Abstract
The measurement of pneumococcal carriage in the nasopharyngeal reservoir is subject to potential confounders that include low-density and multiple-strain colonization. To compare different methodologies, we picked a random sampling of 100 nasopharyngeal specimens recovered from infants less than 2 years of age who were previously assessed for pneumococcal carriage and serotypes by a conventional method that used direct plating from the transport/storage medium (50 specimens were culture negative and 50 specimens were culture positive for pneumococci). We used a broth enrichment approach and a conventional PCR approach (with and without broth enrichment) to determine pneumococcal carriage and serotypes, and the results were compared to the initial conventional culture-based results. Additionally, we used a lytA-targeted real-time PCR for pneumococcal detection. Broth enrichment for both the culture-based and the PCR-based methods enhanced the isolation of pneumococci and detection of serotype diversity, with the most effective serotype deduction method being one that used broth enrichment prior to sequential multiplex PCR. Similarly, we also found that broth enrichment followed by the lytA-specific real-time PCR was the most sensitive for the detection of apparent pneumococcal carriage. The broth enrichment, conventional multiplex PCR, and real-time PCR approaches used in this study were effective in detecting pneumococcal carriage in the 50 specimens that were negative by conventional direct plating from transport medium (range of numbers of positive specimens, 8/50 to 22/50 [16 to 44%]), and the three different serotyping approaches that used broth enrichment increased the number of serotype identifications from the 100 specimens (12 to 29 additional serotype identifications to be positive). A PCR-based approach that employed a broth enrichment step appeared to best enhance the detection of mixed serotypes and low-density pneumococcal carriage.
The principal habitat of the opportunistic pathogen Streptococcus pneumoniae is the nasopharynx, where it normally resides commensally. Current multivalent vaccines against pneumococci target subsets of the 91 known capsular serotypes known to be expressed by this organism (12). Although the biology of pneumococcal carriage is not well understood, it appears that pneumococcal disease occurs as a sequela of nasopharyngeal (NP) carriage (7, 15). Accurate assessment of the serotype distribution associated with pneumococcal colonization, together with knowledge of the serotype distribution associated with pneumococcal disease, could be of great value in the evaluation and formulation of pneumococcal vaccines.
Detection of the NP carriage of pneumococci and serotyping have traditionally relied on conventional culture methods of direct plating of NP specimens from transport or storage media. For example, a recent major study used classical culture isolation and Quellung reaction-based serotyping to reveal significant effects of vaccination on the carriage of vaccine serotype strains (16). Broth enrichment and PCR testing offer potential improvements to the detection and serotyping of pneumococci from nasopharyngeal specimens. Recently, we developed a sequential PCR-based assay for the identification of 29 different serotypes (11) that has more recently expanded to 40 serotypes (http://www.cdc.gov/ncidod/biotech/strep/pcr.htm). Although this scheme is useful for deducing the serotypes of pneumococcal isolates (6, 9), others have found the approach to be useful for determining serotypes directly from clinical and carriage specimens (2, 3, 14). In the study described here, we extended the previous evaluation of the sequential PCR approach for carriage specimens (2) through the use of an expanded scheme that includes 11 additional serospecificities and also through the use of broth enrichment. In addition, we compare the results of culture-based serotyping of carriage specimens obtained with and without broth enrichment. We also demonstrate the sensitivity of a real-time PCR for pneumococcal detection (with and without broth enrichment) compared to the sensitivities of culture and conventional PCR approaches.
MATERIALS AND METHODS
Clinical specimens.
Approximately 6,500 NP calcium alginate swab specimens (Fisherbrand, catalog number 14-959-78; Fisher Scientific, Pittsburg, PA) from individuals 2 years of age or less were collected and transported in 1.0 ml skim milk-tryptone-glucose-glycerol (STGG) medium (10). These NP swabs within STGG medium (NP-STGG specimens) were shipped on wet ice and frozen at −70°C within 4 h. Before they were frozen at −70°C, the NP-STGG specimens were vortexed for 10 to 20 s to disperse the organisms from the swab. These specimens were collected from 2006 to 2008 from children who had previously been immunized with the pneumococcal seven-valent conjugate vaccine (PCV7) and who participated in a study of the long-term effects of vaccination on pneumococcal carriage (8). The specimens were stored for up to 3 years prior to the testing described here. From this large collection, 50 NP swab specimens classified in the study by standard culture methods (the direct culture [DC] method described below) to be pneumococcal culture negative and 50 specimens classified to be pneumococcal culture positive were randomly selected. Each specimen represented a single individual and household, and no individual was represented more than once. All NP-STGG specimens were stored at −70°C after collection, transported to the Centers for Disease Control and Prevention (CDC) Streptococcus Laboratory on dry ice, and kept at −70°C until they were processed.
Broth enrichment for enhanced pneumococcal growth.
Supplemented Todd-Hewitt broth (STHB) consisted of 5 ml of Todd-Hewitt broth containing 0.5% yeast extract combined with 1 ml of rabbit serum. For broth enrichment, after a brief complete thawing and vigorous vortexing of the NP-STGG specimens, 200-μl aliquots were added to 5 ml STHB, and the mixture was incubated for 4 h at 37°C in a CO2 incubator. The enriched STHB culture was streaked for the broth enrichment culture (BEC) method described below. The remaining STHB culture was frozen for further extraction of the DNA used in the broth enrichment sequential multiplex PCR (BEPCR) assay and the lytA gene-specific real-time PCR assay for S. pneumoniae detection using DNA extracted from the enrichment broth growth (BERT), which are also described below.
DNA extracts. (i) Extraction of DNA from isolates.
