Abstract
A semiautomated, repetitive-sequence-based PCR (rep-PCR) instrument (DiversiLab system) was evaluated in comparison with pulsed-field gel electrophoresis (PFGE) to investigate an outbreak of Serratia marcescens infections in a neonatal intensive care unit (NICU). A selection of 36 epidemiologically related and 8 epidemiologically unrelated isolates was analyzed. Among the epidemiologically related isolates, PFGE identified five genetically unrelated patterns. Thirty-two isolates from patients and wet nurses showed the same PFGE profile (pattern A). Genetically unrelated PFGE patterns were found in one patient (pattern B), in two wet nurses (patterns C and D), and in an environmental isolate from the NICU (pattern G). Rep-PCR identified seven different patterns, three of which included the 32 isolates of PFGE type A. One or two band differences in isolates of these three types allowed isolates to be categorized as similar and included in a unique cluster. Isolates of different PFGE types were also of unrelated rep-PCR types. All of the epidemiologically unrelated isolates were of different PFGE and rep-PCR types. The level of discrimination exhibited by rep-PCR with the DiversiLab system allowed us to conclude that this method was able to identify genetic similarity in a spatio-temporal cluster of S. marcescens isolates.
Concerns regarding the emergence of hospital-acquired infections, increasing antimicrobial resistance, and the increase in morbidity, mortality, and costs associated with these infections drive the need for refinement of molecular approaches to aid in the diagnosis and epidemiological analysis of nosocomial infections.
Several methods based on DNA analysis are now available that provide information about the genetic relatedness of isolates of the same species (26). These DNA-based molecular methodologies include pulsed-field gel electrophoresis (PFGE), PCR-based typing methods, and multilocus sequence analysis. Establishing clonality of pathogens can aid in the identification of the source of organisms (environmental or personnel), in distinguishing infectious from non infectious strains, and in distinguishing relapse from reinfection.
PFGE is generally considered one of the most reproducible and highly discriminatory typing techniques available and is considered the “gold standard” molecular approach for the epidemiological analysis of most bacterial pathogens (10, 28). However, PFGE requires specialized equipment and is technically demanding, labor intensive, and relatively slow, as it may take 2 to 5 days to obtain results, depending on the organism and the methods utilized.
Automation and computer-assisted pattern analysis afford several benefits for the molecular epidemiology laboratory: reduction of labor costs, standardization, decreased time to completion, and better comparison between runs. The latter is of special importance when comparing large groups of organisms or comparing organisms submitted to the laboratory over a certain time period. Recently, two semiautomated systems for strain typing have become available on the market: (i) the Ribo-Printer Microbial Characterization System (DuPont Qualicon), which performs ribotyping of strains (8, 9, 11, 14-16, 22), and (ii) the rep-PCR DiversiLab Microbial Typing System (bioMérieux, Italy), which amplifies the regions between the noncoding repetitive sequences in bacterial genomes (12).
The goals of this study were to assess the DiversiLab rep-PCR system as a rapid method for S. marcescens strain identification and differentiation in comparison with PFGE results for strains from an epidemiologically well-characterized outbreak that occurred in the neonatal intensive care unit (NICU) of the Verona University Hospital and to contribute to the discussion of the use of molecular techniques to identify and differentiate bacterial isolates.
MATERIALS AND METHODS
Epidemiologic investigation.
An outbreak of S. marcescens infection occurred in the NICU from August 2007 to December 2007. Microbiology records were reviewed to identify cases of infection and to determine the baseline rate of S. marcescens infection in the NICU prior to the outbreak. Case patients were defined as any neonate hospitalized in the NICU from August 2007 to December 2007 who had one or more clinical or surveillance cultures that yielded S. marcescens. Cultures were performed with samples from the NICU environment, including counters, sinks, bottle warmers, personnel work areas, breast milk, equipment in the breast milk room, infant formula, inhalators, medications, saline ampoules, incubators, eye examination equipment, soaps, lotions, ventilators, oxygenation and humidification devices, and other respiratory care equipment. Each member of the NICU staff was examined for the presence of artificial fingernails, and hand cultures were obtained.
Microbiological method.
