Abstract
A novel family of Burkholderiales bacteria was identified in ileal biopsy specimens from children presenting with symptoms of inflammatory bowel disease. A molecular subtyping approach based on sequencing of a variable region of the bacteria's 23S rRNA genes identified three variants. Pilot analysis identified one variant to be significantly associated with perianal Crohn's disease.
The Burkholderiales order comprises four families of Gram-negative bacteria, some of which harbor species that have been implicated in human diseases such as respiratory infections (Bordetella spp.) and chronic granulomatous disease (Burkholderia cepacia complex spp.) (5, 6, 9, 14). Members of the Burkholderiales have not been previously described in association with gastrointestinal disease. However, Burkholderiales belong to the phylum Proteobacteria, which has been documented to persist in and dominate the gut microbiota of patients with active inflammatory bowel disease (IBD) (4, 11, 12).
IBD includes Crohn's disease (CD) and ulcerative colitis, which are lifelong diseases associated with chronic inflammation anywhere along the gastrointestinal tract. CD is hypothesized to be initiated by an environmental trigger in individuals who are genetically susceptible.
Preliminary subtractive hybridization experiments conducted in our laboratory identified a unique bacterial genome sequence of rRNA genes in ileal tissues from patients with CD. This unique sequence exhibited 89% nucleotide identity to the 23S rRNA genes of Bordetella petrii, a species of the Burkholderiales order. The low sequence homology to 23S rRNA genes of members of the Burkholderiales suggested that this is a new bacterium not previously identified in the human intestine.
Techniques for metagenomic identification and characterization of bacteria have previously used comparative sequence analysis of rRNA genes to infer evolutionary relationships and define bacterial species (7, 8, 10). The objectives of this study were to characterize the new Burkholderiales bacterial species using 23S rRNA gene sequence analysis and to compare its prevalence in ileal tissues from pediatric patients with CD with that in tissues from non-IBD control patients.
Sixty-nine patients (aged 3 to 16 years; mean age, 12 ± 3.45 years) undergoing initial endoscopy at The Royal Children's Hospital, Victoria, Australia, with symptoms suggestive of inflammatory bowel disease (IBD), were prospectively recruited into the study under ethics approval from the Human Ethics Committee of The Royal Children's Hospital (EHRC no. 23003). None of the 36 CD and 33 non-IBD patients had received antibiotics or immunosuppressive drugs prior to endoscopy. The diagnosis of CD was established according to the Montreal classification (13).
Briefly, DNA was extracted from ileal biopsy specimens (3 to 6 mm3) using a standard laboratory protocol (16). The prevalence of Burkholderiales bacteria in CD and non-IBD patients was investigated using PCR targeting a 1.9-kb region of the 23S rRNA gene (Fig. 1). Primer WSbugF (5′ ATG GGT CAG CGA CTT ACG TT) was designed based on the unique 23S rRNA gene sequence obtained by subtractive hybridization, while 2490r (5′ CGA CAT CGA GGT GCC AAA C) was a previously published conserved 23S rRNA gene reverse primer (7). The Expand High Fidelity (EHF) PCR system (Roche) was used to amplify 5 μl of DNA in a 50-μl PCR mixture containing 2.6 U EHF enzyme mix, 1× EHF buffer, 1.5 mM MgCl2, and a mixture of 200 μM deoxynucleoside triphosphates (dNTPs) and 300 nM each primer. PCR conditions were denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 2 min, and a final extension step at 72°C for 7 min. PCR products were analyzed using a 1% Tris-borate-EDTA (TBE) agarose gel and visualized using ethidium bromide and UV illumination. A total of 22 of 36 (61%) CD specimens and 26 of 33 (79%) non-IBD patient specimens were positive for the Burkholderiales 23S rRNA gene.
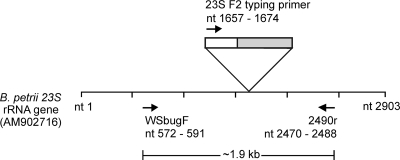
FIG. 1.
