a wild-type “G” nucleotide in position 1484 (Fig. 1, lane 8). Such bands were finally considered to be hybridization artifacts, although one cannot exclude the possibility that they could represent the ongoing development of heteroresistance.
GenoType MTBDRsl test results obtained for the detection of resistance to fluoroquinolones.
Of the 24 fluoroquinolone-resistant (FQ-R) strains, 21 were identified by DNA sequencing to display various mutations in the GyrA QRDR, corresponding either to mutations frequently reported in the literature (mutations in codons 90 and 94) or more rarely encountered mutations (codons 88 and 80) (Table 1). Of the 21 FQ-R strains with mutations in the GyrA QRDR, 17 were detected directly with MTBDRsl by hybridization with the gyrA probes, MUT1 and MUT3A to MUT3D (Table 1). Positive hybridization signals were simultaneously observed in one strain for the D94G mutation and the corresponding wild-type band, WT3 (Fig. 1, lane 6), corresponding to a significant proportion of resistant mutants among a wild-type population. It is worth noting that D94H is described in the MTBDRsl instruction manual (9a) as a mutation predicted from in silico studies and not previously detected in vitro, a prediction that is now confirmed in vivo. Four of the other 21 FQ-resistant strains, corresponding to those with the less frequent mutations G88A, G88C, and T80A-A90E were detected indirectly by lack of hybridization with the wild-type probes, WT1 and WT2 (since codon 80 is not included in the MTBDRsl assay, the mutation at this position was detected only by DNA sequencing). The last 3 FQ-resistant strains (3/24 strains [12%]) had no mutation in GyrA but were instead mutated in GyrB in codons T511P, D472A, or N510D, respectively (Table 1). These 3 strains were not detected by MTBDRsl, since the GyrB QRDR is not included in the assay.
TABLE 1.
GenoType MTBDRsl test results for the detection of fluoroquinolone resistance in 52 M. tuberculosis strainsa
| No. of strains | Fluoroquinolone resistance phenotypeb | Sequencing data [amino acid(s) change or wt] |
No. of strains with the following result by the MTBDRsl test: |
||
|---|---|---|---|---|---|
| GyrA | GyrB | wt | Mutationc | ||
| 24 | R | T80A-A90E | wt | 1 (ΔWT2) | |
| T80A-A90G-D94G | wt | 1 (ΔWT2, ΔWT3, MUTD94G) | |||
| G88A | wt | 2 (ΔWT1) | |||
| G88C | wt | 1 (ΔWT1) | |||
| A90V | wt | 4 (ΔWT2, MUTA90V) | |||
| D94G | wt | 5 (ΔWT3, MUTD94G) | |||
| D94N | wt | 2 (ΔWT3, MUTD94N) | |||
| D94A | wt | 2 (ΔWT3, MUTD94A) | |||
| D94H | wt | 1 (ΔWT3, MUTD94H) | |||
| D94G+wt | wt | 1 (MUTD94G) | |||
| D94G+A90V | wt | 1 (MUTA90V, MUTD94G) | |||
| wt | T511P | 1 | |||
| wt | D472A | 1 | |||
| wt | N510D | 1 | |||
| 28 | S | wt | wt | 26 | |
| wt | S458F | 1 | |||
| T80A-A90G | wt | 1 (ΔWT2) | |||
wt, wild-type.
R, resistant; S, susceptible.
ΔWT, lack of hybridization with the indicated wild-type probe (WT1, WT2, or WT3).
Regarding the 28 strains susceptible to fluoroquinolones, 26 were found by DNA sequencing to have no mutation in the GyrA and GyrB QRDR and were considered to be wild type by the MTBDRsl assay (Table 1). Among the 2 remaining FQ-susceptible strains, one had a wild-type GyrA QRDR and an S458F mutation in GyrB not detected by MTBDRsl, while the other had a double mutation, T80A-A90G in GyrA. The mutation T80A was not detected by MTBDRsl, and A90G was detected indirectly by lack of hybridization with the WT2 probe.