Clinical and environmental cultures were processed using standard practices and routine media. The bacteria were identified by means of an automated Vitek-2 system (bioMérieux, Italy), and antibiotic susceptibility was checked routinely by means of the disc diffusion method, according to the Clinical and Laboratory Standards Institute recommendations (4). The antimicrobial agents included were ampicillin, amoxicillin-clavulanic acid, aztreonam, amikacin, ceftriaxone, ceftazidime, cefuroxime, cefotetan, gentamicin, imipenem, piperacillin, piperacillin-tazobactam, cefepime, and meropenem.
Automated rep-PCR DNA fingerprinting.
DNA was extracted from a 10-μl loopful of a Serratia marcescens colony using an UltraClean Microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA) and following the manufacturer's instructions. The extracted DNA was amplified using a DiversiLab Serratia DNA fingerprinting kit (bioMérieux, Italy), which includes rep-PCR master mix 1, Serratia primers, and kit-specific positive and negative controls, in accordance with the manufacturer's product insert. Briefly, 50 ng of genomic DNA, 2.5 U of AmpliTaq DNA polymerase, and 1.5 μl of 10× PCR buffer (Applied Biosystems, Foster City, CA) were added to the rep-PCR master mix to achieve a total of 25 μl. Thermal cycling parameters for an MJ PTC100 PCR System (Bio-Rad, MI) were as follows: initial denaturation at 94°C for 2 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s, extension at 70°C for 90 s, and a final extension at 70°C for 3 min. The amplified product was stored at −20°C until detection. Analysis of rep-PCR products was implemented using a DiversiLab system in which the amplified fragments of various sizes and intensities are separated and detected using a microfluidics LabChip with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). The isolates' relatedness was analyzed by the DiversiLab software, version 3.4, which uses the Kullback-Leibler distance correlation coefficient to determine distance matrices and the unweighted-pair group method using average linkages to create dendrograms, electropherograms, virtual gel images, and scatterplots. Interpretative criteria provided by the manufacturer categorized isolates as indistinguishable, similar, or different. In general, “different” was defined as <95% similarity and two or more band differences for organisms defined as homogeneous (i.e., those that include a low number of clones in nature) or three band differences for organisms defined as heterogeneous (i.e., those that include an unlimited number of different clones in nature). “Similar” was defined as >95% similarity and one band difference for homogeneous organisms or up to two band differences for heterogeneous organisms. “Indistinguishable” was defined as >97% similarity and no banding differences, including no variation in the intensities of individual bands, although overall intensities may differ. S. marcescens is recognized as a heterogeneous species that probably contains an unlimited number of different clones (3).
PFGE fingerprinting.
Genomic DNA was prepared as described previously (24) but with some modifications. Isolated colonies were harvested from Mueller-Hinton agar plates after overnight incubation at 37°C, and the suspension was adjusted to a concentration of 109 CFU/ml in saline-EDTA buffer (75 mM NaCl, 25 mM EDTA [pH 7.5]). The bacterial suspension was then mixed with an equal volume of 2% low-melting-point agarose (Bio-Rad Laboratories) and allowed to solidify in a 100-μl plug mold (Bio-Rad Laboratories). The DNA block was incubated for 2 h at 37°C in 1 ml of lysis buffer (10 mM Tris-HCl, 100 mM EDTA, 100 mM NaCl, 0.5% Brij 58, 0.2% sodium deoxycholate, 0.5% sodium lauroyl sarcosine, lysozyme [0.5 mg/ml]). Following this step, the lysis buffer was replaced with 1 ml of proteolysis buffer (1% sodium lauroyl sarcosine, 0.5 M EDTA [pH 9.5], proteinase K [500 μg/ml; Sigma]) and this solution was incubated at 52°C overnight. To eliminate the lysed bacterial material and inactivate proteinase K activity, the DNA blocks were washed four times at room temperature in 10 ml of Tris-EDTA buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA). A slice of each plug was cut and incubated with SpeI (Roche, Italy). Restriction fragments of DNA were separated by PFGE with a CHEF-DRII apparatus (Bio-Rad Laboratories) through 1.2% Pulsed Field Certified Agarose (Bio-Rad Laboratories). Electrophoresis was performed at 6 V/cm and 14°C. The run time was 22 h with the pulse time ramping from 5 to 60 s. Tenover's criteria were used to interpret chromosomal DNA restriction patterns and for strain typing (28).
