Abstract
Sin Nombre virus (SNV), Andes virus (ANDV), and Laguna Negra virus (LANV) have been known as the dominant causative agents of hantavirus pulmonary syndrome (HPS). ANDV and LANV, with different patterns of pathogenicity, exist in a sympatric relationship. Moreover, there is documented evidence of person-to-person transmission of ANDV. Therefore, it is important in clinical medicine and epidemiology to know the serotype of a hantavirus causing infection. Truncated SNV, ANDV, and LANV recombinant nucleocapsid proteins (trNs) missing 99 N-terminal amino acids (trN100) were expressed using a baculovirus system, and their applicability for serotyping SNV, ANDV, and LANV infection by the use of enzyme-linked immunosorbent assays (ELISA) was examined. HPS patient sera and natural-reservoir rodent sera infected with SNV, ANDV, and LANV showed the highest optical density (OD) values for homologous trN100 antigens. Since even patient sera with lower IgM and IgG antibody titers were serotyped, the trN100s are therefore considered useful for serotyping with early-acute-phase sera. In contrast, assays testing whole recombinant nucleocapsid protein antigens of SNV, ANDV, and LANV expressed in Escherichia coli detected homologous and heterologous antibodies equally. These results indicated that a screening ELISA using an E. coli-expressed antigen followed by a serotyping ELISA using trN100s is useful for epidemiological surveillance in regions where two or more hantavirus species cocirculate.
Hantaviruses belong to the Hantavirus genus in the Bunyaviridae family. Hantaviruses cause two rodent-borne febrile illnesses in humans, hemorrhagic fever with renal syndrome (HFRS) in the Old World and hantavirus pulmonary syndrome (HPS) in the New World (11, 25). So far, 23 virus species have been registered within the Hantavirus genus. Among the Old World hantaviruses, Hantaan virus (HTNV), Seoul virus (SEOV), Dobrava-Belgrade virus (DOBV), and Puumala virus (PUUV) are commonly associated with HFRS, while the New World species Sin Nombre virus (SNV), New York virus (NYV), Black Creek Canal virus (BCCV), Andes virus (ANDV), and Laguna Negra virus (LANV) regularly cause HPS in the New World (25).
Since 1993, when HPS was first identified in the New World (20), many new hantaviruses with or without human disease have been described throughout North, Central, and South America. ANDV and LANV, with different pathogenicity patterns and with approximately 40% and 15% mortality rates, respectively, exist in a sympatric relationship in Argentina (10, 14). Moreover, there is documented evidence of person-to-person transmission of some kind of ANDV strain (15, 22). However, since the neutralization test (NT), which is the only serological assay available for serotyping, needs specialized techniques and equipment and requires a containment laboratory for virus manipulation (2), serological typing of ANDV and LANV infection has been limited.
Hantavirus virions contain three segmented negative-sense RNAs designated S, M, and L; they encode a nucleocapsid protein (N), enveloped glycoproteins (Gn and Gc), and an RNA-dependent RNA polymerase (L protein), respectively (4). Hantavirus N is the most abundant viral component in both virions and infected cells and can form a stable trimer (7, 12). The N of Old World hantaviruses possesses immunodominant linear epitopes around the first 100 amino acids (aa) of the N terminus (6, 8, 32). These N-terminal epitopes cross-reacted with all of the Old World hantaviruses except PUUV. On the other hand, the variable region at around 230 to 302 aa forms serotype-specific epitopes after multimerization of N (30, 36).
We have developed a baculovirus that expresses truncated recombinant N (trN) lacking 49 aa of the N-terminal end of the N (trN50). trN50 showed decreased reactivity to cross-reactive antibodies but preserved reactivity to serotype-specific antibodies after multimerization of trNs. Use of an enzyme-linked immunosorbent assay (ELISA) system with trN50 successfully differentiated four hantavirus infections with HTNV, SEOV, DOBV, and Thailand virus (THAIV) in HFRS patient and rodent sera. Therefore, it seemed that the ELISA was a faster, safer, and simpler serotyping method than and an effective substitute for the NT (2, 19).
In the present study, we attempted to apply similar N-terminal deletion N antigens for serotyping using ELISA. We first selected SNV, ANDV, and LANV, 3 New World hantaviruses that are major causative agents of HPS, and examined the multimerization of trNs and their antigenic efficacy. We then used the trNs for serotyping of SNV, ANDV, and LANV infections.
MATERIALS AND METHODS
cDNAs and cells.
cDNAs containing coding information for N of SNV strain SN 77734 (5), ANDV (23), and LANV strain 510B (9) were used. High Five cells (Invitrogen, Groningen, Netherlands) were grown in Grace's insect cell culture medium (Invitrogen) supplemented with 10% fetal bovine serum as previously described (2).
MAbs and human and rodent sera.
Monoclonal antibodies (MAbs) to N of HTNV and PUUV were used for antigenic characterization of N by indirect immunofluorescence assay (IFA). MAbs 2E12, 4C3, 4E5, GBO4, ECO2, 1C12, and ECO1 recognize the N-terminal epitope of N. MAbs E5/G6 and F23A1 recognize aa 165 to 173 and aa 291 to 402 of N, respectively. The epitope for MAb C16D11 is unknown (21, 24, 34).
