Abstract
Torovirus, a member of the Coronaviridae family, is a gastrointestinal infectious agent that has been identified in humans, cattle, pigs, and equines. Toroviruses, except equine torovirus, are difficult to propagate in cell culture; indeed, to date, only the Aichi/2004 strain of bovine torovirus (BToV) has been isolated among the human, bovine, and porcine toroviruses. In the present study, four cytopathogenic BToVs were isolated from diarrheal feces of the cattle using the HRT-18 cell line, and their genetic and antigenic properties were compared. The cytopathogenic features of BToV isolates in HRT-18 cells were similar to those of the Aichi/2004 strain. However, none of the isolates showed cytopathogenic effects in the HRT-18 cells of different origin, suggesting that one significant factor contributing to the cytopathogenicity of BToV depends on properties of the HRT-18 cells themselves. All BToVs isolated were able to agglutinate mouse, but not chicken, erythrocytes, while they lacked receptor-destroying enzyme activity. Analysis of the N terminus of the spike gene showed that three isolates, but not the Gifu-2007TI/E strain, were phylogenetically located in cluster 1 and its analogs and revealed high cross-reactivity with each other, as demonstrated by neutralization (NT) and hemagglutination inhibition (HI) assays. The Gifu-2007TI/E strain was classified close to cluster 2 and exhibited relatively low cross-reactivity with these viruses; however, the difference was not sufficient to classify BToVs into serotypes, suggesting that at least two subtypes distinguishable by the structure of the N terminus of the spike gene and that both NT and HI tests may be exist.
The family Coronaviridae consists of enveloped and positive-stranded RNA viruses, specifically coronaviruses and toroviruses (2, 3). Toroviruses identified thus far have been further classified into human, bovine, porcine, and equine toroviruses on the basis of their hosts. Several studies have established associations between human and bovine toroviruses with gastrointestinal and respiratory symptoms (1, 10, 11, 21). However, the research on toroviruses has been limited, compared to that on coronaviruses, primarily because toroviruses, with the exception of equine torovirus, are very difficult to propagate in cell culture (20). In fact, among the human, bovine, and porcine toroviruses, only one strain of bovine torovirus (BToV) has been successfully propagated in cell culture. This strain was isolated from the intestinal contents of a calf with diarrhea in 2004 using the stable cell line HRT-18, derived from a human rectal tumor (13). Although several studies on the genetic characterization of toroviruses have been published, almost all were dependent on analyses of PCR products from biomaterials (5, 9, 10, 12, 18). Thus, antigenic variation among toroviruses and relationships between their genetic and antigenic properties using cultivatable viruses in cell cultures remain to be investigated.
Epidemiological research conducted by our group has suggested that BToV may act as a risk factor for the induction of gastrointestinal and respiratory diseases in cattle in Japan. In addition, the sequences of coding regions for the spike (S) and hemagglutinin-esterase (HE) genes of BToV have been shown to vary among several field viruses (9, 10). Because the products of both genes in coronaviruses are known to correlate closely with their antigenic properties (4), the relationship between genetic and antigenic properties, and antigenic variation of BToV, seem to be important lines of research with respect to understanding immunological and pathogenic aspects of BToV infection and in the development of an effective vaccine.
In the present study, fecal and nasal samples positive for the BToV nucleocapsid (N) gene by reverse transcription-PCR (RT-PCR) were inoculated onto monolayers of HRT-18 cell cultures, and four cytopathogenic BToVs were successfully isolated. The purpose of the present study was to report the isolation of cytopathogenic BToVs in cell culture and their characterization, with respect to their genetic and antigenic properties.
MATERIALS AND METHODS
Samples.
In total, 55 individual samples collected from Japanese cattle positive for the BToV N gene, as determined by the RT-PCR method described previously (9, 10), were used for virus isolation. These samples included some BToV-positive fecal and nasal samples that we have reported previously. The details of the samples were as follows: fecal samples were collected from 48 cattle, 47 showing enteric symptoms and 1 that was asymptomatic, raised on 20 farms located in 12 prefectures between September 2004 and May 2009; nasal samples were obtained from 7 cattle with respiratory symptoms on 6 farms located in 5 prefectures between December 2006 and June 2008. Each sample was diluted 1:10 in Dulbecco modified Eagle medium (DMEM) and centrifuged (3,000 × g, 5 min, room temperature [9]). The supernatants were collected and used for virus isolation in HRT-18 cell cultures.
Virus isolation.