RESULTS
A total of 16 patients admitted to the NICU were identified as infected and/or colonized with S. marcescens from August 2007 to December 2007. The baseline rate of S. marcescens infections/colonizations for the 24 months prior to the outbreak was 0.20 case/month, compared with 4 cases/month during the outbreak. Six patients developed infections; one experienced bacteremia, one conjunctivitis, one conjunctivitis and urinary infection, two urinary infection, and one umbilical wound infection. In 12 colonized patients, S. marcescens was detected in pharyngeal, nasal, and perianal swab samples for surveillance culture. Seven patient cultures from more than one body site grew S. marcescens. In addition, S. marcescens was isolated from the milk of five wet nurses and in three cases the isolates had a PFGE pattern identical to that of the outbreak isolates (Table 1).
TABLE 1.
Description of S. marcescens isolates from NICU and genomic typing results
| Key sample IDa | Isolate source | Date of isolation (mo/day/yr) | PFGE type | rep-PCR type |
|---|---|---|---|---|
| Patient isolates | ||||
| 1 | Blood | 09/21/2007 | A | a |
| 2 | Eye discharge | 09/24/2007 | A | a |
| 3 | Pharyngeal swab | 09/27/2007 | A | a |
| 4 | Pharyngeal swab | 09/27/2007 | A | a |
| 5 | Pharyngeal swab | 09/27/2007 | A | a |
| 6 | Pharyngeal swab | 09/27/2007 | A | a |
| 7 | Pharyngeal swab | 09/28/2007 | A | a |
| 8 | Nasal swab | 09/28/2007 | A | a |
| 9 | Nasal swab | 10/01/2007 | A | c |
| 36 | Stool | 10/04/2007 | A | a |
| 11 | Pharyngeal swab | 10/05/2007 | A | c |
| 12 | Stool | 10/06/2007 | A | c |
| 13 | Stool | 10/09/2007 | A | a |
| 14 | Stool | 10/10/2007 | A | a |
| 15 | Eye discharge | 10/11/2007 | A | c |
| 16 | Pharyngeal swab | 10/11/2007 | A | a |
| 17 | Stool | 10/11/2007 | A | b |
| 18 | Urine | 10/11/2007 | A | b |
| 20 | Stool | 11/06/2007 | A | b |
| 21 | Stool | 11/07/2007 | B | l |
| 22 | Pharyngeal swab | 11/14/2007 | A | b |
| 23 | Pharyngeal swab | 11/15/2007 | A | b |
| 24 | Pharyngeal swab | 11/21/2007 | A | a |
| 25 | Pharyngeal swab | 11/23/2007 | A | b |
| 26 | Pharyngeal swab | 11/23/2007 | A | b |
| 27 | Urine | 11/23/2007 | A | c |
| 28 | Urine | 11/23/2007 | A | c |
| 29 | Umbilical wound | 12/04/2007 | A | b |
| Wet nurse isolates | ||||
| 10A | Milk | 09/04/2007 | A | a |
| 10B | Milk | 09/25/2007 | A | a |
| 10C | Milk | 10/04/2007 | A | a |
| 19 | Milk | 10/24/2007 | A | b |
| 32 | Milk | 10/15/2007 | C | i |
| 33 | Milk | 11/08/2007 | D | e |
| 34 | Milk | 11/12/2007 | A | a |
| NICU environmental isolate | Tap | 10/2/2007 | G | g |
| Epidemiologically unrelated isolates | ||||
| 30 | Blood | 10/07/2007 | E | h |
| 37 | Cutaneous swab | 03/14/2008 | F | f |
| 747c | Bronchial aspirate | 12/30/1999 | O | d |
| 825c | Pus | 01/13/2000 | H | m |
| 1836c | Bronchial aspirate | 01/21/2002 | I | n |
| 1959c | Blood | 04/06/2002 | L | o |
| 2275c | Surgical swab | 11/30/2002 | M | p |
| 823c | Nasal swab | 01/13/2000 | N | q |
Key sample ID reported in Fig. 2.