A schematic representation of the map location of primers used in this study, against a GenBank reference B. petrii 23S rRNA (accession no. AM902716). Primer WSbugF (nt 572 to 591) was designed based on the unique 23S rRNA gene identified previously by subtractive hybridization and amplifies a 1.9-kb region with primer 2490r (nt 2470 to 2488). The typing primer 23S F2 (5′ GGA ACT CGG CAA ATT GAC) binds to the conserved sequence from nt 1657 to nt 1674 and amplifies the variable region from nt 1690 to nt 1810 (in gray). This region contains the 78-bp diagnostic region, nt 1721 to nt 1798.
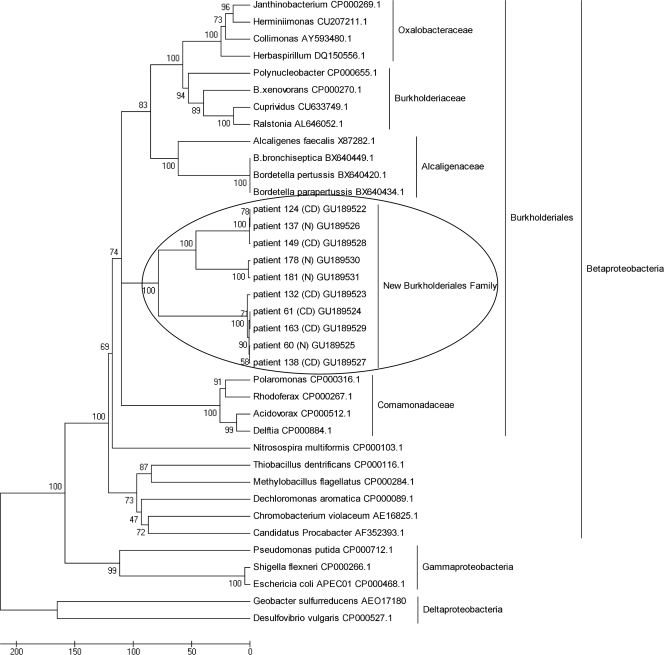
Burkholderiales-positive 1.9-kb 23S rRNA gene PCR products from six CD and four non-IBD patients were cloned and transformed into TOP10 One Shot chemically competent cells (Invitrogen), and sequenced using BigDye Terminator version 3.1. Sequence analysis was verified using Sequencher (Gene Codes Corp., Inc., Ann Arbor, MI), and phylogenetic analysis was conducted using MEGA version 4.0 (15). Figure 2 illustrates how the 1.9-kb 23S rRNA gene sequences from CD and non-IBD patients form a distinct phylogenetic cluster within the Burkholderiales order, separate from the other four existing families. All sequences were highly similar to each other and exhibited 89 to 99% nucleotide identity by pairwise analyses.
FIG. 2.
Neighbor-joining phylogenetic tree, based on partial 23S rRNA gene sequences (1.9 kb), showing positions of strains isolated from patient ileal biopsy specimens (CD patients 124, 149, 132, 61, 163, and 138; non-IBD patients 137, 178, 181, and 60) and related Proteobacteria species from GenBank. Four representative reference genes from each of the Burkholderiales families, Alcaligenaceae, Oxalobacteraceae, Burkholderiaceae, and Comamonadaceae, were retrieved from GenBank for this comparison. Additionally, reference genes of the Gammaproteobacteria and Deltaproteobacteria classes were included to serve as outgroups in the phylogenetic tree. GenBank accession numbers are shown beside each reference name. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Bar, 0.05 number of nucleotide differences.
Based on sequence alignments, a 78-bp variable region (nucleotides [nt] 1721 to 1798 of reference strain Bordetella petrii 23S rRNA genes) was identified. Subtype analysis was performed on sequences from 17 CD and 23 non-IBD patient isolates, which were then categorized into three distinct sequevars: “124-type,” “var124-type,” and “60-type” (Fig. 3). Table 1 details the distribution of CD and non-IBD patients harboring each sequevar. While there appears to be a trend for the 124- and var124-types to be associated with non-IBD patients, this observation was not significant (P = 0.153, two-tailed Fisher's exact test).