Eleven serum samples from HPS patients infected with SNV in the United States were kindly supplied by Brian Hjelle of the University of New Mexico Health Sciences Center. Eleven serum samples from HPS patients infected with ANDV and six serum samples from HPS patients infected with LANV were obtained from Argentina. Thirty-one serum samples from Peromyscus maniculatus infected with SNV and five hantavirus-negative serum samples from Peromyscus maniculatus were obtained from Canada. Twenty-three serum samples from Sigmodontinae rodents (Oligoryzomys longicaudatus, Oligoryzomys flavescens, and Akodon azarae) infected with ANDV and five serum samples from LANV-infected Calomys callosus were obtained from Argentina. Hantavirus-negative human sera were obtained from healthy volunteers. Negative-control rodent sera (Sigmodon hispidus) were kindly supplied by Kimiyuki Tsuchiya of Applied Biology Co. Ltd., Tokyo, Japan. The types of virus in sera from the patients and rodents were determined by detection of the virus genome by reverse transcription-PCR (RT-PCR).
Construction of recombinant baculoviruses expressing whole rNs and trNs.
The gene encoding whole N (aa 1 to 428) and truncated genes encoding truncated recombinant N (aa 50 to 428 [trN50] and aa 100 to 428 [trN100]) were amplified from cDNA of the S segment by the use of PCR. The primers listed below amplified whole and truncated S segments. A 5′ SpeI site and a 3′ XhoI site were introduced into the primers (both sites are shown in italics below). The sequences of the primers (forward and reverse, respectively) were as follows (underlining indicates an added start codon): for SNV whole rN, 5′-gacactagtatgagcaccctcaaagaa-3′ and 5′-tacctcgagttaaagtttaagtttaagtggttc-3′; for ANDV whole rN, 5′-aaaactagtatgagcaacctccaagaa-3′ and 5′-ttactcgagttacagctttaatggc-3′; for LANV whole rN, 5′-taaactagtatgagcaacctccaagaa-3′ and 5′-actctcgagttagagttttaggggttc-3′; for SNV trN50, 5′-tcgactagtatggctgtgtctgcattggag-3′ and 5′-tacctcgagttaaagtttaagtttaagtggttc-3′; for ANDV trN50, 5′-agaactagtatggctgtgtctacactggag-3′ and 5′-ttactcgagttacagctttaatggc-3′; for LANV trN50, 5′-agcactagtatggctgtgtctgcattggag-3′ and 5′-actctcgagttagagttttaggggttc-3′; for SNV trN100, 5′-tcgactagtatggctgtgtctgcattggag-3′ and 5′-tacctcgagttaaagtttaagtttaagtggttc-3′; for ANDV trN100, 5′-cgaactagtatgaatgtcctggatgtcaac-3′ and 5′-ttactcgagttacagctttaatggc-3′; and for LANV trN100, 5′-ctgactagtatgaatgtcctggatgtcaat-3′ and 5′-actctcgagttagagttttaggggttc-3′. After amplification, the DNA fractions were subcloned into pFastBac1 (Invitrogen) and were expressed using a Bac-to-Bac baculovirus expression system (Invitrogen) according to the manufacturer's instructions. Mock baculovirus was made from pFastBac1. The titers of recombinant baculoviruses in the culture supernatant were determined by calculation of 50% tissue culture infective dose (TCID50) values with High Five cells.
Preparation of whole rNs and trNs expressed by baculoviruses.
High Five cells were infected for 3 days with recombinant baculoviruses at a multiplicity of infection (MOI) of 1. Collection and lysis of infected cells were performed using previously published methods (2). Briefly, infected High Five cells were collected in phosphate-buffered saline (PBS) with 2.5 × 106 cells/ml and sonicated. The cell lysate containing recombinant Ns (rNs) was used as an IgG and IgM ELISA antigen. The cell lysate infected with mock baculovirus was used as a negative control. Expression of rNs of SNV, ANDV, and LANV was confirmed by Western blotting (WB) (data not shown) using previously published methods (35). High Five cells expressing whole rNs of PUUV and HTNV were prepared as previously described (2). High Five cells expressing whole rNs and trNs were used for IFA.
Preparation of rNs expressed by E. coli.
The whole rNs of SNV, ANDV, and LANV were expressed in Escherichia coli. DNA fractions containing the entire coding region of N were made by digestion of pFastBac1 with SalI and XhoI. The DNA fractions were subcloned into the pET43b vector (Merck KGaA, Darmstadt, Germany) and transfected into E. coli strain BL21(DE3) (Merck KGaA). A single colony was inoculated into Circle growth medium (MP Biomedicals, Morgan Irvine, CA) containing ampicillin (50 μg/ml) for small-scale culture incubation at 37°C overnight. The culture fluid was then centrifuged, the collected cells were inoculated into 100 ml of Circle growth medium, and isopropyl-β-d-1-thiogalactopyranoside (IPTG) induction was performed according to the procedure for pET system expression.