Virus isolation was carried out using the human rectal tumor cell line HRT-18. The reference Aichi/2004 strain of BToV was used as a positive control. The HRT-18 cells and reference virus were kindly supplied by M. Kuwabara, Central Livestock Hygiene Service Center, Aichi Prefecture, Japan (13). Confluent monolayers of HRT-18 cells, grown in test tubes, were inoculated with 0.1-ml portions of the samples. At 60 min after static absorption at 37°C, all cultures received 1.0 ml of DMEM, were further incubated with rotation at 37°C for 7 days, and were observed for cytopathic effect (CPE). These procedures were further repeated three times. CPE-positive samples were cloned by two rounds of a limiting dilution method and tested for the presence of BToV, bovine coronavirus (BCoV), and bovine rotavirus, as described previously (9, 15).
Measurement of HA and RDE activity.
The hemagglutination (HA) and receptor-destroying enzyme (RDE) titers of isolated virus were measured by the microplate method using mouse (1.0%) and chicken (0.5%) erythrocytes, as described elsewhere (16).
Immunization of guinea pigs.
Antisera against the isolates and reference strain were produced in guinea pigs, as described elsewhere (19). Briefly, concentrated viral suspensions were prepared from culture media by ultracentrifugation, emulsified with oil adjuvant consisting of liquid paraffin supplemented with 10% anhydrous mannitol-oleic acid ester, and used for immunization. The amount of virion contained in the final immunogens was estimated at 107.0 median tissue-culture infective doses (TCID50)/ml. Guinea pigs were injected intramuscularly twice with 0.5 ml and subsequently intraperitoneally with 2.5 ml of the respective immunogens at weekly intervals for 3 weeks. At 2 weeks after final inoculation, sera were collected and used in the experiments.
Genetic analyses.
The isolates were subjected to RNA extraction, followed by nested RT-PCR. The methods used for RNA extraction and nested RT-PCR amplification of the BToV S gene, and primer sets used in the reaction are described elsewhere (9). In addition, the following primers were used to amplify the full-length open reading frame (ORF) of the S gene: fl-F1, ATT TTT GCT GTT GTT GTG AAG (corresponding to nucleotides 22274 to 22294 of AY427798); fl-F2, GTT GCA AGT YTA TGA AAC ACC (corresponding to nucleotides 23155 to 23174 of AY427798); fl-F3, TGC ACT TCA ATG GAT TAT TTT (corresponding to nucleotides 23855 to 23875 of AY427798); fl-F4, AAC ATC TTG GGC TCA GTT TA (corresponding to nucleotides 24917 to 24936 of AY427798); fl-R1, GCA TGG AAA CCT GAT GTA TT (corresponding to nucleotides 22580 to 22561 of AY427798); fl-R2, AAA RCC CAC ACA ACC AGG TA (corresponding to nucleotides 23175 to 23155 of AY427798); fl-R3, TGT GAA GCT GGT GAC ATA AA (corresponding to nucleotides 24075 to 24056 of AY427798); fl-R4, ATA CCT ATC GCA ATA ATG CTC (corresponding to nucleotides 25073 to 25053 of AY427798); and fl-R5, CAG TTG CTG ATA ACT GCT CA (corresponding to nucleotides 25839 to 25820 of AY427798). Nucleotide sequencing and phylogenetic tree analyses were performed as described elsewhere (9).
Virus neutralization (VN) test.
Antisera were serially diluted 2-fold with serum-free DMEM and incubated at 37°C for 60 min with an equal volume of the respective viral suspension containing 200 median TCID50/0.1 ml. Each serum-virus mixture was inoculated onto confluent monolayers of HRT-18 cells grown in test tubes. After 60 min of static absorption at 37°C, all tubes received 1.0 ml of DMEM and were incubated with rotation at 37°C for 7 days. Neutralization antibody titers are expressed as the reciprocal of the highest serum dilution that inhibited 50% of the CPE.
HI test.
Antisera were treated with kaolin and mouse erythrocytes to remove nonspecific hemagglutinins before testing, according to methods described by Hasoksuz et al. (7). Serial 2-fold dilutions of antisera were made in a volume of 25 μl and mixed with an equal volume of 8 HA units of each viral suspension. After a 60-min incubation at 37°C, 25 μl of a 1.0% mouse erythrocyte suspension was added to each suspension. Settling patterns of erythrocytes were read 90 min after incubation at 22°C, and hemagglutination inhibition (HI) antibody titers were expressed as the reciprocal of the highest serum dilution that completely inhibited HA.
Antigenic-relatedness values.