Pneumococcal isolates were identified by the use of the standard optochin and bile solubility tests. The DNAs from purified cultures were extracted for use in the BEC method by suspending a loopful of fresh (overnight) growth from a blood agar plate (BAP) in 300 μl of 0.85% saline. The homogenized suspension was heated at 70°C for 30 min, followed by centrifugation and removal of the supernatant. Fifty microliters of TE (Tris-EDTA) buffer containing 10 μl of mutanolysin (3,000 U/ml) and 8 μl of hyaluronidase (30 mg/ml) was added to the pellet and the mixture was vortexed. After 1 h of incubation at 37°C, the suspensions were heated (100°C) for 10 min and centrifuged, and the supernatants were stored at −20°C until use.
(ii) DNA from STHB-enriched growth and STGG-NP specimens.
DNA extracts for use in the BEPCR, multiplex PCR for S. pneumoniae serotyping by the use of DNA extracted directly from STGG-NP specimens (direct PCR [DPCR]), and BERT assays described below were extracted from thawed 200-μl aliquots of broth (STHB)-enriched culture or NP-STGG specimens by use of a DNeasy blood and tissue kit (Qiagen, Valencia, CA), according to the manufacturer's instructions, and a preincubation step of 1 h in 100 μl prelysis buffer (1.0 M Tris-HCl [2.0 ml], 0.5 M EDTA [0.4 ml], Triton X-100 [1.2 ml], water up to 100 ml) containing 0.02 g/ml lysozyme and 5 U/ml of mutanolysin at 37°C.
Sequential multiplex PCR assay for pneumococcal detection and serotype deduction.
A conventional sequential multiplex PCR able to detect a total of 40 serotypes was performed by using eight sequential reactions (6, 9, 11; see http://www.cdc.gov/ncidod/biotech/strep/pcr.htm for the latest updates). The serotype result obtained by PCR testing is often the same as that obtained by Quellung reaction-based testing (single serotypes, such as serotypes 4, 6C, and 35B). For some results, however, the PCR-based “serotype” describes the common serotype first listed, followed by a minor rarer serotype(s) that, unlike Quellung reaction-based results, cannot currently be resolved by PCR (e.g., serotypes 11A/11D, 15A/15F, and 10F/10C/33C). The one exception to this nomenclature is 15B/15C, for which it is well established that frequent interconversion between these serotypes occurs, such that single cultures frequently yield both serotypes (17). The 13 additional new primers designed for this study are listed in Table 1. These primers either expanded the range of serotypes that could be detected or offered technical improvements (better specificity, efficiency, or resolution) to our previous sequential multiplex PCR assays. Qiagen multiplex PCR kits were used for DNA amplifications under the following conditions: 95°C for 15 min, followed by 35 amplification cycles of 94°C for 30 s, 54°C for 90 s, and 72°C for 60 s. A final hold was performed at 72°C for 10 min (3). Negative and positive serotype-specific controls were coelectrophoresed in each of eight multiplex reactions.
TABLE 1.
New sequential multiplex PCR primers
| Primer | GenBank accession no. | Primer sequence (5′-3′) | Gene target | Nucleotide position | Product size (bp) |
|---|---|---|---|---|---|
| 2-f | CR931633 | TAT CCC AGT TCA ATA TTT CTC CAC TAC ACC | wzy | 10271 | 290 |
| 2-r | ACA CAA AAT ATA GGC AGA GAG AGA CTA CT | 10531 | |||
| 7F/7A-f | CR931643 | TCC AAA CTA TTA CAG TGG GAA TTA CGG | wzy | 14683 | 599 |
| 7F/7A-r | ATA GGA ATT GAG ATT GCC AAA GCG AC | 15256 | |||
| 8-f | CR931644 | GAA GAA ACG AAA CTG TCA GAG CAT TTA CAT | wzy | 11193 | 201 |
| 8-r | CTA TAG ATA CTA GTA GAG CTG TTC TAG TCT | 11364 | |||
| 9V/9A-f | CR931648 | GGG TTC AAA G TC AGA CAG TG A ATC TTA A | wzy | 9966 | 816 |
| 9V/9A-r | CCA TGA ATG A AA TCA ACA TT G TCA GTA GC | 10753 | |||
| 10F/(10C/33C)-f | CR931652 | GGA GTT TAT CGG TAG TGC TCA TTT TAG CA | wzx | 12403 | 248 |
| 10F/(10C/33C)-r | CTA ACA AAT TCG CAA CAC GAG GCA ACA | 12624 | |||
| 13-f | CR931661 | TAC TAA GGT AAT CTC TGG AAA TCG AAA GG | wzx | 14005 | 655 |
| 13-r | CTC ATG CAT TTT ATT AAC CG C TTT TTG TTC | 14630 | |||
| 16F-f | CR931668 | GAA TTT TTC AGG CGT GGG TGT TAA AAG | wzy | 11679 | 717 |
| 16F-r | CAG CAT ATA GCA CCG CTA AGC AAA TA | 12371 | |||
| 21-f | CR931680 | CTA TGG TTA TTT CAA CTC AAT CGT CAC C | wzx | 13247 | 192 |
| 21-r | GGC AAA CTC AGA CAT AGT ATA GCA TAG | 13412 | |||
| 23A-f | CR931683 | TAT TCT AGC AAG TGA CGA AGA TGC G | wzy | 7739 | 722 |
| 23A-r | CCA ACA TGC TTA AAA ACG CTG CTT TAC | 8434 | |||
| 23B-f | CR931684 | CCA CAA TTA G CG CTA TAT TCA TTC AAT CG | wzx | 13227 | 199 |
| 23B-r | GTC CAC GCT GAA TAA AAT GAA GCT CCG | 13399 | |||
| 24/(24A, 24B, 24F)-f | CR931688 | GCT CCC TGC TAT TGT AAT CTT TAA AGA G | wzy | 11701 | 99 |
| 24/(24A, 24B, 24F)-r | GTG TCT TTT ATT GAC TTT ATC ATA GGT CGG | 11770 | |||
| 35A/(35C/42)-f | CR931704 | ATT ACG ACT CCT TAT GTG ACG CGC ATA | wzx | 14394 | 280 |
| 35A/(35C/42)-r | CCA ATC CCA AGA TAT ATG CAA CTA GGT T | 14646 | |||
| 39-f | CR931711 | TCA TTG TAT TAA CCC TAT GCT TTA TTG GTG | wzy | 12289 | 98 |
| 39-r | GAG TAT CTC CAT TGT ATT GAA ATC TAC CAA | 12357 |
Methods for pneumococcal serotyping and detection.