MTBDRsl results for resistance to aminoglycosides/cyclic peptide.
The 9 strains resistant to the 3 second-line aminoglycosides/cyclic peptide KAN, AMK, and CAP had a mutation in the region from positions 1400 to 1500 of the rrs gene known to provide resistance to these drugs: A1401G in 8 strains and G1484T in 1 strain (Table 2). These 9 strains were directly detected by MTBDRsl by hybridization with one of the rrs probes, MUT1 or MUT2. One additional strain resistant to KAN and AMK (but susceptible to CAP) that also had the A1401G mutation was also directly detected by MTBDRsl. In contrast, 5 strains resistant to either CAP (n = 2) or KAN (n = 3) had no mutation in region 1400 of rrs, and were not identified by the MTBDRsl test (Table 2). Analysis of the tlyA gene, recently suggested to be involved in resistance to CAP, indicated that none of the strains resistant to aminoglycosides/cyclic peptide had a mutation in this gene.
TABLE 2.
GenoType MTBDRsl test results for the detection of resistance to aminoglycosides and cyclic peptide in 52 M. tuberculosis strainsa
| No. of strains | Drug resistance phenotypeb |
Sequencing data [nucleotide or amino acid change(s) or wt] |
No. of strains with the following result by the MTBDRsl test: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KAN | AMK | CAP | STR |
rrs [nucleotide change(s) or wt] |
RpsL (amino acid change or wt) | wt | Mutationc | |||
| 1400 region | 510 region | 912 region | ||||||||
| 8 | R | R | R | R | A1401G | wt | wt | K43R | 4 (ΔWT1, MUTA1401G) | |
| A1401G | wt | wt | K88R | 2 (ΔWT1, MUTA1401G) | ||||||
| A1401G | A513C | wt | wt | 1 (ΔWT1, MUTA1401G) | ||||||
| G1484T | wt | A907C | wt | 1 (ΔWT2, MUTG1484T) | ||||||
| 1 | R | R | R | S | A1401G | wt | wt | wt | 1 (ΔWT1, MUTA1401G) | |
| 1 | R | R | S | S | A1401G | wt | wt | wt | 1 (ΔWT1, MUTA1401G) | |
| 2 | S | S | R | R | wt | wt | wt | wt | 2 | |
| 3 | R | S | S | R | wt | wt | wt | wt | 2 | |
| wt | C516T | wt | wt | 1 | ||||||
| 25 | S | S | S | R | wt | wt | wt | wt | 7 | |
| wt | A513C | wt | wt | 1 | ||||||
| wt | wt | wt | K43R | 13 | ||||||
| wt | wt | wt | K88R | 2 | ||||||
| wt | C516T | wt | wt | 1 | ||||||
| wt | wt | A907C+wt | wt | 1 | ||||||
| 12 | S | S | S | S | wt | wt | wt | wt | 12 | |
wt, wild-type.
AMK, amikacin; CAP, capreomycin; KAN, kanamycin; STR, streptomycin; R, resistant; S, susceptible.
ΔWT, lack of hybridization with the wild-type probe (WT1 or WT2).
Among the 25 strains displaying isolated resistance to streptomycin, 18 were identified by DNA sequencing to have mutations responsible for STR resistance in the rpsL gene (K43R and K88R) or in the rrs gene (A513C, C516T, and A907C). None of these displayed mutations in the 1400 region of rrs, either by DNA sequencing or by MTBDRsl testing, in accordance with their susceptibility to KAN, AMK, and CAP. Finally, as expected, the 12 strains susceptible to the 4 aminoglycosides/cyclic peptide exhibited a wild-type pattern by both DNA sequencing and MTBDRsl (Table 2).
Analysis of the MTBDRsl results for detecting resistance to ethambutol.