Examination of the hands of NICU personnel revealed no lesions or artificial fingernails. Hand cultures obtained from NICU personnel did not grow S. marcescens. One environmental culture sample from a sink drain in the filter room yielded a strain of S. marcescens. Education about Serratia and standard infection control procedures was reviewed with NICU personnel. Cleaning and disinfection procedures were reviewed and reinforced with infection control personnel. Contact isolation precautions were implemented for all neonates with S. marcescens infection. NICU personnel were instructed to gather the infected neonates into one area of the unit and to dedicate staff to care only for these patients. All infected and colonized patients were assembled into a cohort. During the outbreak, admission to the NICU was drastically reduced. The first strain stored and analyzed was dated 21 September, and thus, the molecular study related to strains isolated after the restriction of outside admissions. Weekly surveillance cultures of stool and pharyngeal and nasal swabs were taken to detect whether there was any asymptomatic colonization of neonates with S. marcescens. These infection control measures successfully halted the outbreak.
The isolates from the NICU presented the same antimicrobial susceptibility profile: they were susceptible to aztreonam, amikacin, ceftriaxone, ceftazidime, cefotetan, gentamicin, imipenem, piperacillin, piperacillin-tazobactam, cefepime, and meropenem and resistant to ampicillin, amoxicillin-clavulanic acid, and cefuroxime.
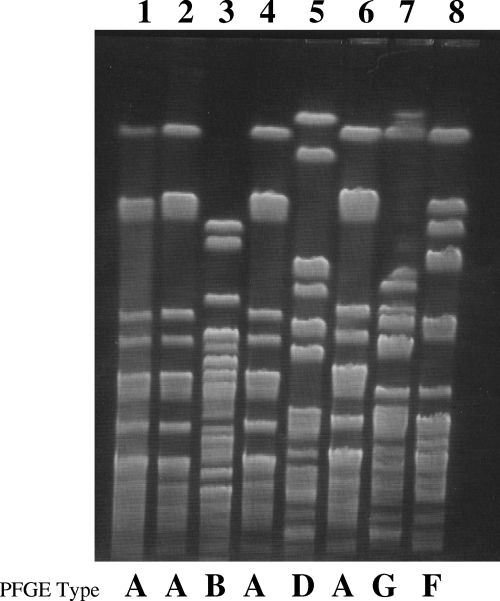
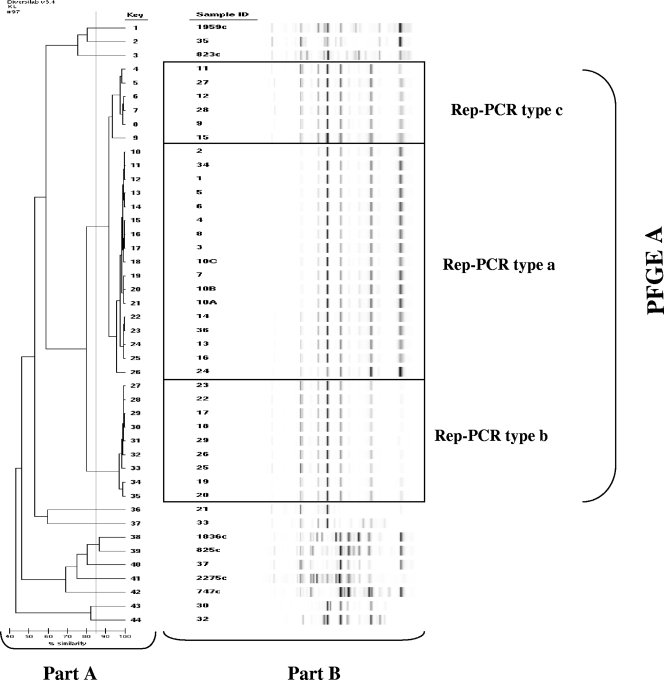
Thirty-six epidemiologically related and eight unrelated isolates were tested by both the PFGE and automated rep-PCR typing methods. Table 1 lists the epidemiological data and the PFGE and rep-PCR patterns of each isolate. By visual inspection of the PFGE profiles, 1 major pattern (type A) and 12 unique genetically unrelated patterns (types B through O) were identified. Isolates from 15 patients and three wet nurses were genetically identical and belonged to type A. Unique unrelated PFGE patterns were identified in one patient (intestinal carriage), in two wet nurses, in an isolate collected from the NICU environment, and in the eight epidemiologically unrelated isolates (adult patients admitted to other wards at a different time of year). A selection of these PFGE types is shown in Fig. 1. The rep-PCR analysis identified three different patterns (types a, b, and c) which included all of the type A PFGE isolates and 12 unique profiles (types d through q) corresponding to unique PFGE types B through O (Table 1 and Fig. 2A). Each rep-PCR pattern clustered together isolates with 95% or more similarity. Since the similarity percentages generated by rep-PCR suggested possible genetic diversity among the isolates belonging to PFGE cluster A, a visual inspection of the sample fingerprints was performed (Fig. 2B). The fingerprints of patterns a and c were similar, whereas pattern b showed differences from patterns c and a in more than two bands. According to the criteria suggested by the manufacturers and the visual inspection only, isolates showing c and a