The cultured cells were collected by centrifugation, resuspended in 5 ml of 0.5 M NaCl binding buffer (0.5 M NaCl, 20 mM imidazole, 20 mM potassium phosphate), and sonicated on ice four times for 15 s each time. Thereafter, the fusion protein was purified using a HisTrap HP column (GE Healthcare, Buckinghamshire, United Kingdom) according to the manufacturer's instructions. A negative antigen that included the Nus-tag protein, made from the pET43b vector but not including the hantavirus gene, was used as a negative control. The first 103 aa of the N-terminal region of HTNV N (HTNV HS103) were prepared as previously described (34). The purity of the recombinant proteins was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (data not shown).
IFA.
To characterize the rNs expressed by the baculovirus, we used a previously described IFA (18, 33). Acetone-fixed smears of High Five cells infected with recombinant baculoviruses were used as antigens.
Detection of multimerized rNs.
To detect multimerization of the rNs expressed by the baculovirus, competitive-sandwich ELISA was performed with MAb E5/G6 recognizing aa 165 to 173 as a capture antibody. Briefly, rNs were captured on the plate with MAb E5/G6 followed by detection with the same MAb E5/G6. A positive reaction with this ELISA indicates that the antigens are forming a multimer (36).
ELISA with whole rNs expressed by E. coli.
By the use of whole rNs expressed by E. coli, 96-well plates were first coated overnight at 4°C with 1 μg/ml of whole rNs in PBS as a capture antigen. After being washed three times with PBS containing 0.05% Tween 20 (PBS-T), the plates were then blocked with PBS containing 3% bovine serum albumin (BSA) for 1 h at 37°C. After blocking, patient and rodent sera were diluted 1:200 with ELISA buffer (PBS containing 0.5% BSA and 0.05% Tween 20) and added to the plates for 1 h at 37°C. After being washed with PBS-T, bound antibody was detected with peroxidase-labeled goat anti-human IgG (H+L) antibody (KPL, Gaithersburg, MD) for patient sera, horseradish peroxidase (HRP)-labeled goat anti-Peromyscus leucopus IgG (H+L) antibody (KPL) for Peromyscus rodent sera, or biotin-labeled mouse anti-Sigmodon hispidus IgG antibody for Sigmodontinae rodent sera for 1 h at 37°C. As another step using biotin-labeled mouse anti-Sigmodon hispidus IgG antibody, streptavidin-HRP conjugate (Prozyme, San Leandro, CA) was subjected to reactions for 30 min at 37°C. After being washed, color reactions were performed with o-phenylenediamine dihydrochloride (OPD) (Sigma-Aldrich, St. Louis, MO) and allowed to develop for 10 to 15 min. Absorbance was measured at 450 nm by using a SpectraMax 340 microplate spectrophotometer (Molecular Device, Sunnyvale, CA). HTNV HS103 antigen was used as a control for the Old World hantavirus experiments for HTNV infections only. HTNV HS103 antigen has 103 aa of the N terminus and includes cross-reactive epitopes.
Serotyping ELISA with trNs expressed by baculovirus.
The serotyping ELISA was carried out as previously described (2, 19). The plates were coated overnight at 4°C with MAb E5/G6 (2 μg/ml in PBS) as a capture antibody. Washing and blocking were carried out in the same manner. After three washes, 10-fold dilutions of rNs were added to the plates for 1 h at 37°C. After washing with PBS-T as described above, 200-fold dilutions of patient and rodent sera were added to the plates and incubated for 1 h at 37°C. The secondary antibody and color development results were the same as those for the ELISA with whole rNs.
IgM ELISA.
The IgM ELISA was carried out as previously described (17). The plates were coated overnight at 4°C with goat anti-human μ-chain antibody (Cappel, Aurora, OH) in 100 mM carbonate buffer as a capture antibody. Washing and blocking were carried out in the same manner. After three washes with PBS-T, 200-fold dilutions of patient sera were added to the plates and incubated for 1 h at 37°C. After washing with PBS-T as described above, 10-fold dilutions of SNV whole rN for baculovirus were added to the plates for 1 h at 37°C. After washes were performed, biotin-labeled E5/G6 MAb was subjected to reactions for 1 h at 37°C. After reaction with streptavidin-HRP conjugate (Prozyme), the color was developed with TMB (3,3′,5,5′-tetramethylbenzidine) for 15 min, and development was stopped with 0.5 M sulfuric acid. Absorbance was measured at 450 nm by using a SpectraMax 340 microplate spectrophotometer (Molecular Device).
RESULTS
Antigenic characterization of rNs expressed by baculovirus.
The IFA tests for antigenic profiling of whole rNs, trN50s, and trN100s expressed in High Five cells were carried out using hantavirus-specific MAbs (Table 1). Whole rNs of New World hantavirus reacted to cross-reactive MAbs (2E12, 4C3, 4E5, GBO4, 1C12, and ECO1) that recognized immunodominant epitopes of the N terminus of N and to cross-reactive MAbs (E5/G6 and F23A1) that recognized aa 165 to 173 and aa 291 to 402 of N, respectively. TrN50s of SNV, ANDV, and LANV mainly reacted with cross-reactive MAbs (E5/G6 and F23A1) but still remained cross-reactive to MAbs (GBO4 and C16D11) recognizing the N-terminal epitopes. In contrast, trN100s of SNV, ANDV, and LANV reacted to only two cross-reactive MAbs, E5/G6 and F23A1.
TABLE 1.