Four methods were used to achieve both pneumococcal identification and serotype identification.
(i) Standard culture method.
The DC method initially used for pneumococcal detection and serotyping consisted of a brief thawing of the NP-STGG specimens and then vortexing and streaking of 10 μl of the specimens onto a sheep BAP (10). After incubation at 37°C in a CO2 incubator for 18 to 24 h, pneumococcal colonies suspected of being alpha-hemolytic were subcultured before they were tested for optochin susceptibility and bile solubility. Positive isolates were subsequently serotyped by the Quellung reaction with CDC antisera. Isolates found to be serotype 6A by the Quellung reaction were subsequently subjected to the serotype 6C-specific PCR assay, as described previously (5). Only one colony that represented each colony morphology encountered was picked.
(ii) BEC method.
The BEC method entailed streaking of a 10-μl loop of STHB-enriched culture for bacterial colony isolation on a BAP. For STHB-enriched specimens, five suspected pneumococcal colonies were tested for optochin susceptibility and bile solubility, even when only one type of colony morphology was evident. Individual isolates identified as S. pneumoniae were initially typed by the sequential multiplex PCR method. Eight specimens yielded less than five suspected pneumococcal colonies, which were also initially typed by the sequential multiplex PCR assay. Sequential multiplex PCR-based results for serotypes 7F/7A, 10F/10C/33C, 11A/11D, 12F/12A/44/46, 15A/15F, 22F/22A, 33F/33A/37, 35A/35C/42, and 35F/47 were further resolved to the individual serotype level by use of the Quellung reaction (these complex designations are listed with the commonly observed serotype listed first; e.g., for serotype 7F/7A, serotype 7F is commonly found, while 7A is extremely rare in our experience).
(iii) BEPCR method.
The BEPCR method employed 5.0 μl of DNA extracted directly from an STHB-enriched culture as the template in the sequential multiplex PCR assay.
(iv) DPCR method.
The DPCR method employed 5.0 μl of DNA extracted directly from the NP-STGG specimens in the sequential multiplex PCR assay.
Real-time PCR assay with broth-enriched and STGG-NP specimen DNA extracts for pneumococcal detection (BERT and lytA gene-specific real-time PCR for S. pneumoniae detection from DNA extracted directly from STGG-NP swab specimen [DRT] methods).
A pneumococcus-specific real-time PCR targeting the lytA gene was performed as described previously (5) with DNA extracts prepared from enriched STHB cultures (BERT method) or prepared directly from NP-STGG specimens (DRT method). Negative samples were defined as those with cycle threshold (CT) values of greater than 35.
Statistics.
P values were calculated by using the EpiInfo (version 3.4.3) program and Mantel-Haenszel chi-square testing of proportions. When cells had values of less than 5, two-tailed Fisher exact tests were used.
RESULTS
As described in the Materials and Methods, we used a sample of 100 STGG-NP specimens that had previously been screened for the presence of pneumococci by direct streaking of these vortexed specimens upon BAP plates (DC method in Table 2). We randomly selected from a large collection of specimens 50 specimens that were culture negative for pneumococci and 50 specimens that were culture positive. Only 1 of the 50 culture-positive specimens was found to contain more than one serotype (types 3 and 23B in specimen 83) by the DC method. Two of the culture-positive specimens (specimens 71 and 94) were nonserotypeable by the DC method.
TABLE 2.
Cumulative serotype and pneumococcus identification comparison of different methods with 100 preselected specimens tested by the DC method
| STGG-NP specimen no(s). | Serotype detected by the following methoda: |
No. of specimens |
||||
|---|---|---|---|---|---|---|
| DC | BEC | BEPCR | DPCR | DRT positives | BERT positive | |
| 1-34 | Neg | Neg | Neg | Neg | 1 | 9 |
| 35-37 | Neg | Neg | 021 | 021 | 3 | 3 |
| 38 | Neg | 021 | Neg | Neg | 0 | 1 |
| 39 | Neg | 35B, NTa | 35B | 35B | 1 | 1 |
| 40 | Neg | 35B | Neg | Neg | 0 | 0 |
| 41 | Neg | Neg | Neg | 35B | 1 | 0 |
| 42 | Neg | 034 | 034, 10F/10C/33C, 35A/35C/42 | 034, 10F/10C/33C, 35A/35C/42, 15B/15C | 1 | 1 |
| 43 | Neg | 15C, NT | 10F/10C/33C, 15B/15C | 10F/10C/33C | 1 | 1 |
| 44 | Neg | 23A | 23A | Neg | 0 | 1 |
| 45 | Neg | 17F, NT | 17F | Neg | 0 | 1 |
| 46 | Neg | Neg | Neg | 17F | 1 | 1 |
| 47 | Neg | 23B | 23B | neg | 0 | 1 |
| 48 | Neg | Neg | Neg | 06C | 1 | 1 |
| 49 | Neg | Neg | Neg | 22F/22A | 1 | 1 |
| 50 | Neg | Neg | Neg | 15B/15C | 1 | 0 |
| 51-55 | 06C | 06C | 06C | 06C | 5 | 5 |
| 56 | 06C | 06C, 15A | 06C, 15A/15F | 06C, 15A/15F | 1 | 1 |
| 57-58 | 06C | 06C | 06C | Neg | 0 | 2 |
| 59 | 021 | 021 | 021, 06C, 23B | 021, 06C, 23B | 1 | 1 |