Antigenic similarities among the BToV isolates were indicated by the percent antigenic-relatedness values (R), calculated from the results of cross-VN and HI tests using the following formula, as described previously (7, 17):
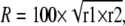 |
where r1 and r2 are the heterologous titer of strain 1/homologous titer of strain 2 and the heterologous titer of strain 2/homologous titer of strain 1, respectively.
RESULTS
Virus isolation.
Four fecal samples of the 55 fecal and nasal samples investigated revealed CPE similar to that of the reference Aichi/2004 strain after serial passage in HRT-18 cell cultures. Agents causing CPE were identified as BToV by RT-PCR and immunofluorescence using antiserum prepared to the reference Aichi/2004 strain of BToV (data not shown). They were designated as the Miyagi-2006TI/E, Gifu-2007TI/E, Hokkaido-2008TI/E, and Gifu-2009TI/E strains. Notably, the strains could be isolated and caused CPE only in HRT-18 cells provided by M. Kuwabara, Central Livestock Hygiene Service Center, Aichi Prefecture, Japan, while BToV isolation was unsuccessful when cells maintained in our laboratory were used as a substrate. As shown in Table 1, the cytopathic BToVs were isolated from feces obtained from four cattle raised in three prefectures between February 2006 and March 2009; all four cattle had gastrointestinal problems, while their breeds and ages differed. In addition, the BCoV N gene was coincidentally detected in the original fecal materials of Gifu-2007TI/E and Gifu-2009TI/E strains, but BCoV was undetectable after their serial passages in HRT-18 cell cultures.
TABLE 1.
Origin and general properties of the BToV strains isolated from cattle
| BToV strain | Origin of the sample |
Host |
|||||
|---|---|---|---|---|---|---|---|
| Material | Sampling period | Prefecture | Other pathogen detected | Body condition (symptoms) | Age (mo) | Breed | |
| Miyagi-2006TI/E | Loose stool | Feb. 2006 | Miyagi | Gastrointestinal | Calf | Beef | |
| Gifu-2007TI/E | Muddy diarrhea stool | Apr. 2007 | Gifu | BCoV | Gastrointestinal and respiratory | 17 | Beef, female |
| Hokkaido-2008TI/E | Watery diarrhea stool | Mar. 2008 | Hokkaido | Gastrointestinal and respiratory | 1 | Beef, female | |
| Gifu-2009TI/E | Loose stool | Mar. 2009 | Gifu | BCoV | Gastrointestinal | 40 | Dairy, female |
HA and RDE activity.
Four BToV isolates, as well as the reference Aichi/2004 strain, showed similar HA activity, with titers ranging from 16 to 128 to mouse erythrocytes at both 4 and 37°C (Table 2). However, none of them agglutinated chicken erythrocytes at either temperature. Moreover, all of the isolates lacked RDE activity against both mouse and chicken erythrocytes.
TABLE 2.
Hemagglutination and receptor-destroying enzyme activities of BToV strains
| Strain | HA titer at: |
RDE titera |
||||
|---|---|---|---|---|---|---|
| 4°C |
37°C |
|||||
| Mouse | Chicken | Mouse | Chicken | Mouse | Chicken | |
| Miyagi-2006TI/E | 32 | <2 | 16 | <2 | <5 | <5 |
| Gifu-2007TI/E | 64 | <2 | 64 | <2 | <5 | <5 |
| Hokkaido-2008TI/E | 128 | <2 | 128 | <2 | <5 | <5 |
| Gifu-2009TI/E | 32 | <2 | 32 | <2 | <5 | <5 |
| Aichi/2004 | 32 | <2 | 32 | <2 | <5 | <5 |
Expressed as the reciprocal of the highest dilution of virus causing complete disappearance of HA patterns at 4°C after 2 h of incubation at 37°C. RDE, receptor-destroying enzyme.
Genetic analysis.
As indicated in Table 3, the similarities of the 594-bp nucleotide sequences and 197 amino acid (aa) residues of the S gene N-terminal region were ≥91.6 and ≥91.9% among the isolates and the reference Aichi/2004 strain, respectively. These similarities were comparable with the results determined previously in comparing BToVs derived from feces and nasal swabs (9, 10). Interestingly, the nucleotide sequences of Hokkaido-2008TI/E and Gifu-2009TI/E strains were completely identical, although they were derived from the clinical specimens collected at geographically distinct areas and at different times.
TABLE 3.