Antigenic characterization of rNs expressed by recombinant baculovirus with MAbs in the IFA testa
| Origin | MAb | Epitope | IFA resultb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole rN |
trN50 (New World hantavirus) |
trN100 (New World hantavirus) |
||||||||||||
| Old World hantavirus |
New World hantavirus |
|||||||||||||
| PUUV | HTNV | SEOV | SNV | ANDV | LANV | SNV | ANDV | LANV | SNV | ANDV | LANV | |||
| PUUV | 2E12 | N terminus | + | ± | ± | + | + | + | − | − | − | − | − | − |
| 4C3 | N terminus | + | + | + | + | + | + | − | − | − | − | − | − | |
| 4E5 | N terminus | + | + | ± | + | + | + | − | − | − | − | − | − | |
| GBO4c | N terminus | + | + | + | + | + | + | ± | + | ± | − | − | − | |
| HTNV | ECO2 | aa 1-33 | − | + | + | − | − | − | − | − | − | − | − | − |
| 1C12 | N terminus | + | + | + | + | + | + | − | − | − | − | − | − | |
| ECO1c | aa 34-103 | + | + | + | + | + | + | − | − | − | − | − | − | |
| C16D11 | Unknown | + | + | + | − | + | + | − | + | + | − | − | − | |
| E5/G6 | aa 165-173 | + | + | + | + | + | + | + | + | + | + | + | + | |
| F23A1 | aa 291-402 | − | + | + | + | + | + | + | + | + | + | + | + | |
PUUV, Puumala virus; HTNV, Hantaan virus; SEOV, Seoul virus.
Symbols: +, positive IFA result of >1:1 (1:100 for ascitic fluid samples); ±, scarcely positive IFA result; −, negative IFA result.
The sample was ascitic fluid.
Detection of multimerization of rNs.
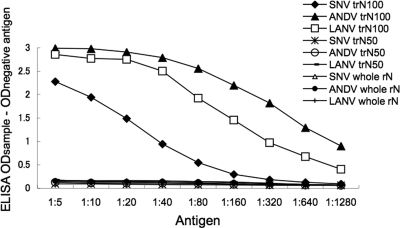
Since E5/G6 reacted to rNs in the IFA, E5/G6 was used in competitive-sandwich ELISA. As shown in Fig. 1, there was no reaction to trN50s and whole rNs. This implied that the trN50s and whole rNs captured by E5/G6 could not react with E5/G6 as a detector due to competition. Thus, trN50s and whole rNs of SNV, ANDV, and LANV were found as monomers. On the other hand, there were strong reactions to trN100s, indicating that trN100s of SNV, ANDV, and LANV existed as multimers. As the serotype-specific epitopes were considered to be formed after multimerization of trNs, we selected trN100s as ELISA antigens for serotyping ELISA.
FIG. 1.
Multimerization of rNs. Antigens were captured and detected with MAb E5/G6. Each antigen was diluted 1:5 to 1:1,280 and subjected to capture ELISA. The OD value was corrected by the negative control. The ELISA was performed three times in duplicate, and the means of the OD values were plotted.
Reactivities of whole rNs and trN100s with infected sera.
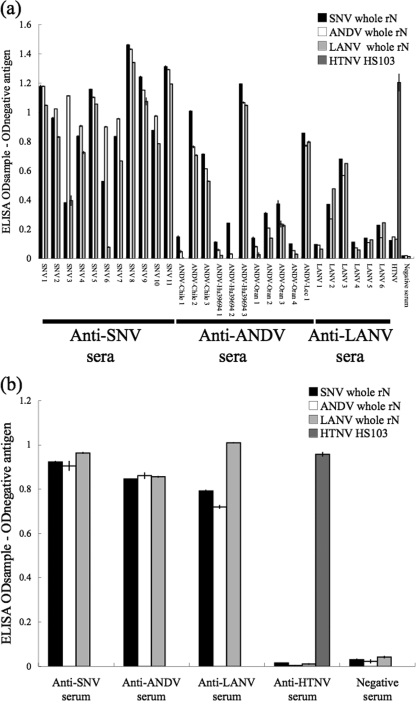
To examine the applicability of the recombinant antigens in ELISA, the ELISA optical density (OD) values determined with whole rNs and trN100s in 28 human and 59 rodent serum samples were compared. As shown in Fig. 2, when tested using whole rN antigens, SNV, ANDV, and LANV infections were not differentiated because of strong cross-reaction. However, HTNV-infected sera reacted strongly only to HTNV antigen of the Old World hantavirus.
FIG. 2.
(a) Reaction of whole rNs with hantavirus-positive human sera. (b) Reaction of whole rNs with representative hantavirus-positive rodent sera (anti-SNV serum, Peromyscus maniculatus; anti-ANDV serum, Oligoryzomys longicaudatus; anti-LANV serum, Calomys callosus; anti-HTNV serum, Rattus norvegicus). The total number of hantavirus-positive rodent sera was 59. HTNV-infected patient or rat sera were used as the control for Old World hantavirus infection. The OD value was corrected using the negative-control value. The ELISA was performed three times in duplicate, and the bars show the mean values.