profiles could be classified as similar (fingerprints that show differences in one or two bands), whereas isolates with pattern b could be classified as different but highly related (more than 80% similarity). The reason why indistinguishable or similar banding profiles gave rise to the low similarity index was investigated by comparing the electropherograms directly using the pattern overlay function of the DiversiLab software. Small difference in the fluorescence intensities of individual peaks were revealed by the analysis, since the software weighted more intense peaks more heavily. These results indicated that types a, b, and c should be classified as highly related and confirmed the monoclonal origin of the outbreak. In comparison with PFGE, rep-PCR exhibited a higher discrimination power and evidenced the emergence of subclones within a cluster of highly related isolates.
FIG. 1.
Representative PFGE patterns of S. marcescens isolates recovered during the NICU outbreak. The same strain (PFGE type A, lanes 1, 2, 4, and 6) was recovered in 32 isolates from patients and wet nurses. Genetically unrelated PFGE patterns B, D, G, and F are also shown (lanes 3, 5, 7, and 8). Lane 1, sample ID 1; lane 2, sample ID 17; lane 3, sample ID 21; lane 4, sample ID 27; lane 5, sample ID 33; lane 6, sample ID 10A; lane 7, sample ID 35; lane 8, sample ID 37.
FIG. 2.
A dendrogram (A) and virtual gel images (B) representing the rep-PCR fingerprint patterns of the S. marcescens isolates described in Table 1, generated by the DiversiLab software, version 3.4. Clonal isolates are boxed.
The reproducibility of rep-PCR patterns was assessed on three different genomic extracts of three isolates and on 10 runs of the same genomic extract. The results showed virtually indistinguishable patterns within each of the three isolates and within the different runs (data not shown). A high level of reproducibility of rep-PCR patterns generated by DiversiLab was also reported by Healy et al. (12).
DISCUSSION
S. marcescens is a well-recognized, hospital-acquired pathogen, and outbreaks in neonatal units have been previously described (6, 9, 17, 19, 30). Usually, one clone is associated with an outbreak and in the study by Jang et al., for instance, it was easy to determine an epidemiological link (17). However, it has also been reported that S. marcescens outbreaks may be polyclonal and this may be due to a mixture of new external clones, as well as evolutionary changes in a common nosocomial strain (3, 5).
The manual rep-PCR technique has been shown to be useful for typing prokaryotic and eukaryotic organisms, including those of epidemiological significance (1, 2, 3, 15, 27, 31) and the semiautomated rep-PCR system that has been introduced recently (12) was employed for typing Staphylococcus aureus, enterococci, Neisseria meningitidis, Acinetobacter, Aspergillus, and Candida albicans (2, 7, 13, 18, 21, 23, 25, 29). Differences in discriminatory power between PFGE and semiautomated rep-PCR are dependent on the bacterial species being studied. Rep-PCR was found to be less discriminatory than PFGE for S. aureus and enterococci (21, 23, 25, 29) and concordant for Acinetobacter and Escherichia coli (2, 7, 20).
