Abstract
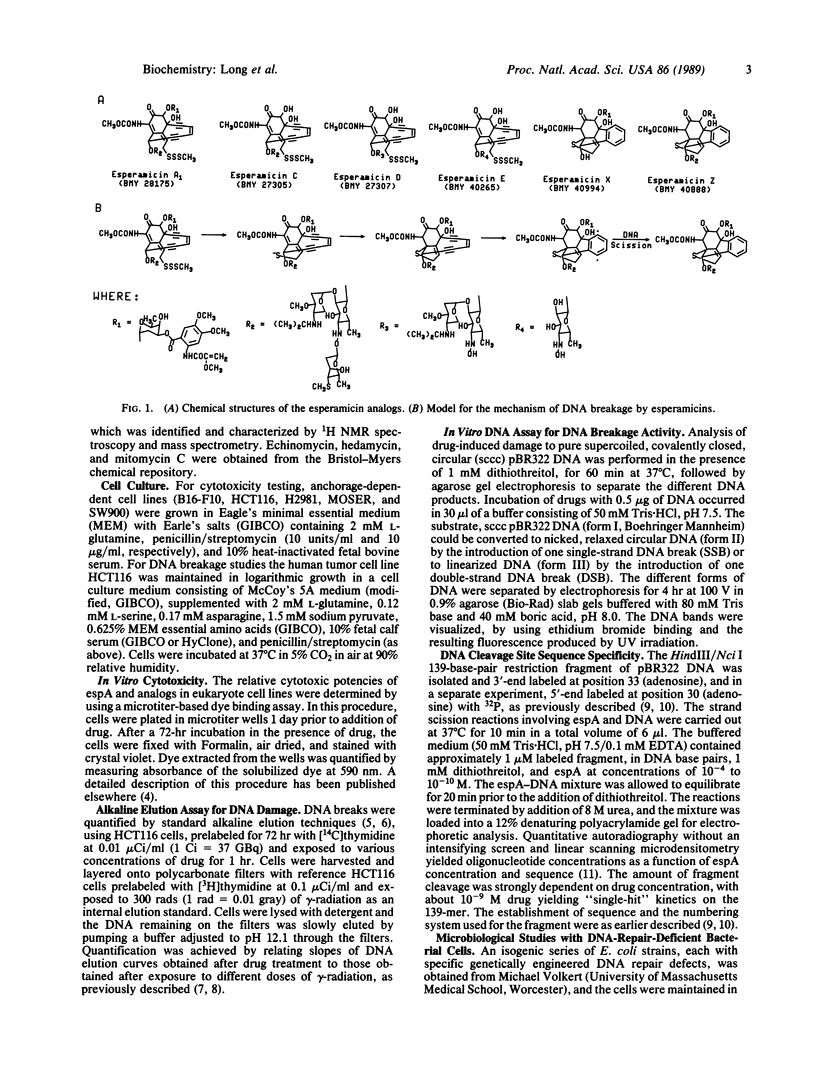
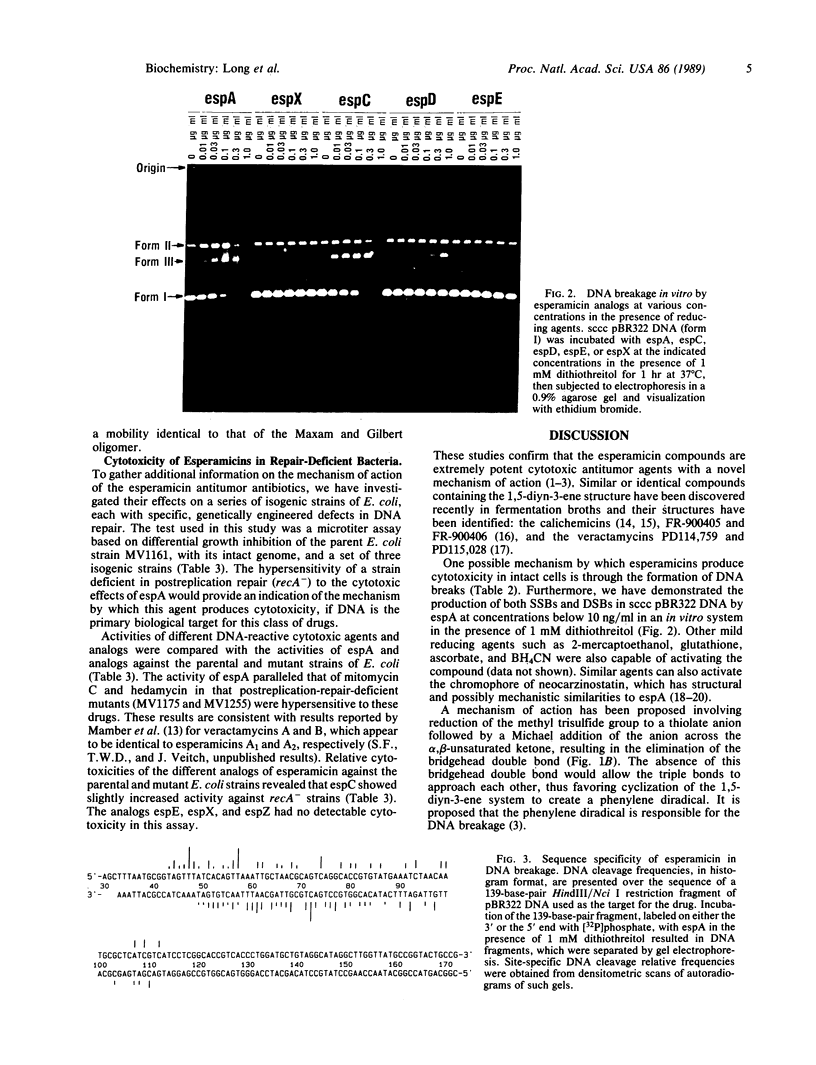
The esperamicins represent a class of antitumor antibiotics characterized by an unusual chemical core structure and extremely potent cytotoxicity. The mechanism by which these drugs produce cytotoxicity was investigated and found to be related to the formation of single- and double-strand DNA breaks. Using five structurally related analogs, we defined a structure-activity relationship for cytotoxicity in various eukaryotic and DNA-repair-deficient prokaryotic cell lines, for DNA breakage in a human colon carcinoma cell line, and for DNA breakage in vitro in pBR322 DNA. Mild reducing agents such as dithiothreitol greatly increased the DNA breakage potency of these analogs in vitro. Results suggest that the pendant aromatic chromophore of esperamicin A1 may contribute to the uptake of the drug into cells but may also hinder double-strand DNA break formation. Little DNA breakage specificity was observed for the drug in a 139-base-pair fragment of pBR322 DNA. Evidence supports a previously proposed mechanism whereby esperamicins may produce the observed DNA breaks through reduction of the methyl trisulfide group to a thiolate anion followed by a Michael addition of the anion across the alpha,beta-unsaturated ketone. This addition may result in the saturation of the bridgehead double bond, thus allowing the two triple bonds to approach each other, causing cyclization of the diyn-ene to form a phenylene diradical. It is likely that this diradical is the active form of the drug responsible for single- and double-strand DNA breakage produced by this class of antitumor agents.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bromley S. D., Ward B. W., Dabrowiak J. C. Cationic porphyrins as probes of DNA structure. Nucleic Acids Res. 1986 Nov 25;14(22):9133–9148. doi: 10.1093/nar/14.22.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catino J. J., Francher D. M., Edinger K. J., Stringfellow D. A. A microtitre cytotoxicity assay useful for the discovery of fermentation-derived antitumor agents. Cancer Chemother Pharmacol. 1985;15(3):240–243. doi: 10.1007/BF00263894. [DOI] [PubMed] [Google Scholar]