| 60 | 021 | 021 | 021 | 021 | 1 | 1 |
| 61 | 021 | 021 | 021 | Neg | 0 | 1 |
| 62-63 | 15A | 15A | 15A/15F | 15A/15F | 2 | 2 |
| 64 | 15A | 15A, NT | 15A/15F, 11A/11D, 031 | 15A/15F, 11A/11D | 1 | 1 |
| 65-66 | 19A | 19A | 19A | 19A | 2 | 2 |
| 67-68 | 23A | 23A | 23A | 23A | 2 | 2 |
| 69 | 23A | 23A | 23A, 004, 35B | 23A | 1 | 1 |
| 70 | 23A | 23A | 23A | Neg | 1 | 1 |
| 71 | NT | 23A, NT | 23A | 23A | 1 | 1 |
| 72-73 | 11A | 11A | 11A/11D | 11A/11D | 2 | 2 |
| 74 | 11A | 11A | 11A/11D, 10F/10C/33C | 11A/11D | 1 | 1 |
| 75 | 11A | 11A, NT | 11A/11D, 10F/10C/33C, 15B/15C | 11A/11D, 15B/15C | 1 | 1 |
| 76 | 15B | 15B, 15C | 15B/15C, 16F | 15B/15C | 1 | 1 |
| 77 | 15C | 15C | 15B/15C | 15B/15C | 1 | 1 |
| 78 | 15C | 15B, 15C | 15B/15C | 15B/15C | 1 | 1 |
| 79 | 10A | 10A | 10A, 35A/35C/42 | 10A, 35A/35C/42 | 1 | 1 |
| 80-82 | 10A | 10A | 10A | 10A | 3 | 3 |
| 83 | 003, 23B | 003, 23B | 003, 23B | 003, 23B | 1 | 1 |
| 84 | 003 | 003 | 003 | 003 | 1 | 1 |
| 85-86 | 35A | 35A | 35A/35C/42 | 35A/35C/42 | 2 | 2 |
| 87 | 35A | 35A, 021 | 35A/35C/42, 10A, 020, 021 | 35A/35C/42, 10A, 21 | 1 | 1 |
| 88-89 | 23B | 23B | 23B | 23B | 2 | 2 |
| 90 | 23B | 23B | 23B | Neg | 0 | 1 |
| 91-92 | 33F | 33F | 33F/33A/37 | 33F/33A/37 | 2 | 2 |
| 93 | 034 | 034 | 034 | 034 | 1 | 1 |
| 94 | NT | 034 | 034 | 034 | 1 | 1 |
| 95 | 07F | 07F | 7F/7A | 7F/7A | 1 | 1 |
| 96 | 12F | 12F | 12F/12A/44/46 | 12F/12A/44/46 | 1 | 1 |
| 97 | 17F | 17F | 17F | 17F | 1 | 1 |
| 98 | 22F | 22F | 22F/22A | 22F/22A | 1 | 1 |
| 99 | 35B | 35B | 35B | 35B | 1 | 1 |
| 100 | 35F | 35F | 35F/47F | 35F/47F | 1 | 1 |
| No. of positive specimens (no. of serotype identifications) | 50 (49) | 58 (61) | 59 (78) | 56 (68) | 58 (NAa) | 72 (NA) |
Neg, negative result; NT, nontypeable; NA, not applicable.
Assessment of BEC, BEPCR, and DPCR methods for detection of pneumococcus.
Of the 50 preselected DC-negative NP-STGG specimens, 34 were negative by all serotype identification methods used and 16 were positive by at least one of the other three methods used for serotyping (Table 2, NP-STGG specimens 35 to 50). The enhancement effect of broth enrichment for culture-based and PCR-based pneumococcal detection/serotyping was apparent. These three alternative methods each detected significantly more additional pneumococcus-positive specimens than DC (0 of 50 negative specimens by DC, 8 [16%] additional positive specimens by BEC [P = 0.006], 9 [18%] additional positive specimens by BEPCR [P = 0.003], 11 [12%] additional positive specimens by DPCR [P < 0.0001]). The serotyping results obtained by the BEC method were in complete agreement with the serotypes deduced by PCR (Table 2). There were no instances of pneumococcus-negative BEC or BEPCR results that corresponded to positive DC results (Table 2), while DPCR showed the inferior result of being negative for five specimens that were DC positive (P = 0.06 compared with the result of DC; Table 2). All eight of the BEC-positive, DC-negative specimens yielded a single serotype result, while two to four serotypes were observed in two of the DC-negative specimens by the two PCR-based methods.
Detection of multiple serotypes.
Of the 50 randomly chosen DC-positive specimens (Table 2, STGG-NP specimens 51 to 100), 47 revealed a single serotype, 2 yielded nontypeable pneumococci, and only 1 specimen (specimen 83) revealed two serotypes that were also revealed by the other three methods. The results from the broth enrichment methods (both BEC and BEPCR) perfectly overlapped the positive serotyping results from the DC method and extended the DC method results by the detection of additional serotypes (Table 2). The BEC method detected three instances of dual serotypes (specimens 56, 83, and 87), while the BEPCR method detected two serotypes in six specimens (specimens 43, 56, 74, 76, 79, and 83). In addition, the BEPCR method detected three serotypes in five specimens (specimens 42, 59, 64, 69, and 75) and four serotypes in a single specimen (specimen 87).
Of the two specimens that yielded nontypeable isolates by the DC method, by use of the BEC method, one was found to contain type 23A as well as nontypeable isolates and the other yielded type 34 isolates. These two serotypes were in agreement with the results found by the BEPCR and DPCR methods. The PCR-based type 23A and 34 results were verified by testing of the isolates by the Quellung reaction.
Although it was the only serotyping method that yielded negative results (for a total of five specimens) among the DC-positive set, the DPCR method detected two serotypes in five specimens (specimens 56, 64, 75, 79, and 83), three serotypes in two specimens (specimens 59 and 87), and four serotypes in a single specimen (specimen 42) (Table 2).
Efficiency in detecting individual serotypes.