Our study is the first to assess the reliability with which the DiversiLab rep-PCR system, in comparison with PFGE, identified the genetic similarity of S. marcescens isolates during a well-characterized outbreak which occurred in a NICU. We clearly documented the spread of infections due to S. marcescens in the NICU by tracing S. marcescens isolation in colonized and infected patients, as well as in wet nurses, and by matching clinical and molecular typing data. The PFGE analysis clearly suggested that the outbreak was monoclonal since 32 of the 38 isolates collected during the outbreak showed identical fingerprints. The rep-PCR results apparently suggested a higher level of genetic variability, and both the dendrogram and the similarity matrix generated by the DiversiLab software (Fig. 2) indicated that not all of the samples in the cluster of PFGE type A isolates showed the same level of similarity. In particular, the rep-PCR type b isolates showed a lower degree of similarity than did those of types a and c. Visual inspection of fingerprints and analysis of normalized sample graphs resolved this discrepancy and suggested that type b isolates could be classified as similar to type a and c isolates. It is possible that in S. marcescens the marker used for the rep-PCR analysis (the region between the noncoding repetitive sequences in bacterial genomes) is genetically less stable than the one used for PFGE (the target sequence of the SpeI restriction enzyme). As a consequence, the variability shown by rep-PCR likely represented evolutionary changes of the same clone that could not be detected by PFGE. Interestingly, rep-PCR allowed us to track the molecular evolution of the epidemic clone. Type a was isolated in September, at the beginning of the outbreak, and gave rise to type c in October. Type b emerged in November and circulated in the NICU together with types a and c until the end of the outbreak in December. The origin of the outbreak could be attributed to the use of breast milk samples collected from wet nurses and contaminated with S. marcescens. The babies of two of these wet nurses were colonized by isolates sharing the same type a isolates from the milk, but it was not possible to document the supply of milk from these wet nurses to other infected babies since the breast milk was pooled.
Automated systems for microorganism typing constitute a compelling tool for the identification and management of nosocomial infections. They assess outbreaks in real time, provide comprehensive surveillance or epidemiological data, and possess data-archiving capability, all of which are required to build libraries and share data among laboratories.
The main limitation of automated systems is the lack of validated interpretative criteria. Without criteria for bacterial strain typing interpretation, no method can be used alone for studies of ongoing outbreaks. Typing results make it possible to categorize isolates as (i) identical, (ii) highly related but not identical, (iii) moderately related, and (iv) unrelated. Since 1995, criteria for bacterial strain typing interpretation have been available but validated only for interpreting chromosomal DNA restriction patterns produced by PFGE (28). In this case, validation was done by investigators who correlated epidemiological data from several outbreaks with strain typing results produced by PFGE. If a system provides results that are too discriminatory, the potential association between the isolates is completely missed and the mistaken conclusion that multiclonal outbreaks have occurred with multiple sources of infecting isolates is generated. This conclusion could be correct if the diversity reflects poor genetic relatedness and not the rapid genetic evolution of the same clone that could occur in a very short time period. Conversely, a method with low discriminatory power could generate the wrong conclusion that outbreaks are monoclonal and thus that only one source is involved.
Close cooperation between clinicians and microbiologists is important for resolving an epidemic event. Microbiologists play a primary role in the study of isolates circulating in a hospital. At the same time, microbiologists must also be able to rapidly confirm or exclude the possibility of an epidemic cluster by fingerprinting of isolates. In conclusion, our data suggest that the automated rep-PCR system is a reliable tool for epidemiological investigation. The DiversiLab fingerprinting rep-PCR provides an option to clinical laboratories to perform molecular typing and will benefit infection control procedures with the rapid introduction of control measures and significantly improve patient care; however, the study of more outbreak events is required to validate it as a method to be used alone in ongoing outbreaks.
Acknowledgments
We thank Mariella Ceriani, Carmen Ceriani, and Ennio Mortani for their invaluable help in performing all the epidemiological work.
Footnotes
Published ahead of print on 17 March 2010.