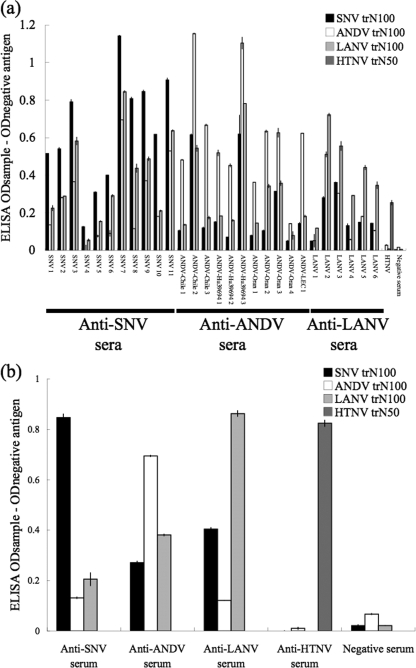
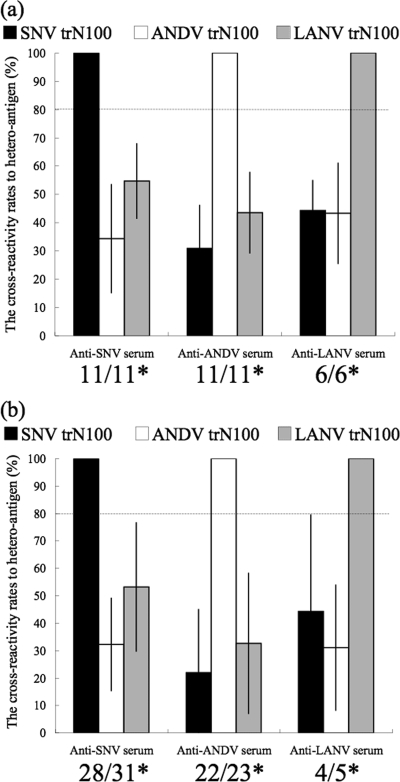
On the other hand, the trN100s of SNV, ANDV, and LANV showed serotype-specific reaction patterns. The ELISA ODs of heterologous antigens were less than half of those of homologous reactions for most of the sera tested (Fig. 3). The mean reactive rate of OD values of serum specimens for heterologous antigens was 37.9%, with 22.9% as the standard deviation (SD) of those for homologous antigens. Therefore, as the mean of the reactive rate for homologous and heterologous antigens plus twice the SD of the OD values was 83.7%, the cutoff value to distinguish homologous and heterologous reactions was tentatively determined to be 80% (Fig. 4). All of the 28 patient sera and 54 of the 59 rodent sera were serotyped. Although 5 of the rodent sera showed stronger cross-reactivity, the OD values were higher for homologous antigens, except for that of 1 Peromyscus maniculatus anti-SNV serum sample.
FIG. 3.
(a) Reaction of trN100s with hantavirus-positive human sera. (b) Reaction patterns of trN100s with representative hantavirus-positive rodent sera (the serum samples represented in Fig. 3b were the same as those in Fig. 2b) in ELISA. A total of 28 patient serum samples and 59 rodent serum samples were used to assess our serotyping ELISA system. HTNV-infected patient or rat sera were used as the control for Old World hantavirus infection. The OD value was corrected using the negative-control value. The ELISA was performed three times in duplicate, and the bars show the mean values.
FIG. 4.
The mean OD values for rates of reaction to heterologous antigens (y axis). (a) Patterns of cross-reactions of hantavirus-positive human sera to heteroantigens. (b) Patterns of cross-reactions of hantavirus-positive rodent sera to heteroantigens. The mean OD value for rates of reaction of serum specimens to heterologous antigens was 37.9%, with an SD of 22.9% for those to homologous antigens. Therefore, as the mean of the rates of reaction for homologous and heterologous antigens plus twice the SD of the OD values was 83.7%, the cutoff value to distinguish homologous and heterologous reactions was tentatively determined to be 80%. The dotted lines indicate the cutoff value to identify the serotype. *, the numbers indicate the number of serotyped samples (left) and the total number of samples (right).
Detection of IgG and IgM antibodies to whole rN in patient sera.
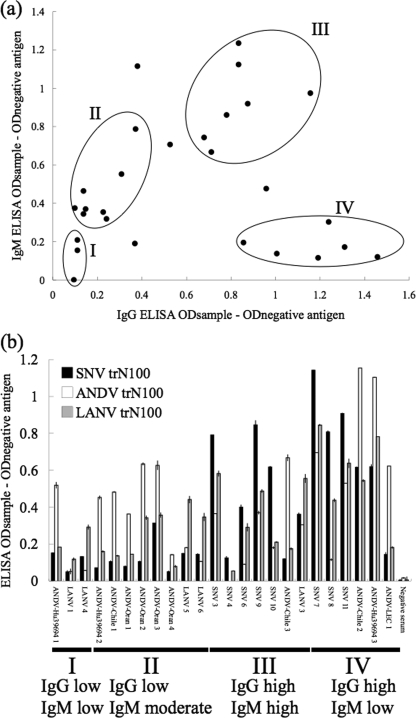
To investigate phase when HPS patient sera were obtained, IgG and IgM antibodies in HPS patient sera were examined by using whole rN of SNV. As shown in Fig. 5a, they were roughly divided into 4 clusters (clusters I, II, III, and IV) based on the levels of IgG and IgM antibodies: low IgG and low IgM, low IgG and moderate IgM, high IgG and high IgM, and high IgG and low IgM. The results shown in Fig. 3a were rearranged in the order of sera classified into each cluster (Fig. 5b). As shown, irrespective of titers of the IgM and IgG antibody, they were serotyped.