Among the 28 strains resistant to ETB, 14 had the frequently reported mutation M306V and were unambiguously identified by MTBDRsl (Table 3). Two other ETB-R strains had an M306I mutation in EmbB. The first (ATG→ATA) was directly detected by MTBDRsl, and the other (ATG→ATC) was indirectly detected by the lack of hybridization with the wild-type probe (Table 3). In contrast, the 12 remaining ETB-resistant strains (43%) showed no mutation in embB by DNA sequencing and yielded wild-type patterns using the MTBDRsl assay. In total, the MTBDRsl strip assay detected 16/28 (57%) strains resistant to ETB. Among the 24 strains susceptible to ETB, 22 had no mutation at codon 306 of the embB gene and exhibited a wild-type pattern with the MTBDRsl assay. Unexpectedly, the last 2 ETB-susceptible strains, as confirmed by repeated independent tests, had either an M306V mutation detected directly by the probe embB MUT1B or an M306I mutation determined by the base change ATG→ATT detected indirectly with MTBDRsl by lack of hybridization with the WT probe (Table 3).
TABLE 3.
GenoType MTBDRsl test results for the detection of ethambutol resistance in 52 M. tuberculosis strainsa
| No. of strains | Ethambutol resistance phenotypeb | No. of strains with the following result for the embB gene by sequencing: |
No. of strains with the following result by the MTBDRsl test: |
|||
|---|---|---|---|---|---|---|
| wt | M306V | M306I | wt | Mutationc | ||
| 28 | R | 14 | 14 (ΔWT, MUTM306V) | |||
| 1 | 1 (ΔWT, MUTM306I)d | |||||
| 1 | 1 (ΔWT)e | |||||
| 12 | 12 | |||||
| 24 | S | 22 | 22 | |||
| 1 | 1 (ΔWT, MUTM306V) | |||||
| 1 | 1 (ΔWT)f | |||||
wt, wild-type.
R, resistant; S, susceptible.
ΔWT, lack of hybridization with the the wild-type probe.
M306I, base exchange (ATG→ATA) at codon 306.
M306I, base exchange (ATG→ATC [not detected directly]) at codon 306.
M306I, base exchange (ATG→ATT [not detected directly]) at codon 306.
Sensitivity and specificity.
The sensitivity and specificity of the MTBDRsl test were calculated by comparing the results from the DNA strip assay to those of the in vitro phenotypic testing taken as a reference. The sensitivity values of the MTBDRsl test to detect resistance to different drugs were as follows: 87% (21/24) for fluoroquinolones, 77% (10/13) for kanamycin, 100% (10/10) for amikacin, 80% (9/11) for capreomycin, and 57% (16/28) for ethambutol. The specificity values of the MTBDRsl test to detect susceptibility to different drugs were as follows: 96% (27/28) for fluoroquinolones, 100% (39/39) for kanamycin, 100% (42/42) for amikacin, 98% (40/41) for capreomycin, and 92% (22/24) for ethambutol. The sensitivity and specificity values of DNA sequencing were identical to the values worked out for MTBDRsl for aminoglycosides/cyclic peptide and ethambutol but higher for fluoroquinolones. For fluoroquinolones, sequencing the gyrB gene improved the sensitivity (24/24 [100%]).
DISCUSSION
When MDR TB is detected, the main therapeutic issue that must be addressed is to determine the susceptibility of the strain to the remaining first-line drugs, i.e., streptomycin, ethambutol, and pyrazinamide, and to second-line drugs, particularly aminoglycosides/cyclic peptide (KAN, AMK, and CAP) and fluoroquinolones. Since in vitro testing is particularly cumbersome and difficult to interpret for second-line drugs, there is a major interest in rapid molecular detection methods for resistance to these drugs. In the present study, we assessed the capacity of the GenoType MTBDRsl test to detect mutations linked to resistance to ETB and second-line drugs on patterns of MDR TB showing a wide range of combined additional resistance to FQ, AMK, KAN, CP, ETB, and STR, for which the number of available drugs can be very limited, particularly in the case of XDR TB.