Nucleotide and amino acid identities of the S gene: N-terminal 594-bp ORFa
| Strain | Nucleotide and amino acid identitiesb (%) |
||||
|---|---|---|---|---|---|
| Miyagi-2006TI/E | Gifu-2007TI/E | Hokkaido-2008TI/E | Gifu-2009TI/E | Aichi/2004 | |
| Miyagi-2006TI/E | 91.6 | 96.3 | 96.3 | 97.6 | |
| Gifu-2007TI/E | 92.9 | 91.6 | 91.6 | 92.4 | |
| Hokkaido-2008TI/E | 99.0 | 91.9 | 100.0 | 97.5 | |
| Gifu-2009TI/E | 99.0 | 91.9 | 100.0 | 97.5 | |
| Aichi/2004 | 99.5 | 92.4 | 98.5 | 98.5 | |
The reference BToVs used for sequence comparison and accession numbers were as follows: Aichi/2004 (AB285127), BRV-1 (AY427798), BRV-2 (AF076621), and B145 (AJ575388).
Nucleotide identities are indicated in the upper right-hand portion of the table. Amino acid identities are indicated in the lower left-hand portion of the table.
Analysis of the full-length ORF of the S gene showed that the ORF of the isolates and the reference Aichi/2004 strain encoded a 1,584-aa residue protein, as did the registered sequences of the BRV-1 and B145 that were detected in the feces of cattle from North America and Europe, respectively (5, 18). Among the four isolates and the reference Aichi/2004 strain, the nucleotide similarities of the S gene ORF were ≥95.4% for the nucleotide sequence and ≥96.1% at the amino acid level (Table 4). In a comparison with all registered sequences, the similarities of the nucleotide sequence and amino acid residues of the S gene ORF were ≥91.5 and ≥90.3%, respectively. Representative sequence data have been deposited in the DNA Data Bank of Japan and assigned the accession numbers AB526862 to AB526866.
TABLE 4.
Nucleotide and amino acid identities of the S gene: full-length ORFa
| Strain | Nucleotide identityb (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Miyagi-2006TI/E | Gifu-2007TI/E | Hokkaido-2008TI/E | Gifu-2009TI/E | Aichi/2004 | BRV-1 | BRV-2 | B145 | |
| Miyagi-2006TI/E | 95.4 | 98.4 | 98.5 | 98.5 | 95.8 | 93.1 | 97.1 | |
| Gifu-2007TI/E | 96.2 | 95.4 | 95.5 | 95.7 | 96.4 | 91.5 | 95.0 | |
| Hokkaido-2008TI/E | 98.7 | 96.1 | 99.9 | 98.2 | 95.8 | 93.0 | 97.1 | |
| Gifu-2009TI/E | 98.8 | 96.1 | 99.6 | 98.2 | 95.9 | 92.9 | 97.1 | |
| Aichi/2004 | 98.7 | 96.3 | 98.5 | 98.5 | 95.9 | 93.0 | 97.1 | |
| BRV-1 | 97.0 | 97.5 | 96.8 | 96.9 | 97.2 | 93.2 | 95.8 | |
| BRV-2 | 91.9 | 90.3 | 91.9 | 91.8 | 92.0 | 91.6 | 92.9 | |
| B145 | 97.9 | 96.3 | 97.7 | 97.9 | 97.7 | 97.2 | 91.6 | |
The reference bovine toroviruses (BToVs) used for sequence comparison and accession numbers were as follows: Aichi/2004 (AB285127), BRV-1 (AY427798), BRV-2 (AF076621), and B145 (AJ575388).
Nucleotide identities are indicated in the upper right-hand portion of the table. Amino acid identities are indicated in the lower left-hand portion of the table.