REFERENCES
- 1.Cangelosi, G. A., R. J. Freeman, K. N. Lewis, D. Livingston-Rosanoff, K. S. Shah, S. J. Milan, and S. V. Goldberg. 2004. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J. Clin. Microbiol. 42:2685-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carretto, E., D. Barbarini, C. Farina, A. Grosini, P. Nicoletti, E. Manso, and the APSI Acinetobacter Study Group, Italy. 2008. Use of DiversiLab semiautomated repetitive-sequence-based polymerase chain reaction for epidemiologic analysis on Acinetobacter baumannii isolates in different Italian hospitals. Diagn. Microbiol. Infect. Dis. 60:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Casolari, C., M. Pecorari, G. Fabio, S. Cattani, C. Venturelli, L. Piccini, M. G. Tamassia, W. Gennari, A. M. Sabbatini, G. Leporati, P. Marchegiano, F. Rumpianesi, and F. Ferrari. 2005. A simultaneous outbreak of Serratia marcescens and Klebsiella pneumoniae in a neonatal intensive care unit. J. Hosp. Infect. 61:312-320. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.David, M. D., T. M. Weller, P. Lambert, and A. P. Fraise. 2006. An outbreak of Serratia marcescens on the neonatal unit: a tale of two clones. J. Hosp. Infect. 63:27-33. [DOI] [PubMed] [Google Scholar]
- 6.Fleisch, F., U. Zimmermann-Baer, R. Zbinden, G. Bischoff, R. Arlettaz, K. Waldvogel, D. Nadal, and C. Ruef. 2002. Three consecutive outbreaks of Serratia marcescens in a neonatal intensive care unit. Clin. Infect. Dis. 34:767-773. [DOI] [PubMed] [Google Scholar]
- 7.Fontana, C., M. Favaro, S. Minelli, M. C. Bossa, G. P. Testore, F. Leonardis, S. Natoli, and C. Favalli. 2008. Acinetobacter baumannii in intensive care unit: a novel system to study clonal relationship among the isolates. BMC Infect. Dis. 8:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung, C. P., M. W. Ho, F. D. Wang, K. Tsai, C. E. Liu, and L. K. Siu. 2001. Investigation of an outbreak caused by methicillin resistant Staphylococcus aureus in a cardiovascular surgery unit by ribotyping, randomly amplified polymorphic DNA and pulsed-field gel electrophoresis. APMIS 109:474-480. [DOI] [PubMed] [Google Scholar]
- 9.Giles, M., S. Tabrizi, E. Grabsch, N. Friedman, E. Gillespie, D. Kotsanas, H. Li, T. Korman, and A. Daley. 2007. A comparison of three typing methods for Serratia marcescens during an outbreak across four neonatal intensive care units. Aust. Infect. Control 12:20-24. [Google Scholar]
- 10.Goering, R. 2004. Pulsed-field gel electrophoresis, p. 185-196. In D. H. Persing F. C. Tenover, J. Versalovic, Y.-W. Tang, E. R. Unger, D. A. Relman, and T. J. White (ed.), Molecular microbiology: diagnostic principles and practice. American Society for Microbiology, Washington, DC.
- 11.Hammerum, A. M., V. Fussing, F. M. Aarestrup, and H. C. Wegener. 2000. Characterization of vancomycin-resistant and vancomycin-susceptible Enterococcus faecium isolates from humans, chickens and pigs by RiboPrinting and pulsed-field gel electrophoresis. J. Antimicrob. Chemother. 45:677-680. [DOI] [PubMed] [Google Scholar]
- 12.Healy, M., J. Huong, T. Bittner, M. Lising, S. Frye, S. Raza, R. Schrock, J. Manry, A. Renwick, R. Nieto, C. Woods, J. Versalovic, and J. R. Lupski. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Healy, M., K. Reece, D. Walton, J. Huong, K. Shah, and D. P. Kontoyiannis. 2004. Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 42:4016-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollis, R. J., J. L. Bruce, S. J. Fritschel, and M. A. Pfaller. 1999. Comparative evaluation of an automated ribotyping instrument versus pulsed-field gel electrophoresis for epidemiological investigation of clinical isolates of bacteria. Diagn. Microbiol. Infect. Dis. 34:263-268. [DOI] [PubMed] [Google Scholar]
- 15.Hulton, C. S., C. F. Higgins, and P. M. Sharp. 1991. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol. Microbiol. 5:825-834. [DOI] [PubMed] [Google Scholar]