Among the 24 fluoroquinolone-resistant strains included in the present study, 21 (87.5%) had mutations in GyrA, which were all detected by the MTBDRsl test, confirming the capacity of the test to identify FQ resistance caused by mutations in the GyrA QRDR. However, it is worth mentioning that a clinical isolate phenotypically susceptible to FQ and having the double mutation T80A-A90G was identified as being resistant by the MTBDRsl assay. This discrepant result can be explained because the T80A substitution is linked to a significant decrease in the level of resistance to FQ and, when combined with A90G, confers hypersusceptibility to ofloxacin, with MIC values up to 4-fold lower than those observed for a wild-type strain (2). Conversely, the A90E mutation found in combination with T80A in a clinical isolate resistant to FQ was shown, when present alone, to confer high-level resistance to FQ, and its association with T80A still results in FQ resistance (A. Aubry, unpublished data). Therefore, the corresponding clinical isolate was correctly identified as resistant by the MTBDRsl test. Taken together, these data highlight that detection, either directly by DNA sequencing or indirectly by MTBDRsl (e.g., by lack of hybridization with the wild-type probes), of GyrA or GyrB mutations whose phenotypic consequences are not established should be interpreted with caution.
A significant finding of our study was the presence of mutations in the GyrB QRDR in 3 (12.5%) fluoroquinolone-resistant isolates. The two mutations, D472A and N510D, were previously reported to represent the most frequent GyrB mutations causing FQ resistance in M. tuberculosis (2, 27). The third mutation, T511P, along with other mutations at positions 510, 511, and 512, were reported in Mycobacterium smegmatis and M. tuberculosis FQ-resistant strains selected in vitro (33). These mutations represent a significant limitation of the line probe test compared with DNA sequencing. Their inclusion in the MTBDRsl DNA strip would significantly improve the capacity of the test to detect FQ resistance in the clinical setting. Finally, it is of interest to note that the GyrB S458F mutation, detected only by DNA sequencing in the present study, was found in a strain susceptible to FQ (Table 1). This previously unreported mutation, located far from the GyrB QRDR, was shown by site-directed mutagenesis to have no effect on the FQ pattern (Aubry, unpublished).
The nucleotide changes in the region from positions 1400 to 1500 of the rrs gene found in 10 strains were all detected by the MTBDRsl test, indicating that the assay performs well in detecting the presence of these mutations. All these mutants were resistant to the aminoglycosides/cyclic peptide KAN, AMK, and CAP, except for one strain, which remained susceptible to capreomycin with an A1401G mutation. A low level of CAP resistance not detected in conventional phenotypic tests could explain this discrepancy. Conversely, 5 strains susceptible to AMK but resistant to either KAN only or to CAP only were not detected by MTBDRsl and did not carry nucleotide changes in the region from positions 1400 to 1500 of rrs. Therefore, it is likely that other yet-uncharacterized molecular mechanisms are the cause of the observed monoresistance to KAN or CAP in these strains. Among other plausible causes of isolated resistance to capreomycin in M. tuberculosis, mutations inactivating the tlyA gene encoding a 2′-O-methyltransferase were previously reported by Feuerriegel et al. (9). However, such mutations were not detected by DNA sequencing in our study, and further investigations are needed to elucidate the causes of monoresistance to KAN or CAP.