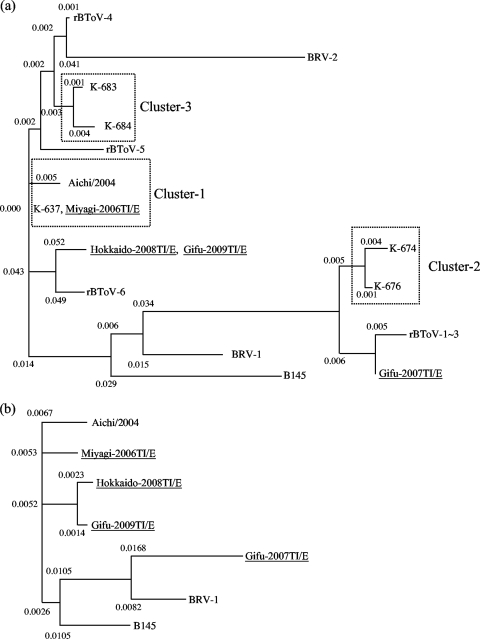
Phylogenetic analysis of the 197-aa region of the S gene N-terminal revealed that Miyagi-2006TI/E, the sequence of which was identical to that of K-637, grouped into cluster 1, which also included the Aichi/2004 strain (9). The Hokkaido-2008TI/E and Gifu-2009TI/E strains were located in a position close to cluster 1 and were most closely related to rBToV-6, detected previously in a nasal swab collected from a Japanese cow (10). In contrast, Gifu-2007TI/E was distant from the other BToV isolates and close to cluster 2, which consisted of K-674 and K-676, originally detected in feces of Japanese cattle. Gifu-2007TI/E was most closely related to rBToV-1-3, which were found in nasal swabs collected from Japanese cattle between December 2006 and June 2007 (10) (Fig. 1a). Phylogenetic analysis of the full-length amino acid sequence of the S gene ORF revealed that the BToV isolates—Miyagi-2006TI/E, Hokkaido-2008TI/E, and Gifu-2009TI/E—and the reference Aichi/2004 strain were relatively closer to one another than to Gifu-2007TI/E. Similarly to the analysis for the 197-aa region of the S gene N terminal, the Gifu-2007TI/E strain was located more distal to the other isolates than BRV-1 and B145 (Fig. 1b).
FIG. 1.
Neighbor-joining phylogenetic tree showing the relationships for the deduced sequences of the spike gene. (a and b) N-terminal 197 amino acids (a) and full-length amino acid sequences (b) from bovine torovirus isolates and reference strains. The numbers represent the distance to the nearest node. The additional sequences refer to Table 2 for comparison. Their accession numbers are as follows: K-637 (AB270909), K-674 (AB270914), K-676 (AB270916), K-683 (AB270918), K-684 (AB270919), rBToV-2 (AB371899), rBToV-4 (AB448741), and rBToV-6 (AB448745).
Cross-neutralization tests.
The results of cross-VN tests with the four isolates and the reference BToV Aichi/2004 strain are shown in Table 5. Although some antigenic variation was evident among the viruses investigated, they were considered to be antigenically related to each other, with R values of 19.8 to 100%. Furthermore, the comparisons among strains Miyagi-2006TI/E, Hokkaido-2008TI/E, and Gifu-2009TI/E and reference BToV Aichi/2004 strains, which share more than 98.5% sequence identity in the 197-aa region of the S gene N-terminal, yielded relatively high R values, 56.6 to 100%. In contrast, the Gifu-2007TI/E strain with lower similarity, <92.9% of the 197-aa region, appeared to be unique compared to the other viruses; R values between this strain and the other viruses were relatively low (19.8 to 31.3%; Table 3a).
TABLE 5.
Result of virus neutralization and HI tests using guinea pig antisera against BToV strain Aichi/2004 and the four newly isolated strains
| Test and strain | Antibody titer using guinea pig antiserum (R%)a |
||||
|---|---|---|---|---|---|
| Miyagi-2006TI/E | Gifu-2007TI/E | Hokkaido-2008TI/E | Gifu-2009TI/E | Aichi/2004 | |
| Neutralization test | |||||
| Miyagi-2006TI/E | 51,200 (100) | 10,240 | 40,960 | 20,480 | 12,800 |
| Gifu-2007TI/E | 32,000 (25.0) | 102,400 (100) | 32,000 | 12,800 | 8,000 |
| Hokkaido-2008TI/E | 12,800 (63.2) | 6,400 (28.0) | 25,600 (100) | 12,800 | 6,400 |
| Gifu-2009TI/E | 25,600 (70.7) | 6,400 (19.8) | 40,960 (100) | 20,480 (100) | 6,400 |
| Aichi/2004 | 20,480 (56.6) | 12,800 (31.3) | 20,480 (70.7) | 12,800 (62.5) | 10,240 (100) |
| HI test | |||||
| Miyagi-2006TI/E | 20,480 (100) | 6,400 | 32,000 | 12,800 | 6,400 |
| Gifu-2007TI/E | 640 (14.0) | 10,240 (100) | 800 | 400 | 400 |
| Hokkaido-2008TI/E | 25,600 (88.4) | 20,480 (17.7) | 51,200 (100) | 12,800 | 8,000 |
| Gifu-2009TI/E | 20,480 (69.9) | 12,800 (17.7) | 51,200 (89.4) | 16,000 (100) | 6,400 |
| Aichi/2004 | 20,480 (44.2) | 12,800 (19.8) | 32,000 (62.5) | 12,800 (63.2) | 12,800 (100) |
For the neutralization test results, the neutralization antibody titers are indicated, and percent antigenic-relatedness values (R%) are indicated in parentheses. For the HI test, HI antibody titers are indicated, and percent antigenic relatedness values (R) are indicated in parentheses. Identity values are indicated in boldface.