FIG. 5.
(a) Patterns of IgM and IgG ELISA OD values for human cases. The sera were divided into four clusters (clusters I to IV) by the patterns of IgM and IgG. The OD value was corrected using the negative-control value. (b) Patterns of reactions of trN100s with hantavirus-positive human sera in ELISA were arranged by phase. Only ANDV-Chile 2 and ANDV-Hu39694 3 have been known to be convalescent-phase sera. The rest of the serum samples of those two patients were recorded as acute-phase samples, but details are unknown.
DISCUSSION
At present, NT is the only method for differentiating serotypes of New World hantavirus infections (31). In this study, we showed that ELISA using N-terminally truncated trN100 antigens of three HPS-associated hantavirus species (SNV, ANDV, and LANV) was able to serologically differentiate serum specimens from SNV-, ANDV-, and LANV-infected patients or rodents (Fig. 3). On the other hand, whole rN antigens showed strong cross-reactivity with heterologous sera (Fig. 2). Therefore, screening by ELISA using whole rNs followed by serotyping using trN100s can be recommended as a rapid and practical system for hantavirus seroepidemiology and diagnosis.
To date, MAbs to N of New World hantaviruses have not been readily available (30). Consequently, antigenic characterization of rNs was indirectly confirmed using MAbs to Old World hantavirus by IFA. As we expected from our previous study (34), most of the MAbs that recognized immunodominant epitopes of the N terminus of N for Old World hantaviruses reacted to whole rNs of SNV, ANDV, and LANV but not to trN100s (Table 1). These results indicate that the first 100 aa of the N terminus of N possess immunodominant, cross-reactive epitopes, as previously reported (6, 8, 32). Therefore, it is thought that the structure of N of New World hantavirus is similar to that of N of Old World hantavirus.
It has been shown that the Old World hantavirus N built whole rN-whole rN or trN50-trN50 multimers and that both multimers were able to form multimerization-dependent serotype-specific epitopes (36). In this study, the trN50s and whole rNs of New World hantavirus were found as monomeric N in sandwich ELISA (Fig. 1). In contrast, the trN100s of SNV, ANDV, and LANV were detected as multimers (Fig. 1). These results support those of previous studies indicating that the first 100 aa of the N terminus did not contribute to N-N interaction but rather were inhibitory for homotypic interaction (12, 36). Multimerization of SNV N was confirmed by a yeast two-hybrid method (1). Thus, we believe that intact Ns of SNV, ANDV, and LANV form multimers in both virions and infected cells. It was considered that such a structural error of rNs was caused by limitations of the baculovirus expression system used in this study. In spite of the uncertainties of the expression system, trN100s were useful as serotyping antigens. Interestingly, the trN50 and whole rN of BCCV were detected as multimers in E5/G6-capture E5/G6-detected ELISA (data not shown). Thus, the functional roles of the N-terminal regions of New World hantaviruses N differ with respect to the capacity to form multimers.
In human cases, it is generally possible to detect viral RNA by RT-PCR in peripheral blood mononuclear cells and in serum samples during the febrile prodrome and early in the course of the cardiopulmonary phase. However, it is difficult to detect viral RNA in blood by the use of RT-PCR after the cardiopulmonary phase (13, 16, 28). Sera in which IgG and IgM antibody titers are low can be considered to represent a prodrome phase, according to previous data (3, 26, 27, 29). On the basis of the IgM and IgG antibody titers, the patient sera were divided into 4 clusters (Fig. 5a). Since clusters I and II contain serum specimens showing low or moderate IgM titers with low IgG antibody titers, the sera might have been obtained early after the onset of infection. Nevertheless, the HPS patient sera in clusters I and II were serotyped in this study (Fig. 5b). Therefore, the trN100-based typing ELISA is useful not only for latter-phase sera but also for early-acute-phase patient sera. Early differential diagnosis might contribute to the treatment effect in the future.
In conclusion, serologic diagnosis by ELISA with trN100s is rapid, simple, and safe in comparison to NT. This method is expected to be useful for clinical diagnosis and epidemiological surveillance in regions where two or more hantaviruses cocirculate. Although the percentage of amino acid sequence identity between ANDV N and LANV N used in this study was high (90.0%), trN100s were able to reduce cross-reactivity to heterologous sera. Thus, it might be possible to apply this system to other hantaviruses.
Acknowledgments
This work was supported by the Global COE program Establishment of International Collaboration Center for Zoonosis Control from the Japanese Ministry of Education, Culture, Sports, Science and Technology. This work was also partially supported by Grants-in-Aid for Scientific Research and the Development of Scientific Research from the Japanese Ministry of Health, Labor and Welfare (grant 200829018A). This study was supported in part by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, MEXT, Japan.
We also acknowledge Stewart Chisholm of the Stewart English School (S.E.S.) for revising the grammar in the final draft.
Footnotes
Published ahead of print on 24 March 2010.