The BEPCR method cumulatively detected 24 different serotypes in the 100 NP-STGG specimens (Table 3). All culture-based positive results, which accounted for 19 serotypes, were reflected among these results (serotypes 15B and 15C were considered to be the same serotype, since they interconvert [17]). We found that BEPCR was the most effective method for the detection of individual serotypes (Table 3). For example, for the 10 serotype 6C-positive specimens, the BEPCR method was positive for serotype 6C in 9 of these specimens, while the other 3 methods were positive for serotype 6C in 8 of these specimens. Similarly, the BEPCR method gave the highest number of specimens positive for serotypes 21, 23B, and 10F/10C/33C. In addition, the BEPCR method was the only approach that provided positive results for serotypes 4, 20, and 31 in individual specimens.
TABLE 3.
Cumulative number of serotypes detected in the 100 study specimens by four different methods
| Serotype(s) | No. of positive specimensa |
||||
|---|---|---|---|---|---|
| Totalb | BEPCR | DPCR | BEC | DC | |
| 6C | 10 | 9 | 8 (1) | 8 | 8 |
| 21 | 8 | 7 (3) | 6 (3) | 5 (1) | 3 |
| 15B/Cc | 7 | 5 (1) | 6 (2) | 4 (1) | 3 |
| 23A | 6 | 6 (1) | 4 | 6 (1) | 4 |
| 23B | 6 | 6 (1) | 4 | 5 (1) | 4 |
| 11A/Dd | 5 | 5 | 5 | 4 | 4 |
| 10A | 5 | 5 | 5 | 4 | 4 |
| 35A/35C/42 | 5 | 5 (1) | 5 (1) | 3 | 3 |
| 35B | 5 | 3 (1) | 4 (2) | 3 (2) | 1 |
| 10F/10C/33C | 4 | 4 (2) | 2 (2) | 0 | 0 |
| 15A/15F | 4 | 4 | 4 | 4 | 3 |
| 34 | 3 | 3 (1) | 3 (1) | 3 (1) | 1 |
| 17F | 2 | 2 (1) | 1 (1) | 2 (1) | 1 |
| 22F/22A | 2 | 1 | 2 (1) | 1 | 1 |
| 19A | 2 | 2 | 2 | 2 | 2 |
| 33F/33A/37 | 2 | 2 | 2 | 2 | 2 |
| 3 | 2 | 2 | 2 | 2 | 2 |
| 4 | 1 | 1 | 0 | 0 | 0 |
| 7F/7A | 1 | 1 | 1 | 1 | 1 |
| 12F/12A/44/46 | 1 | 1 | 1 | 1 | 1 |
| 16F | 1 | 1 | 0 | 0 | 0 |
| 20 | 1 | 1 | 0 | 0 | 0 |
| 31 | 1 | 1 | 0 | 0 | 0 |
| 35F/47 | 1 | 1 | 1 | 1 | 1 |
| Total no. of positive specimens | 59 | 59 | 56 | 58 | 50 |
| Total no. of serotype identifications | 78 | 68 | 61 | 49 | |
| Cumulative no. of serotypes identified | 24 | 20 | 19 | 19 | |
The number found in DC-negative specimens, if any, is provided in parentheses.
Cumulative number of pneumococcus-positive specimens that were positive by one or more of the four methods used for serotype identification.
Serotype 15B/15C is considered a single serotype for this work. In some instances, the DC or BEC approach identified either or both serotype 15B and serotype 15C. The BEPCR and DPCR methods were not capable of distinguishing serotypes 15B and 15C.
For the BEC and DC methods, the complex serospecificities corresponded to the first serotype listed (e.g., for 11A/D, serotype 11A). The PCR-based approaches used are not capable of resolving these serotypes.
DC specimens that were randomly preselected revealed 49 serotypeable isolates and 2 nontypeable isolates. Overall, the BEPCR method was the most effective for serotype detection, yielding 78 positive serotype results in comparison to 68 and 61 positive serotype results for the DPCR and BEC methods, respectively (Tables 2 and 3). Of the two PCR-based serotyping methods used, only the DPCR method gave negative results for specimens (5 specimens) among the 50 specimens that were positive by the DC method. In addition, the DPCR method was negative for five additional specimens that were negative by the DC method but that were positive with the BEC method. While the DPCR method was negative for a total of 10 specimens that yielded culture-based (DC or BEC) positive results, the BEPCR method was negative in only 2 instances in which there were culture-based positive serotype results (2 specimens that were positive only by the BEC approach [1 specimen with serotype 21 and 1 specimen with serotype 35B] (Table 2).
One-to-one ratio of serotype to colony morphology by BEC method.
For the DC method, only one representative colony from each pneumococcal colony morphology encountered was serotyped. In contrast, for the BEC method, 5 individual colonies from each STGG-NP specimen from 50 of the 58 positive specimens were obtained and serotyped. For eight specimens, only one to four individual colonies were available after streaking of the specimens for the sequential multiplex assay. We found that the selection of multiple colonies exhibiting identical morphologies had no effect upon the outcome. There were no examples of a single specimen yielding multiple serotypes in association with a single colony morphology (data not shown), with the exception that in two specimens the BEC method exhibited both serotype 15B and serotype 15C in association with colonies with the same morphology. There was only one DC-positive specimen used for this study (specimen 83 in Table 2), and that specimen revealed two different colony morphologies, which corresponded to serotypes 3 and 23B, respectively. This identical serotype combination was also obtained by the other three methods. Two colony morphologies were evident from three specimens by the BEC method; and these corresponded to types 23B and 3 (specimen 83), types 6C and 15A (specimen 56), and types 35A and 21 (specimen 87).
Complex PCR-based serospecificities correspond to the most frequently encountered invasive serotypes.