Cross-HI tests.
The results of cross-HI tests well coincided with those of cross-VN tests (Table 5). R values calculated from HI antibody titers ranged between 44.2 and 89.4% among the Miyagi-2006TI/E, Hokkaido-2008TI/E, Gifu-2009TI/E, and reference Aichi/2004 strains. In contrast, the Gifu-2007TI/E strain yielded relatively low R values, ranging from 14.0 to 19.8% between the other viruses.
DISCUSSION
Toroviruses have been difficult to propagate in cell culture. Indeed, to date, only one strain has been isolated among human, bovine, and porcine toroviruses (13). In the present study, however, cytopathogenic BToVs were isolated from fecal samples collected from four cattle between February 2006 and March 2009. They all showed intestinal symptoms and some manifested respiratory signs, a finding suggestive of that BToV played some roles as a predisposing factor.
The HRT-18 cell line used in the present study was provided by M. Kuwabara, who succeeded in first isolating a cytopathogenic BToV strain, Aichi/2004 (13). Unexpectedly, all of the BToVs isolated in the present study lacked susceptibility to HRT-18 cells maintained in our laboratory and used previously. In contrast, the BCoV strains isolated previously in the HRT-18 cells of our laboratory failed to propagate in or exhibited only weak susceptibility to those provided by M. Kuwabara. The fecal materials from which the Gifu-2007TI/E and Gifu-2009TI/E strains were isolated were coincidentally positive for BCoVs, but only BToVs were propagated after repeated passage in cell culture. This seems to indicate that BToVs were preferentially propagated under the particular conditions, including the source of the HRT-18 cells used. In addition, in the virus isolation process, several bovine enteroviruses, which had been considered as being nonreplicable in HRT-18 cells, were isolated from several samples besides four BToV isolated samples (our unpublished data). Based on these results, one significant factor that allowed the BToV isolation appears to be the particular properties of the HRT-18 cell line used. Although the cytological differences between the two lines of HRT-18 cells remain unclear, an unexpected cellular mutation that affects susceptibility to viruses might occur during long-term maintenance of the cells in the separate laboratories.
BToV, as well as certain other viruses, is known to possess HA activity to animal erythrocytes via hemagglutinin present on the surface of the viral envelope. BCoV, another member of the Coronaviridae family having similar genomic structures to BToV, is well known to show several HA and RDE reaction patterns with mouse and chicken erythrocytes (6, 7), whereas those of BToVs have not been investigated to date. In the present study, five BToVs, four novel isolates and the Aichi/2004 strain, were compared for HA and RDE reaction patterns to mouse and chicken erythrocytes, and we found that all viruses tested revealed similar HA and RDE properties to each other; they all agglutinated only mouse erythrocytes at both 4 and 37°C but lacked RDE activity. Because the number and source of BToVs tested were limited to five fecal specimens, the presence of BToV strains with different HA and RDE properties cannot be ruled out. However, that the five BToVs analyzed here were isolated at distinct times and in different geographic areas suggests that the HA and RDE properties observed are representative of most, if not all, BToVs.
Thus far, 33 fecally and six nasally derived BToVs have been sequenced directly from the biomaterials (5, 9, 10, 12, 18), and their identities in the 197-aa N-terminal sequence of the S gene have been estimated at >89%. Similarly, the BToV isolates investigated in the present study showed ≥91.9% identity with each other, while the identity increased to ≥98.5% when the Gifu-2007TI/E strain was excluded from the comparison. The full-length sequences of the S gene ORF, however, have been reported for only three BToVs (BRV-1, BRV-2, and B145) thus far. Here, the sequences of the full-length S gene determined for the novel BToV isolates were likewise found to be well conserved among the strains compared to three viruses mentioned above; sequence similarities, however, were somewhat lower than those determined for the 197-aa N-terminal region, except for the Gifu-2007TI/E strain. This strain showed lower sequence identities with the other three viruses in the 197-aa N-terminal region (91.9 to 92.9%) than those in the full-length S gene (96.1 to 96.3%).