In the present study, more than a third (12/28) of ETB-resistant strains had no mutation in codon 306, accounting for the low sensitivity of the test for this drug (57%). There are some discrepancies in the literature between the phenotypic and genotypic characterization of ETB-resistant strains. On one hand, several authors found a strong correlation between ETB resistance and the presence of mutations in codon M306 and suggested that these mutations can be regarded as a marker for ETB resistance in diagnostic assays (14, 16, 29). On the other hand, it was reported that mutations in codon 306 in embB can also be found in ETB-susceptible strains (2/24 in the present study), and are present in only 30% to 68% of all ETB-resistant clinical isolates. Therefore, they were also suggested to represent a marker of the predisposition for clinical isolates to develop antibiotic resistance rather than a specific marker for resistance to ETB (10, 23, 25). Taken together, these data highlight the fact that the molecular basis of ETB resistance in M. tuberculosis is still insufficiently understood to allow detection of ETB resistance as well as ETB susceptibility to rely only on genotypic testing (sensitivity of 57% and negative predictive value of 65%).
In conclusion, the GenoType MTBDRsl assay is a promising test for rapidly detecting the most common mutations involved in resistance to fluoroquinolones and aminoglycosides/cyclic peptide in MDR isolates, and concomitantly XDR strains among MDR isolates. However, because the mechanisms of resistance to antituberculous drugs are complex and still not fully understood, the present version of the MTBDRsl test shows some limitations in detecting resistance to ethambutol and, to a lesser extent, monoresistance to aminoglycosides, as well as resistance to fluoroquinolones. Compared with the results of the first study on MTBDRsl published recently (12) (which did not include kanamycin resistance testing), our results are more optimistic for amikacin, since they suggest that the MTBDRsl assay performs very well in predicting resistance or susceptibility to amikacin among MDR strains (the positive and negative predictive values are both 100%). Conversely, the results are less optimistic for capreomycin, fluoroquinolones, and ethambutol. Indeed, the results of the assay are difficult to interpret for ETB, an important component of MDR treatment, because about a third of resistant strains are not detected, whereas about a tenth of susceptible strains are detected as being resistant. For KAN, CAP, and FQ, the accuracy of the assay in predicting susceptibility seems to be somewhat limited by mutations not included in the test (particularly in gyrB) or as-yet uncharacterized resistance mechanisms (particularly those related to aminoglycosides/cyclic peptide monoresistance). This result makes the decision to consider the strain susceptible, as in the case of the wild-type result, less reliable, particularly in the context of patients with MDR TB for whom there is an absolute need to determine a fully active treatment (32). To illustrate this point, the MTBDRsl test failed to detect 2/8 XDR strains (25%) among the MDR strains in our study. One of these two strains showed an isolated resistance to capreomycin (wild-type rrs), and the other had an isolated resistance to KAN (wild-type rrs) and a gyrB amino acid substitution conferring resistance to fluoroquinolones (the failure to detect FQ resistance would have a serious impact, since the use of fluoroquinolones, when still active, is considered a positive prognostic factor in the treatment of MDR TB) (6). Finally, it is worth noting that some of our MDR strains that were resistant to second-line aminoglycosides/cyclic peptide were still susceptible to STR. The emergence of XDR strains of M. tuberculosis has renewed an interest in streptomycin; hence, this aminoglycoside could again play a valuable role in the treatment of MDR or XDR tuberculosis (in our study, 2/8 XDR strains remained susceptible to streptomycin). The inclusion of streptomycin resistance determinants on the MTBDRsl DNA strip (rpsL codons 43 and 88 and rrs positions 513, 516, and 907) would, therefore, represent another significant improvement in the test.
Acknowledgments
This work was supported by a grant from the Ministère de la Recherche (program grant UPRES EA 1541).
Footnotes
Published ahead of print on 24 March 2010.