- Dabrowiak J. C., Skorobogaty A., Rich N., Vary C. P., Vournakis J. N. Computer assisted microdensitometric analysis of footprinting autoradiographic DATA. Nucleic Acids Res. 1986 Jan 10;14(1):489–499. doi: 10.1093/nar/14.1.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan P. B. Design of sequence-specific DNA-binding molecules. Science. 1986 Apr 25;232(4749):464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- Fry D. W., Shillis J. L., Leopold W. R. Biological and biochemical activities of the novel antitumor antibiotic PD 114,759 and related derivatives. Invest New Drugs. 1986;4(1):3–10. doi: 10.1007/BF00172009. [DOI] [PubMed] [Google Scholar]
- Goldberg I. H. Free radical mechanisms in neocarzinostatin-induced DNA damage. Free Radic Biol Med. 1987;3(1):41–54. doi: 10.1016/0891-5849(87)90038-4. [DOI] [PubMed] [Google Scholar]
- Kappen L. S., Goldberg I. H. Deoxyribonucleic acid damage by neocarzinostatin chromophore: strand breaks generated by selective oxidation of C-5' of deoxyribose. Biochemistry. 1983 Oct 11;22(21):4872–4878. doi: 10.1021/bi00290a002. [DOI] [PubMed] [Google Scholar]
- Kiyoto S., Nishikawa M., Terano H., Kohsaka M., Aoki H., Imanaka H., Kawai Y., Uchida I., Hashimoto M. New antitumor antibiotics, FR-900405 and FR-900406. II. Production, isolation, characterization and antitumor activity. J Antibiot (Tokyo) 1985 Jul;38(7):840–848. doi: 10.7164/antibiotics.38.840. [DOI] [PubMed] [Google Scholar]
- Konishi M., Ohkuma H., Saitoh K., Kawaguchi H., Golik J., Dubay G., Groenewold G., Krishnan B., Doyle T. W. Esperamicins, a novel class of potent antitumor antibiotics. I. Physico-chemical data and partial structure. J Antibiot (Tokyo) 1985 Nov;38(11):1605–1609. doi: 10.7164/antibiotics.38.1605. [DOI] [PubMed] [Google Scholar]
- Long B. H., Musial S. T., Brattain M. G. Single- and double-strand DNA breakage and repair in human lung adenocarcinoma cells exposed to etoposide and teniposide. Cancer Res. 1985 Jul;45(7):3106–3112. [PubMed] [Google Scholar]
- Lown J. W., Sondhi S. M., Ong C. W., Skorobogaty A., Kishikawa H., Dabrowiak J. C. Deoxyribonucleic acid cleavage specificity of a series of acridine- and acodazole-iron porphyrins as functional bleomycin models. Biochemistry. 1986 Sep 9;25(18):5111–5117. doi: 10.1021/bi00366a020. [DOI] [PubMed] [Google Scholar]
- Mamber S. W., Okasinski W. G., Pinter C. D., Tunac J. B. Bacterial DNA reactivity of the novel antitumor antibiotics PD 114,759 and PD 115,028 (veractamycins A and B). J Antibiot (Tokyo) 1987 Jul;40(7):1044–1049. doi: 10.7164/antibiotics.40.1044. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]