The difference between porcine respiratory coronavirus and porcine transmissible gastroenteritis virus in tissue tropism has been considered as being attributable to the existence of a broad deletion sequence in the S gene of the former virus (14). No informative evidence on variation in the S gene capable of distinguishing respiratory tract- from intestine-derived BToVs has been reported (10). In a previous study, we compared the sequences of the N-terminal region of the BToV S gene amplified directly from nasal and fecal samples and found no significant variation in the region analyzed (10). However, our analysis was limited to the N terminus of the S gene. The full-length S gene ORF of four isolates and foreign BToVs encoded 1,584 aa, with the exception of BRV-2, in which the ORF encoded 1,583 aa. Accordingly, BToV seems to be genetically well conserved among intestinally derived strains. To examine the relationship between tissue tropism and S gene structures of BToVs, isolating BToVs from respiratory materials and comparing their complete S gene sequences with those of intestinally derived BToVs would be of great interest.
The BToV isolates were phylogenetically classified into cluster 1 or its analogs, with the exception of the Gifu-2007TI/E strain, which belonged closer to cluster 2. In a previous report, we found that BToVs belonging to cluster 1 or its surroundings were predominant (9). According to the S gene sequences reported by Kirisawa et al. (12); however, 16 of 17 BToVs analyzed showed identity >99.0% with the Gifu-2007TI/E strain, suggesting that they are possibly placed around cluster 2. These findings suggest that BToVs of both genotypes and their surroundings are widespread in Japan.
All BToV isolates were antigenically related to each other, as revealed by both NT and HI tests, although the degree of cross-reactivity was different between the isolates. Moreover, the overall R values of both tests were well correlated with each other, although they were somewhat lower in the HI than NT tests. With respect to phylogenetic characteristics, the cross-reactivity between the Miyagi-2006TI/E and Aichi/2004 of cluster 1, and the its neighboring viruses Hokkaido-2008TI/E and Gifu-2009TI/E was remarkably high compared to the Gifu-2007TI/E strain, located around cluster 2. Unfortunately, no BToV belonging to cluster 3 was isolated in the present study, but investigating antigenicity in this context would be of interest. R values below 25% are generally regarded as an indication of a significant antigenic difference between two viruses, and a difference greater than 20-fold in both directions (R < 5%) is used as a serotype criterion in several gastroenteric viruses (8, 17, 23). Thus, it seems unlikely that BToVs comprise multiple serotypes, at least based on the results of NT and HI tests using four viruses in the present study. BToVs seem to consist of a single serotype, although the serological properties of BToVs belongings to cluster 1 and surroundings and cluster 2 were markedly different. Nonetheless, our results suggest that at least two subtypes distinguishable by NT and HI tests, and probably related to the structures of N terminus of the S gene, may be present in BToVs. This is based on the finding that the sequence diversity between the Gifu-2007TI/E strain and the other isolates was greater in the N-terminal region than in the full-length S gene.
We believe the present study to be the first report dealing with antigenic characterization of BToVs by both NT and HI tests, and we could partially determine their serological characteristics. Antigenic and genetic investigations using more BToVs cultivatable in cell culture, especially those derived from respiratory specimens, are expected to yield more variable information for understanding serological properties of the BToVs.
Our preliminary investigation suggests that the HE gene of the isolates consisted of a nonfunctional ORF that probably resulted from sequential insertion and/or deletion during the isolation process in tissue culture. Although the mechanism and frequency of these mutations remain unclear, the high mutability seems to be a unique characteristic for the HE gene, compared to the coding regions of the N and S genes, which revealed no or less divergence between isolated viruses and the original fecal materials (data not shown). This is not always relevant to antigenic diversity of BToV, however, because isolates classified within cluster 1 and its surroundings showed strong cross-reactivity in NT and HI tests. The details of the frequency, mechanism, and significance of these mutations in the HE gene are now under investigation.
A few studies have been conducted to determine pathogenicity and antigenicity of BToV using infected diarrheal stools (22). In the present study, we successfully isolated four novel BToV strains in tissue cultures following the report of Kuwabara et al., who first reported a cytopathogenic BToV in 2007 (13). We believe the cytopathogenic viruses isolated will provide important tools for studying the pathological role and immunology of BToVs in enteric and respiratory diseases.