REFERENCES
- 1.Alangaden, G. J., B. N. Kreiswirth, A. Aouad, M. Khetarpal, F. R. Igno, S. L. Moghazeh, E. K. Manavathu, and S. A. Lerner. 1998. Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry, A., N. Veziris, E. Cambau, C. Truffot-Pernot, V. Jarlier, and L. M. Fisher. 2006. Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes. Antimicrob. Agents Chemother. 50:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brossier, F., N. Veziris, C. Truffot-Pernot, V. Jarlier, and W. Sougakoff. 2006. Performance of the genotype MTBDR line probe assay for detection of resistance to rifampin and isoniazid in strains of Mycobacterium tuberculosis with low- and high-level resistance. J. Clin. Microbiol. 44:3659-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brossier, F., N. Veziris, V. Jarlier, and W. Sougakoff. 2009. Performance of MTBDRplus for detecting high/low levels of Mycobacterium tuberculosis resistance to isoniazid. Int. J. Tuber. Lung Dis. 13:260-265. [PubMed] [Google Scholar]
- 5.Canetti, G., N. Rist, and J. Grosset. 1963. Measurement of sensitivity of the tuberculous bacillus to antibacillary drugs by the method of proportions. Methodology, resistance criteria, results and interpretation. Rev. Tuberc. Pneumol. (Paris) 27:217-272. [PubMed] [Google Scholar]
- 6.Chan, E. D., V. Laurel, M. J. Strand, J. F. Chan, M. L. Huynh, M. Goble, and M. D. Iseman. 2004. Treatment and outcome analysis of 205 patients with multidrug-resistant tuberculosis. Am. J. Respir. Crit. Care Med. 169:1103-1109. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, A. F., W. W. Yew, E. W. Chan, M. L. Chin, M. M. Hui, and R. C. Chan. 2004. Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dauendorffer, J. N., I. Guillemin, A. Aubry, C. Truffot-Pernot, W. Sougakoff, V. Jarlier, and E. Cambau. 2003. Identification of mycobacterial species by PCR sequencing of quinolone resistance-determining regions of DNA gyrase genes. J. Clin. Microbiol. 41:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feuerriegel, S., H. S. Cox, N. Zarkua, H. A. Karimovich, K. Braker, S. Rüsch-Gerdes, and S. Niemann. 2009. Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob. Agents Chemother. 53:3353-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Hain Lifescience GmbH. 2009. GenoType MTBDRsl: instruction manual. Hain Lifescience GmbH, Nehren, Germany.
- 10.Hazbón, M. H., M. Bobadilla del Valle, M. I. Guerrero, M. Varma-Basil, I. Filliol, M. Cavatore, R. Colangeli, H. Safi, H. Billman-Jacobe, C. Lavender, J. Fyfe, L. García-García, A. Davidow, M. Brimacombe, C. I. León, T. Porras, M. Bose, F. Chaves, K. D. Eisenach, J. Sifuentes-Osornio, A. Ponce de León, M. D. Cave, and D. Alland. 2005. Role of embB codon 306 mutations in Mycobacterium tuberculosis revisited: a novel association with broad drug resistance and IS6110 clustering rather than ethambutol resistance. Antimicrob. Agents Chemother. 49:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillemann, D., S. Rüsch-Gerdes, and E. Richter. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 45:2635-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillemann, D., S. Rüsch-Gerdes, and E. Richter. 2009. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 47:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honoré, N., and S. T. Cole. 1994. Streptomycin resistance in mycobacteria. Antimicrob. Agents Chemother. 38:238-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isola, D., M. Pardini, F. Varaine, S. Niemann, S. Rüsch-Gerdes, L. Fattorini, G. Orefici, F. Meacci, C. Trappetti, M. Rinaldo Oggioni, the LONG-DRUG Study Group, and G. Orrù. 2005. A pyrosequencing assay for rapid recognition of SNPs in Mycobacterium tuberculosis embB306 region. J. Microbiol. Methods 62:113-120. [DOI] [PubMed] [Google Scholar]
- 15.Jain, A., R. Mondal, S. Srivastava, R. Prasad, K. Singh, and R. C. Ahuja. 2008. Novel mutations in embB gene of ethambutol resistant isolates of Mycobacterium tuberculosis: a preliminary report. Indian J. Med. Res. 128:634-639. [PubMed] [Google Scholar]