- 16.Inglis, T. J., A. Clair, J. Sampson, L. O'Reilly, S. Vandenberg, K. Leighton, and A. Watson. 2003. Real-time application of automated ribotyping and DNA macrorestriction analysis in the setting of a listeriosis outbreak. Epidemiol. Infect. 131:637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang, T. N., C. P. Fung, T. L. Yang, S. H. Shen, C. S. Huang, and S. H. Lee. 2001. Use of pulsed-field gel electrophoresis to investigate an outbreak of Serratia marcescens infection in a neonatal intensive care unit. J. Hosp. Infect. 48:13-19. [DOI] [PubMed] [Google Scholar]
- 18.Li, X., N. Brown, A. S. Chau, J. L. Lopez-Ribot, M. T. Ruesga, G. Quindos, C. A. Mendrick, R. S. Hare, D. Loebenberg, B. DiDomenico, and P. M. McNicholas. 2004. Changes in susceptibility to posaconazole in clinical isolates of Candida albicans. J. Antimicrob. Chemother. 53:74-80. [DOI] [PubMed] [Google Scholar]
- 19.Miranda, G., C. Kelly, F. Solorzano, B. Leanos, R. Coria, and J. Evans Patterson. 1996. Use of pulsed-field gel electrophoresis typing to study an outbreak of infection due to Serratia marcescens in a neonatal intensive care unit. J. Clin. Microbiol. 34:3138-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitout, J. D. D., L. Campbell, D. L. Church, P. W. Wang, D. S. Guttman, and D. B. Gregson. 2009. Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J. Clin. Microbiol. 47:1212-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pounder, J. I., C. K. Shutt, B. J. Schaecher, and G. L. Woods. 2006. Clinical evaluation of repetitive sequence-based polymerase chain reaction using the DiversiLab System for strain typing of vancomycin-resistant enterococci. Diagn. Microbiol. Infect. Dis. 54:183-187. [DOI] [PubMed] [Google Scholar]
- 22.Price, C. S., H. Huynh, S. Paule, R. J. Hollis, G. A. Noskin, M. A. Pfaller, and L. R. Peterson. 2002. Comparison of an automated ribotyping system to restriction endonuclease analysis and pulsed-field gel electrophoresis for differentiating vancomycin-resistant Enterococcus faecium isolates. J. Clin. Microbiol. 40:1858-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross, T. L., W. G. Merz, M. Farkosh, and K. C. Carroll. 2005. Comparison of an automated repetitive sequence-based PCR microbial typing system to pulsed-field gel electrophoresis for analysis of outbreaks of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5642-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi, Z., P. Liu, Y. Lau, Y. Lin, and B. Hu. 1997. Use of pulsed-field gel electrophoresis to investigate an outbreak of Serratia marcescens. J. Clin. Microbiol. 35:325-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shutt, C. K., J. I. Pounder, S. R. Page, B. J. Schaecher, and G. L. Woods. 2005. Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains. J. Clin. Microbiol. 43:1187-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, A., R. V. Goering, S. Simjee, S. L. Foley, and M. J. Zervos. 2006. Application of molecular techniques to the study of hospital infection. Clin. Microbiol. Rev. 19:512-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snelling, A. M., P. Gerner-Smidt, P. M. Hawkey, J. Heritage, P. Parnell, C. Porter, A. R. Bodenham, and T. Inglis. 1996. Validation of use of whole-cell repetitive extragenic palindromic sequence-based PCR (REP-PCR) for typing strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii complex and application of the method to the investigation of a hospital outbreak. J. Clin. Microbiol. 34:1193-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. V. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenover, F. C., E. A. Gay, S. Frye, S. J. Eells, M. Healy, and J. E. McGowan, Jr. 2009. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulse-field gel electrophoresis. J. Clin. Microbiol. 47:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Ogtrop, M. L., D. van Zoeren-Grobben, E. M. Verbakel-Salomons, and C. P. van Boven. 1997. Serratia marcescens infections in neonatal departments: description of an outbreak and review of the literature. J. Hosp. Infect. 36:95-103. [DOI] [PubMed] [Google Scholar]
- 31.Versalovic, J., V. Kapur, E. O. Mason, Jr., U. Shah, T. Koeuth, J. R. Lupski, and J. M. Musser. 1993. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J. Infect. Dis. 167:850-856. [DOI] [PubMed] [Google Scholar]