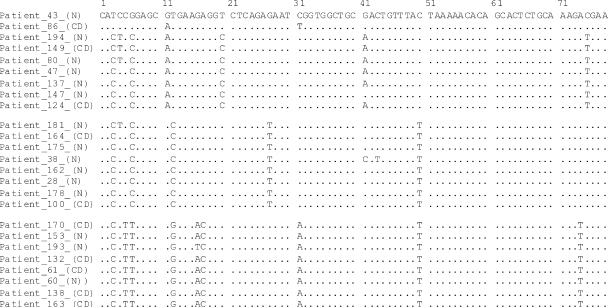
FIG. 3.
Multiple sequence alignment of 78-bp partial 23S rDNA sequences from 25 patients: 10 CD and 15 non-IBD (N) patients. The patient 43 sequence is taken as the consensus. There are three distinct variants: “124-type,” which includes patients 86, 194, 149, 80, 47, 137, 147, and 124; “var124-type,” which encompasses patients 181, 164, 175, 38, 162, 28, 178, and 100; and “60-type” for patients 170, 153, 193, 132, 61, 60, 138, and 163. Patient 43 is distinct from each of the three variants but has the fewest differences with the var124-type.
TABLE 1.
Distribution of Burkholderiales sequevars in CD and non-IBD patients
| Patient group | No. of patients in sequevar |
||
|---|---|---|---|
| 124-type | var124-type | 60-type | |
| CD | 4 | 6 | 7 |
| Non-IBD | 10 | 9 | 4 |
To determine the association between each variant type and disease behavior and location, we analyzed the phenotypic details available from 12 CD patients (Table 2). Perianal involvement (p) was present in six of seven patients with the 60-type Burkholderiales strain but not in any of the five patients with the 124-type strain (P = 0.015, Fisher's exact test). There was also a tendency for the 60-type patients (six of seven) to have ileocolonic disease either with or without upper gastrointestinal involvement (L3±L4), while only two of five 124-type patients had ileocolonic disease (L3). This observation is consistent with those of other studies suggesting that the altered mucosal environment in CD patients could select for the growth of particular strains of bacteria (3, 16). Future studies of a larger cohort of CD patients will be necessary to determine whether this association is significant.
TABLE 2.
Crohn's disease patient phenotypes and the corresponding variants of the newly described Burkholderiales strains (60-type, 124-type, and var124-type)a
| Variant | Patient no. | Location(s) of diseaseb | Behavior of diseasec |
|---|---|---|---|
| 60-type | 61 | L3 | B2p |
| 132 | L3 | B1 | |
| 138 | L3 | B1p | |
| 163 | L4 | B1p | |
| 170 | L3+L4 | B1p | |
| 195 | L3 | B1p | |
| 197 | L3+L4 | B1p | |
| 124-type | 86 | L1 | B1 |
| 124 | L3+L4 | B1 | |
| 149 | L3+L4 | B1 | |
| var124-type | 91 | L2 | B1 |
| 100 | L1+L4 | B1 |
All patients were 16 years or younger at the time of diagnosis (phenotype A1). Phenotypes are based on the Montreal classification system (13).
L1, terminal ileum; L2, colon; L3, ileocolon; L4, upper gastrointestinal tract; L1+L4, terminal ileum plus upper gastrointestinal tract; L3+L4, ileocolon plus upper gastrointestinal tract.
B1, nonstricturing, nonpenetrating; B2, stricturing; p, perianal disease.
This study identified a novel family of bacteria within the Burkholderiales order and Proteobacteria phylum. The new Burkholderiales group comprises three sequevars, 60-type, 124-type, and var124-type, that can be distinguished by sequence analysis of a 78-bp hypervariable region of the 23S rRNA gene. Preliminary data suggest an association of the 60-type with perianal Crohn's disease and detail the importance of stratifying patients into specific disease subgroups to allow better assessment of microbial associations (1). Future studies characterizing the single nucleotide polymorphism (SNP) expression profile of genes commonly associated with CD could precisely stratify patients and potentially identify gene mutations that make a patient susceptible to particular infectious triggers.