Acknowledgments
We thank Emi Shirahase and Tomoe Nakai of Kyoto Biken Laboratories, Inc. (Kyoto, Japan) for collecting the data used in this study. We also thank Masaki Kuwabara of the Livestock Hygiene Service Center in Aichi Prefecture for providing the Aichi/2004 strain and the HRT-18 cells.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Beards, G. M., C. Hall, J. Green, T. H. Flewett, F. Lamouliatte, and P. Du Pasquier. 1984. An enveloped virus in stools of children and adults with gastroenteritis that resembles the Breda virus of calves. Lancet i:1050-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brian, D. A., and R. S. Baric. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 4.Clark, M. A. 1993. Bovine coronavirus. Br. Vet. J. 149:51-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draker, R., R. L. Roper, M. Petric, and R. Tellier. 2006. The complete sequence of the bovine torovirus genome. Virus Res. 115:56-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukutomi, T., H. Tsunemitsu, and H. Akashi. 1999. Detection of bovine coronaviruses from adult cows with epizootic diarrhea and their antigenic and biological diversities. Arch. Virol. 144:997-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasoksuz, M., S. L. Lathrop, K. L. Gadfield, and L. J. Saif. 1999. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am. J. Vet. Res. 60:1227-1233. [PubMed] [Google Scholar]
- 8.Hubálek, Z. 1982. Numerical comparative serology: the methods. J. Appl. Bacteriol. 52:307-318. [DOI] [PubMed] [Google Scholar]
- 9.Ito, T., N. Okada, and S. Fukuyama. 2007. Epidemiological analysis of bovine torovirus in Japan. Virus Res. 126:32-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito, T., N. Okada, M. Okawa, S. Fukuyama, and M. Shimizu. 2009. Detection and characterization of bovine torovirus from the respiratory tract in Japanese cattle. Vet. Microbiol. 136:366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamieson, F. B., E. E. Wang, C. Bain, J. Good, L. Duckmanton, and M. Petric. 1998. Human torovirus: a new nosocomial gastrointestinal pathogen. J. Infect. Dis. 178:1263-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirisawa, R., A. Takeyama, M. Koiwa, and H. Iwai. 2007. Detection of bovine torovirus in fecal specimens of calves with diarrhea in Japan. J. Vet. Med. Sci. 69:471-476. [DOI] [PubMed] [Google Scholar]
- 13.Kuwabara, M., K. Wada, Y. Maeda, A. Miyazaki, and H. Tsunemitsu. 2007. First isolation of cytopathogenic bovine torovirus in cell culture from a calf with diarrhea. Clin. Vaccine Immunol. 14:998-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laude, H., K. Van Reeth, and M. Pensaert. 1993. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet. Res. 24:125-150. [PubMed] [Google Scholar]
- 15.Okada, N., and Y. Matsumoto. 2002. Bovine rotavirus G and P types and sequence analysis of the VP7 gene of two G8 bovine rotaviruses from Japan. Vet. Microbiol. 84:297-305. [DOI] [PubMed] [Google Scholar]
- 16.Sato, K., Y. Inaba, H. Kurogi, E. Takahashi, K. Satoda, T. Omori, and M. Matsumoto. 1977. Hemagglutination by calf diarrhea coronavirus. Vet. Microbiol. 2:83-87. [Google Scholar]
- 17.Shimizu, M., H. Watanabe, K. Satou, and S. Murakami. 1989. Antigenic diversity of bovine viral diarrhea-mucosal disease (BVD-MD) viruses recently isolated from persistently infected cattle and mucosal disease, and serologic survey on bovine sera using antigenically different BVD-MD viruses. J. Vet. Med. Sci. 51:1115-1122. [DOI] [PubMed] [Google Scholar]
- 18.Smits, S. L., A. Lavazza, K. Matiz, M. C. Horzinek, M. P. Koopmans, and R. J. de Groot. 2003. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 77:9567-9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsunemitsu, H., and L. J. Saif. 1995. Antigenic and biological comparisons of bovine coronaviruses derived from neonatal calf diarrhea and winter dysentery of adult cattle. Arch. Virol. 140:1303-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss, M., F. Steck, and M. C. Horzinek. 1983. Purification and partial characterization of a new enveloped RNA virus (Berne virus). J. Gen. Virol. 64:1849-1858. [DOI] [PubMed] [Google Scholar]
- 21.Woode, G. N., J. F. Pohlenz, N. E. Gourley, and J. A. Fagerland. 1984. Astrovirus and Breda virus infections of dome cell epithelium of bovine ileum. J. Clin. Microbiol. 19:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woode, G. N., L. J. Saif, M. Quesada, N. J. Winand, J. F. Pohlenz, and N. K. Gourley. 1985. Comparative studies on three isolates of Breda virus of calves. Am. J. Vet. Res. 46:1003-1010. [PubMed] [Google Scholar]
- 23.Wyatt, R. G., H. B. Greenberg, W. D. James, A. L. Pittman, A. R. Kalica, J. Flores, R. M. Chanock, and A. Z. Kapikian. 1982. Definition of human rotavirus serotypes by plaque reduction assay. Infect. Immun. 37:110-115. [DOI] [PMC free article] [PubMed] [Google Scholar]