Nine of the 23 PCR-based “serotypes” (serotypes 7F/7A, 10F/10C/33C, 11A/11D, 12F/12A/44/46, 15A/15F, 22F/22A, 33F/33A/37, 35A/35C/42, and 35F/47) found by the BEPCR and DPCR methods in the present study actually corresponded to combinations of serotypes (Tables 2 and 3), and the first serotype listed is the most common serotype of the combination found for invasive isolates recovered within the United States. For eight of these nine complex results obtained by the BEPCR and DPCR methods, corroborative culture-based results by the Quellung reaction revealing simple serotypes were obtained and corresponded to the most common invasive serotypes found in specimens with these combinations of serotypes (serotypes 7F, 11A, 12F, 15A, 22F, 33F, 35A, and 35F) during population-based surveillance in the United States (data not shown).
Serotype distribution.
According to the cumulative results revealed by the use of the four methods (Tables 2 and 3), a total of 24 serotypes were encountered among 65 specimens. All 24 serotypes were detected (within 59 specimens) by the BEPCR method. Recently discovered serotype 6C was the most frequent serotype encountered (10 specimens). Eleven other serotypes were detected among four to eight specimens. Only a single serotype (serotype 4) included in PCV7 was detected in a single specimen, circumstantially consistent with the prior vaccination of these individuals.
Real-time lytA-targeted PCR for STGG-NP specimens with and without broth enrichment.
Similar to the culture-based and conventional PCR-based testing results, a 4-h broth enrichment step appeared to enhance the sensitivity of the real-time lytA PCR assay (P = 0.04 compared with the results of BERT and DRT among DC-negative specimens; Table 2).
All 50 specimens that were DC positive for pneumococci were also positive by the real-time pneumococcus-specific lytA PCR assay performed with DNA extracted from broth-enriched STGG-NP specimens (BERT method). In addition, the BERT method was positive for 22 DC-negative specimens (P < 0.001). While the lytA PCR assay performed directly with STGG-NP specimen DNA extracts was positive for 12 DC-negative specimens, it gave negative results for 4 DC-positive specimens. Overall, there were a total of 72 BERT-positive specimens but only 58 DRT-positive specimens (Table 2). In general, the positive real-time lytA PCR results appeared to be consistent with the positive culture-based (DC or BEC) results, in the sense that 57 of 58 culture-positive specimens were BERT positive, and 51 of those specimens yielded CT values within the range of 15 to 29 (average CT, 23 among the 57 positive specimens), while the 15 culture-negative specimens that were BERT positive yielded CT values within the range of 31 to 35 (data not shown). Similarly consistent results were found when the results of the DRT method were compared to the culture-based findings.
DISCUSSION
The factors involved in the accurate measurement of carriage and carriage serotypes are still poorly understood. For this reason, studies that estimate invasiveness on the basis of the ability of pneumococcal strains to progress from NP carriage to invasive disease are still subjective, at least to some degree. Our findings indicate that current carriage methods are not optimally sensitive for the assessment of pneumococcal carriage and the serotype distribution and that the use of broth enrichment and PCR offer obvious improvements. Fully 44% of the DC-negative specimens were pneumococcus positive by the BERT method. All 50 of the DC method-positive specimens were positive by the BERT method. In addition, seven of the eight specimens that were DC method negative and BEC method positive were also positive by the BERT method. The majority (26/42; 62%) of specimens that were pneumococcus negative by the two culture-based methods were also BERT method negative (Table 2) and displayed no CT values (data not shown). Taken together, it is likely that BERT method-positive specimens correlate with the carriage of viable pneumococci. This likelihood calls into question the serotype distribution shown in Table 3, since the BERT method potentially detects strains that are poorly detected by the other methods used in this study. With this possibility in mind, we have developed individual real-time serotype identification assays for 17 key pneumococcal serotypes and are progressing toward a real-time sequential PCR assay for serotype deduction. At this point, it is impossible for us to speculate on the biological significance of the high incidence of previously undetected pneumococcus-positive carriage specimens in this study set. We do believe that these results give some indication of the poorly understood complexity of the pneumococcal carriage reservoir.
Three methods (BEC, BEPCR, and DPCR) appeared to be roughly equivalent in their abilities to detect pneumococcus-positive specimens overall (Table 2, 58, 59, and 56 specimens positive by the BEC, BEPCR, and DPCR methods, respectively); however, the DPCR method was inferior, in that it was the only method used that yielded negative results for DC-positive specimens (a total of 5 of 50 specimens). PCR-based approaches were superior overall to the corresponding culture-based methods in detecting pneumococcal serotypes. DPCR gave about 1.4-fold more positive serotype identifications than the corresponding DC method (Table 3; 68 and 49 serotype identifications, respectively). Similarly, the BEPCR method yielded about 1.3-fold more serotype identifications than the BEC method (78 and 61 serotype identifications, respectively). In large part, this was due to the ability of the two PCR approaches to identify multiple serotypes within the same specimen. Only 3 specimens yielded multiple serotypes (only two each) by use of the BEC approach, while DPCR and BEPCR yielded up to 4 serotypes in 8 and 12 specimens, respectively. It seems likely that the relative inferiority of culture-based approaches to the PCR-based methodology for the detection of multiple serotypes lies in large part to the masking of lower-density serotypes by the predominant serotype when specimens are streaked for colony isolation on BAPs. In addition, it also appears possible that the BEPCR method is more sensitive at detecting single unmixed serotypes present at lower densities, since it was positive for three specimens that yielded negative results by the other three methods (Table 3). However, this aspect is still unclear, since overall, conventional PCR was not much more effective for the detection of pneumococcus-positive specimens (59 specimens positive by BEPCR compared to 58 specimens positive by the corresponding BEC method).