- 16.Johnson, R., A. M. Jordaan, L. Pretorius, E. Engelke, G. van der Spuy, C. Kewley, M. Bosman, P. D. van Helden, R. Warren, and T. C. Victor. 2006. Ethambutol resistance testing by mutation detection. Int. J. Tuber. Lung Dis. 10:68-73. [PubMed] [Google Scholar]
- 17.Kliiman, K., and A. Altraja. 2009. Predictors of poor treatment outcome in multi- and extensively drug-resistant pulmonary TB. Eur. Respir. J. 33:1085-1094. [DOI] [PubMed] [Google Scholar]
- 18.Kocagöz, T., C. J. Hackbarth, I. Unsal, E. Y. Rosenberg, H. Nikaido, and H. F. Chambers. 1996. Gyrase mutations in laboratory-selected, fluoroquinolone-resistant mutants of Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 40:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krüüner, A., P. Jureen, K. Levina, S. Ghebremichael, and S. Hoffner. 2003. Discordant resistance to kanamycin and amikacin in drug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:2971-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matrat, S., N. Veziris, C. Mayer, V. Jarlier, C. Truffot-Pernot, J. Camuset, E. Bouvet, E. Cambau, and A. Aubry. 2006. Functional analysis of DNA gyrase mutant enzymes carrying mutations at position 88 in the A subunit found in clinical strains of Mycobacterium tuberculosis resistant to fluoroquinolones. Antimicrob. Agents Chemother. 50:4170-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maus, C. E., B. B. Plikaytis, and T. M. Shinnick. 2005. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:3192-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maus, C. E., B. B. Plikaytis, and T. M. Shinnick. 2005. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mokrousov, I., T. Otten, B. Vyshnevskiy, and O. Narvskaya. 2002. Detection of embB306 mutations in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis from Northwestern Russia: implications for genotypic resistance testing. J. Clin. Microbiol. 40:3810-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plinke, C., S. Rüsch-Gerdes, and S. Niemann. 2006. Significance of mutations in embB codon 306 for prediction of ethambutol resistance in clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 50:1900-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safi, H., B. Sayers, M. H. Hazbón, and D. Alland. 2008. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob. Agents Chemother. 52:2027-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreevatsan, S., X. Pan, K. E. Stockbauer, D. L. Williams, B. N. Kreiswirth, and J. M. Musser. 1996. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob. Agents Chemother. 40:1024-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takiff, H. E., L. Salazar, C. Guerrero, W. Philipp, W. M. Huang, B. Kreiswirth, S. T. Cole, W. R. Jacobs, Jr., and A. Telenti. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Telenti, A., W. J. Philipp, S. Sreevatsan, C. Bernasconi, K. E. Stockbauer, B. Wieles, J. M. Musser, and W. R. Jacobs, Jr. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 3:567-570. [DOI] [PubMed] [Google Scholar]
- 29.Van Rie, A., R. Warren, I. Mshanga, A. M. Jordaan, G. D. van der Spuy, M. Richardson, J. Simpson, R. P. Gie, D. A. Enarson, N. Beyers, P. D. van Helden, and T. C. Victor. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J. Clin. Microbiol. 39:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization. 2001. Guidelines for drug susceptibility testing for second-line anti-tuberculosis drugs for DOTS-Plus. WHO/CDS/TB/2001.288. World Health Organization, Geneva, Switzerland.
- 31.World Health Organization. 2008. Anti-tuberculosis drug resistance in the world. Fourth global report. WHO/HTM/TB/2008.394. World Health Organization, Geneva, Switzerland.
- 32.World Health Organization. 2008. Guidelines for the programmatic management of drug-resistant tuberculosis: emergency update 2008. WHO/HTM/TB/2008.402. World Health Organization, Geneva, Switzerland.
- 33.Zhou, J., Y. Dong, X. Zhao, S. Lee, A. Amin, S. Ramaswamy, J. Domagala, J. M. Musser, and K. Drlica. 2000. Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J. Infect. Dis. 182:517-525. [DOI] [PubMed] [Google Scholar]