REFERENCES
- 1.Alfadhli, A., Z. Love, B. Arvidson, J. Seeds, J. Willey, and E. Barklis. 2001. Hantavirus nucleocapsid protein oligomerization. J. Virol. 75:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araki, K., K. Yoshimatsu, M. Ogino, H. Ebihara, A. Lundkvist, H. Kariwa, I. Takashima, and J. Arikawa. 2001. Truncated hantavirus nucleocapsid proteins for serotyping Hantaan, Seoul, and Dobrava hantavirus infections. J. Clin. Microbiol. 39:2397-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bharadwaj, M., R. Nofchissey, D. Goade, F. Koster, and B. Hjelle. 2000. Humoral immune responses in the hantavirus cardiopulmonary syndrome. J. Infect. Dis. 182:43-48. [DOI] [PubMed] [Google Scholar]
- 4.Borges, A. A., G. M. Campos, M. L. Moreli, R. L. Souza, V. H. Aquino, F. P. Saggioro, and L. T. Figueiredo. 2006. Hantavirus cardiopulmonary syndrome: immune response and pathogenesis. Microbes Infect. 8:2324-2330. [DOI] [PubMed] [Google Scholar]
- 5.Botten, J., K. Mirowsky, D. Kusewitt, M. Bharadwaj, J. Yee, R. Ricci, R. M. Feddersen, and B. Hjelle. 2000. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus). Proc. Natl. Acad. Sci. U. S. A. 97:10578-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elgh, F., A. Lundkvist, O. A. Alexeyev, G. Wadell, and P. Juto. 1996. A major antigenic domain for the human humoral response to Puumala virus nucleocapsid protein is located at the amino-terminus. J. Virol. Methods 59:161-172. [DOI] [PubMed] [Google Scholar]
- 7.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Petterson, A. Plyusin, and C. Schmaljohn. 2000. Bunyaviridae, p. 599-621. In C. M. F. M. H. V. van Regenmortel, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: the classification and nomenclature of viruses. Seventh report of the international committee on taxonomy of viruses. Academic Press, San Diego, CA.
- 8.Gött, P., L. Zoller, G. Darai, and E. K. Bautz. 1997. A major antigenic domain of hantaviruses is located on the aminoproximal site of the viral nucleocapsid protein. Virus Genes 14:31-40. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, A. M., M. D. Bowen, T. G. Ksiazek, R. J. Williams, R. T. Bryan, J. N. Mills, C. J. Peters, and S. T. Nichol. 1997. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology 238:115-127. [DOI] [PubMed] [Google Scholar]
- 10.Jonsson, C. B., J. Hooper, and G. Mertz. 2008. Treatment of hantavirus pulmonary syndrome. Antiviral Res. 78:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kariwa, H., K. Yoshimatsu, and J. Arikawa. 2007. Hantavirus infection in East Asia. Comp. Immunol. Microbiol. Infect. Dis. 30:341-356. [DOI] [PubMed] [Google Scholar]
- 12.Kaukinen, P., V. Kumar, K. Tulimaki, P. Engelhardt, A. Vaheri, and A. Plyusnin. 2004. Oligomerization of hantavirus N protein: C-terminal alpha-helices interact to form a shared hydrophobic space. J. Virol. 78:13669-13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lednicky, J. A. 2003. Hantaviruses: a short review. Arch. Pathol. Lab. Med. 127:30-35. [DOI] [PubMed] [Google Scholar]
- 14.Levis, S., J. Garcia, N. Pini, G. Calderon, J. Ramirez, D. Bravo, S. St. Jeor, C. Ripoll, M. Bego, E. Lozano, R. Barquez, T. G. Ksiazek, and D. Enria. 2004. Hantavirus pulmonary syndrome in northwestern Argentina: circulation of Laguna Negra virus associated with Calomys callosus. Am. J. Trop. Med. Hyg. 71:658-663. [PubMed] [Google Scholar]
- 15.Martinez, V. P., C. Bellomo, J. San Juan, D. Pinna, R. Forlenza, M. Elder, and P. J. Padula. 2005. Person-to-person transmission of Andes virus. Emerg. Infect. Dis. 11:1848-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertz, G. J., B. Hjelle, M. Crowley, G. Iwamoto, V. Tomicic, and P. A. Vial. 2006. Diagnosis and treatment of new world hantavirus infections. Curr. Opin. Infect. Dis. 19:437-442. [DOI] [PubMed] [Google Scholar]
- 17.Miyamoto, H., H. Kariwa, K. Araki, K. Lokugamage, D. Hayasaka, B. Z. Cui, N. Lokugamage, L. I. Ivanov, T. Mizutani, M. A. Iwasa, K. Yoshimatsu, J. Arikawa, and I. Takashima. 2003. Serological analysis of hemorrhagic fever with renal syndrome (HFRS) patients in far eastern Russia and identification of the causative hantavirus genotype. Arch. Virol. 148:1543-1556. [DOI] [PubMed] [Google Scholar]
- 18.Morii, M., K. Yoshimatsu, J. Arikawa, G. Zhou, H. Kariwa, and I. Takashima. 1998. Antigenic characterization of Hantaan and Seoul virus nucleocapsid proteins expressed by recombinant baculovirus: application of a truncated protein, lacking an antigenic region common to the two viruses, as a serotyping antigen. J. Clin. Microbiol. 36:2514-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura, I., K. Yoshimatsu, B. H. Lee, M. Okumura, M. Taruishi, K. Araki, H. Kariwa, I. Takashima, and J. Arikawa. 2008. Development of a serotyping ELISA system for Thailand virus infection. Arch. Virol. 153:1537-1542. [DOI] [PubMed] [Google Scholar]