The use of broth enrichment appeared to play a significant role in enhancing the identification of pneumococcus-positive specimens and detecting pneumococcal serotypes by both the culture- and PCR-based approaches. Contrary to the view expressed in a previous report (2), in which the investigators omitted a broth enrichment step to avoid the masking of minority serotypes, we found the reverse to be true, in that broth enrichment increased the total number of serotype identifications made (compare the 78 and 68 total serotype identifications for the BEPCR and DPCR methods, respectively) and resulted in the revealing of a greater extent of overall serotype diversity (compare the 23 and 20 cumulative serotypes for the BEPCR and DPCR approaches, respectively). From our experience, the length of the broth enrichment step should be limited to no more than 6 h to prevent excessive competition that masks more poorly growing and minority pneumococcal strains. For the identification of pneumococcus-positive specimens, the effect of broth enrichment is most apparent when the numbers of DRT-positive and BERT-positive specimens are compared (69% carriage positive by the BERT method compared to 58% by the DRT method). For the identification of individual serotypes, the use of broth enrichment also significantly increased the total number of serotype identifications in both culture-based approaches since the BEC method increased the number of individual serotype identifications found by the DC method by 1.24-fold (61 and 49, respectively; Table 3). One important observation was that the serotype determination of multiple (up to five) colonies from the same specimen that exhibited the same morphology added no benefit. For all pneumococci recovered by the BEC method, there was a 1:1 correlation of the serotype to the colony morphology. The BEC method resulted in the detection of two serotypes (excluding the codetection of serotypes 15B and 15C) in two situations in which the DC method detected only one. This was due to the fact that in both of these instances we detected two different colony morphologies, while only one morphology was detected for the same specimens by the DC method. This indicates that broth enrichment adds an advantage to the detection of multiple colony morphologies, probably through enhancement of the growth of minority strains to the level of detection upon subculture to BAP plates.
There are some distinct technical disadvantages inherent to PCR-based serotype deduction that must still be overcome. During the present study, all specimens yielding PCR-based serotyping results also yielded a corresponding control band for the cpsA gene target, the sequence of which is highly conserved among pneumococcal capsular polysaccharide biosynthetic (cps) loci (data not shown). While we have obtained this result for the vast majority of pneumococcal isolates that we have encountered, this is, unfortunately, not an absolute correlation, since approximately 1 to 2% of the PCR-serotypeable isolates were PCR negative for the cpsA control. We have found that serotypes 38 and 25F are generally negative for the cpsA band. Serotype 14 isolates have rarely been negative for this control as well. We intend to replace cpsA with an alternative, more reliable positive control. Although it is logical to formulate serotype-specific primer sets on the basis of variable cps locus genes that are predicted to encode serotype-specific functions, another potential confounder is the fact that the exact genetic signatures within these genes that dictate substrate differences among related serotypes are largely unknown. For example, even though it appears to be probable that the different wzy genes (which encode serotype-specific oligosaccharide repeat unit polymerases) expressed between serotypes 19A and 19F provide the basis of the structural differences between the serotype 19A and 19F capsules (1), we previously encountered one serotype 19F isolate that had a more serotype 19A-like wzy gene that yielded a false-positive sequential multiplex PCR result for serotype 19A (13).
Another potential disadvantage of PCR-based serotype deduction assays is that the known capsular biosynthetic loci from other closely related sets of serotypes are virtually indistinguishable, which currently prevents their resolution. In many of these instances, this disadvantage could be insignificant if only the most common serotype is taken into account. For example, of the 1,173 pneumococcal isolates corresponding to the nine complex PCR-based serotypes that were found during this study (7F/7A, 10F/10C/33C, 11A/D, 12F/12A/44/46, 15A/15F, 22F/22A, 33F/33A/37, 35A/35C/42, and 35F/47) and that were recovered during population-based invasive disease surveillance in the United States during 2007 (see http://www.cdc.gov/ncidod/dbmd/abcs/), only 1 isolate (a single type 15F isolate [unpublished observations]) did not correspond to serotype 7F, 11A, 12F, 15A, 22F, 33F, 35A, or 35F (no serotype 10F, 10C, or 33C invasive isolates were recovered). The Quellung reaction-based results obtained by the DC and BEC methods also revealed only the common invasive serotypes corresponding to eight of the nine complex serotypes found by the PCR methodology in this study (there were no positive pneumococcal culture results for the serotype 10F/10C/33C-positive specimens found by the PCR-based methods). Although these minor serotypes have rarely been encountered among invasive disease isolates recovered in the United States, it remains to be seen if the same serotypes are also rare or nonexistent in carriage among various populations. A final potentially misleading aspect of PCR-based serotype deduction is the fact that the relatively rare strains that do not express capsule may be PCR positive for specific serotypes by this assay and would be nontypeable by testing by the Quellung reaction.
Although this work focused on different approaches for positive pneumococcal detection and serotyping, it also draws attention to the continuing effort to accurately assess the dynamic seroepidemiology of this pathogen. It was interesting that PCV7-targeted serotypes 6A and 6B were not encountered, while recently discovered serotype 6C was the most frequent serotype encountered among this population of previously vaccinated infants. This serotype previously remained undetected among our conventionally serotyped serotype 6A isolates, and only recently, we have found through the use of a PCR assay that this is currently the predominant serogroup 6 serotype among invasive isolates recovered in the United States in the post-PCV7 period (4).
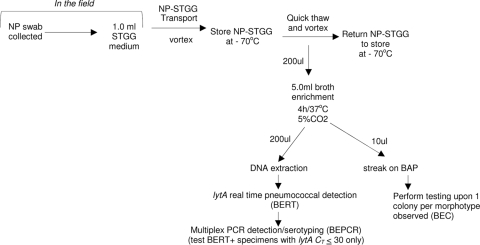
On the basis of our findings, we recommend the use of an initial broth enrichment step, as described for this study, for resolution of carriage serotypes from stored NP-STGG specimens. We recommend, then, the extraction of DNA from broth-enriched growth and performance of the BERT assay. DNA extracts corresponding to BERT assay-positive specimens should be subjected to the BEPCR assay (Fig. 1, bottom left flowchart). Of the total of 75 pneumococcus-positive specimens found through the use of all the approaches seen in Table 2, only 3 were not positive by the BERT assay. In addition, when testing of carriage isolates is required (by the Quellung reaction, susceptibility testing, etc.), we recommend the testing of only one colony of each colony morphology (Fig. 1, bottom right flowchart). To some degree, the DPCR and BEPCR approaches are complementary. For example, the DPCR approach provided six serotype calls for DC-negative specimens that were missed by the BEPCR approach. Even though BEPCR was superior to DPCR in the overall assessment (Table 3), the use of both approaches is worth considering in the future when a less laborious and more multiplexed technology is available.