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank with accession numbers GU189522 to GU189531.
Acknowledgments
W. H. Sim is a recipient of a Melbourne International Research Scholarship from the University of Melbourne. C. D. Kirkwood is an NHMRC CDA Fellow (334 364). This research was supported by research grants from the CASS foundation, the Lynne Quayle Charitable Trust, and Murdoch Childrens Research Institute.
Footnotes
Published ahead of print on 17 March 2010.
REFERENCES
- 1.Ahmad, T. 2004. Phenotype-determining genes in inflammatory bowel disease. Novartis Found. Symp. 263:17-40, 211-218. [DOI] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Conte, M. P., S. Schippa, I. Zamboni, M. Penta, F. Chiarini, L. Seganti, J. Osborn, P. Falconieri, O. Borrelli, and S. Cucchiara. 2006. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 55:1760-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank, D. N., A. L. St. Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guide, S. V., F. Stock, V. J. Gill, V. L. Anderson, H. L. Malech, J. I. Gallin, and S. M. Holland. 2003. Reinfection, rather than persistent infection, in patients with chronic granulomatous disease. J. Infect. Dis. 187:845-853. [DOI] [PubMed] [Google Scholar]
- 6.Harrington, A. T., J. A. Castellanos, T. M. Ziedalski, J. E. Clarridge III, and B. T. Cookson. 2009. Isolation of Bordetella avium and novel Bordetella strain from patients with respiratory disease. Emerg. Infect. Dis. 15:72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt, D. E., V. Klepac-Ceraj, S. G. Acinas, C. Gautier, S. Bertilsson, and M. F. Polz. 2006. Evaluation of 23S rRNA PCR primers for use in phylogenetic studies of bacterial diversity. Appl. Environ. Microbiol. 72:2221-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig, W., and K. H. Schleifer. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol. Rev. 15:155-173. [DOI] [PubMed] [Google Scholar]
- 9.Mahenthiralingam, E., A. Baldwin, and C. G. Dowson. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104:1539-1551. [DOI] [PubMed] [Google Scholar]
- 10.Peplies, J., F. O. Glockner, R. Amann, and W. Ludwig. 2004. Comparative sequence analysis and oligonucleotide probe design based on 23S rRNA genes of Alphaproteobacteria from North Sea bacterioplankton. Syst. Appl. Microbiol. 27:573-580. [DOI] [PubMed] [Google Scholar]
- 11.Peterson, D. A., D. N. Frank, N. R. Pace, and J. I. Gordon. 2008. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3:417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sartor, R. B. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134:577-594. [DOI] [PubMed] [Google Scholar]
- 13.Silverberg, M. S., J. Satsangi, T. Ahmad, I. D. Arnott, C. N. Bernstein, S. R. Brant, R. Caprilli, J. F. Colombel, C. Gasche, K. Geboes, D. P. Jewell, A. Karban, E. V. Loftus, Jr., A. S. Pena, R. H. Riddell, D. B. Sachar, S. Schreiber, A. H. Steinhart, S. R. Targan, S. Vermeire, and B. F. Warren. 2005. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can. J. Gastroenterol. 19(Suppl. A):5-36. [DOI] [PubMed] [Google Scholar]
- 14.Spilker, T., A. A. Liwienski, and J. J. LiPuma. 2008. Identification of Bordetella spp. in respiratory specimens from individuals with cystic fibrosis. Clin. Microbiol. Infect. 14:504-506. [DOI] [PubMed] [Google Scholar]
- 15.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 16.Wagner, J., K. Short, A. G. Catto-Smith, D. J. Cameron, R. F. Bishop, and C. D. Kirkwood. 2008. Identification and characterisation of Pseudomonas 16S ribosomal DNA from ileal biopsies of children with Crohn's disease. PLoS One 3:e3578. [DOI] [PMC free article] [PubMed] [Google Scholar]