- 20.Nichol, S. T., C. F. Spiropoulou, S. Morzunov, P. E. Rollin, T. G. Ksiazek, H. Feldmann, A. Sanchez, J. Childs, S. Zaki, and C. J. Peters. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914-917. [DOI] [PubMed] [Google Scholar]
- 21.Niklasson, B., M. Jonsson, A. Lundkvist, J. Horling, and E. Tkachenko. 1991. Comparison of European isolates of viruses causing hemorrhagic fever with renal syndrome by a neutralization test. Am. J. Trop. Med. Hyg. 45:660-665. [DOI] [PubMed] [Google Scholar]
- 22.Padula, P. J., A. Edelstein, S. D. Miguel, N. M. Lopez, C. M. Rossi, and R. D. Rabinovich. 1998. Hantavirus pulmonary syndrome outbreak in Argentina: molecular evidence for person-to-person transmission of Andes virus. Virology 241:323-330. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds, S., E. Galanis, M. Krajden, M. Morshed, D. Bowering, W. Abelson, and T. R. Kollmann. 2007. Imported fatal hantavirus pulmonary syndrome. Emerg. Infect. Dis. 13:1424-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruo, S. L., A. Sanchez, L. H. Elliott, L. S. Brammer, J. B. McCormick, and H.-S. Fisher. 1991. Monoclonal antibodies to three strains of hantaviruses: Hantaan, R22, and Puumala. Arch. Virol. 119:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt, J., H. Meisel, S. G. Capria, R. Petraityte, A. Lundkvist, B. Hjelle, P. A. Vial, P. Padula, D. H. Kruger, and R. Ulrich. 2006. Serological assays for the detection of human Andes hantavirus infections based on its yeast-expressed nucleocapsid protein. Intervirology 49:173-184. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, J., H. Meisel, B. Hjelle, D. H. Kruger, and R. Ulrich. 2005. Development and evaluation of serological assays for detection of human hantavirus infections caused by Sin Nombre virus. J. Clin. Virol. 33:247-253. [DOI] [PubMed] [Google Scholar]
- 28.Terajima, M., J. D. Hendershot III, H. Kariwa, F. T. Koster, B. Hjelle, D. Goade, M. C. DeFronzo, and F. A. Ennis. 1999. High levels of viremia in patients with the Hantavirus pulmonary syndrome. J. Infect. Dis. 180:2030-2034. [DOI] [PubMed] [Google Scholar]
- 29.Tischler, N. D., H. Galeno, M. Rosemblatt, and P. D. Valenzuela. 2005. Human and rodent humoral immune responses to Andes virus structural proteins. Virology 334:319-326. [DOI] [PubMed] [Google Scholar]
- 30.Tischler, N. D., M. Rosemblatt, and P. D. Valenzuela. 2008. Characterization of cross-reactive and serotype-specific epitopes on the nucleocapsid proteins of hantaviruses. Virus Res. 135:1-9. [DOI] [PubMed] [Google Scholar]
- 31.Vaheri, A., O. Vapalahti, and A. Plyusnin. 2008. How to diagnose hantavirus infections and detect them in rodents and insectivores. Rev. Med. Virol. 18:277-288. [DOI] [PubMed] [Google Scholar]
- 32.Yamada, T., B. Hjelle, R. Lanzi, C. Morris, B. Anderson, and S. Jenison. 1995. Antibody responses to Four Corners hantavirus infections in the deer mouse (Peromyscus maniculatus): identification of an immunodominant region of the viral nucleocapsid protein. J. Virol. 69:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimatsu, K., J. Arikawa, and H. Kariwa. 1993. Application of a recombinant baculovirus expressing hantavirus nucleocapsid protein as a diagnostic antigen in IFA test: cross reactivities among 3 serotypes of hantavirus which causes hemorrhagic fever with renal syndrome (HFRS). J. Vet. Med. Sci. 55:1047-1050. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimatsu, K., J. Arikawa, M. Tamura, R. Yoshida, A. Lundkvist, B. Niklasson, H. Kariwa, and I. Azuma. 1996. Characterization of the nucleocapsid protein of Hantaan virus strain 76-118 using monoclonal antibodies. J. Gen. Virol. 77(Pt. 4):695-704. [DOI] [PubMed] [Google Scholar]
- 35.Yoshimatsu, K., J. Arikawa, R. Yoshida, H. Li, Y. C. Yoo, H. Kariwa, N. Hashimoto, M. Kakinuma, T. Nobunaga, and I. Azuma. 1995. Production of recombinant hantavirus nucleocapsid protein expressed in silkworm larvae and its use as a diagnostic antigen in detecting antibodies in serum from infected rats. Lab. Anim. Sci. 45:641-646. [PubMed] [Google Scholar]
- 36.Yoshimatsu, K., B. H. Lee, K. Araki, M. Morimatsu, M. Ogino, H. Ebihara, and J. Arikawa. 2003. The multimerization of hantavirus nucleocapsid protein depends on type-specific epitopes. J. Virol. 77:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]