FIG. 1.
Flowchart suggested for use for determination of serotype diversity and characterization of pneumococcal isolates from nasopharyngeal carriage specimens.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Aanensen, D. M., A. Mavroidi, S. D. Bentley, P. R. Reeves, and B. G. Spratt. 2007. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7856-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonio, M., I. Hakeem, K. Sankareh, Y. B. Cheung, and R. A. Adegbola. 2009. Evaluation of sequential multiplex PCR for direct detection of multiple serotypes of Streptococcus pneumoniae from nasopharyngeal secretions. J. Med. Microbiol. 58:296-302. [DOI] [PubMed] [Google Scholar]
- 3.Azzari, C., M. Moriondo, G. Indolfi, C. Massai, L. Becciolini, M. de Martino, and M. Resti. 2008. Molecular detection methods and serotyping performed directly on clinical samples improve diagnostic sensitivity and reveal increased incidence of invasive disease by Streptococcus pneumoniae in Italian children. J. Med. Microbiol. 57:1205-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho, M. D. G., F. C. Pimenta, R. E. Gertz, Jr., H. H. Joshi, A. A. Trujillo, L. Keys, J. Findley, I. S. Moura, I. H. Park, S. K. Hollingshead, T. Pilishvili, C. G. Whitney, M. H. Nahm, B. W. Beall, and the Active Bacterial Core Surveillance Team. 2009. PCR-based quantitation and clonal diversity of the current prevalent invasive serogroup 6 pneumococcal serotype, 6C, in the United States in 1999 and 2006 to 2007. J. Clin. Microbiol. 47:554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho, M. D. G., M. L. Tondella, K. McCaustland, L. Weidlich, L. McGee, L. W. Mayer, A. Steigerwalt, M. Whaley, R. R. Facklam, B. Fields, G. Carlone, E. W. Ades, R. Dagan, and J. S. Sampson. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J. Clin. Microbiol. 45:2460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dias, C. A., L. M. Teixeira, M. da G. Carvalho, and B. Beall. 2007. Sequential multiplex PCR for determining capsular serotypes of pneumococci recovered from Brazilian children. J. Med. Microbiol. 56:1185-1188. [DOI] [PubMed] [Google Scholar]
- 7.Gray, B. M., G. Converse III, and H. Dillon, Jr. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage and infections during the first 24 months of life. J. Infect. Dis. 142:923-933. [DOI] [PubMed] [Google Scholar]
- 8.Millar, E. V., K. L. O'Brien, J. R. Scott, D. Jackson, C. G. Whitney, R. Reid, M. Santosham, and the American Indian LTNP Study Group. Near elimination of vaccine-type pneumococcal carriage by pneumococcal conjugate vaccine in a community at high risk of carriage and disease, abstr. P3-090, p. 335. Abstr. 6th Int. Symp. Pneumococci Pneumococcal Dis.
- 9.Morais, L., M. D. G. Carvalho, A. Roca, B. Flannery, I. Mandomando, M. Soriano-Gabarró, B. Sigauque, P. Alonso, and B. Beall. 2007. Sequential multiplex PCR for identifying pneumococcal capsular serotypes from South-Saharan African clinical isolates. J. Med. Microbiol. 56:1181-1184. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien, K. L., M. A. Bronsdon, R. Dagan, P. Yagupsky, J. Janco, J. Elliott, C. G. Whitney, Y. H. Yang, L. G. Robinson, B. Schwartz, and G. M. Carlone. 2001. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J. Clin. Microbiol. 39:1021-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pai, R., R. E. Gertz, and B. Beall. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44:124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, I. H., D. Pritchard, R. Cartee, A. Brandao, M. Brandileone, and M. Nahm. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pimenta, F. C., R. E. Gertz, Jr., A. Roundtree, J. Yu, M. H. Nahm, R. R. McDonald, M. D. G. Carvalho, and B. W. Beall. 2009. Rarely occurring 19A-like cps locus from a serotype 19F pneumococcal isolate indicates continued need of serology-based quality control for PCR-based serotype determinations. J. Clin. Microbiol. 47:2353-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha, S. K., G. L. Darmstadt, A. H. Baqui, B. Hossain, M. Islam, D. Foster, H. Al-Emran, A. Naheed, S. E. Arifeen, S. P. Luby, M. Santosham, and D. Crook. 2008. Identification of serotype in culture negative pneumococcal meningitis using sequential multiplex PCR: implication for surveillance and vaccine design. PLoS One 3:e3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syrjaren, R. K., K. Auranen, T. Leino, T. Kilpi, and P. Mäkelä. 2005. Pneumococcal acute otitis media in relation to pneumococcal nasopharyngeal carriage. Pediatr. Infect. Dis. J. 24:801-806. [DOI] [PubMed] [Google Scholar]
- 16.van Gils, E. J., R. H. Veenhoven, E. Hak, G. D. Rodenburg, D. Bogaert, E. P. Ijzerman, J. P. Bruin, L. van Alphen, and E. A. Sanders. 2009. Effect of reduced-dose schedules with 7-valent pneumococcal conjugate vaccine on nasopharyngeal pneumococcal carriage in children: a randomized controlled trial. JAMA 302:159-167. [DOI] [PubMed] [Google Scholar]
- 17.van Selm, S., L. M. M. A. van Cann, M. A. Kolkman, B. A. van der Zeijst, and J. P. van Putten. 2003. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect. Immun. 71:6192-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]