Abstract
We examined if multilocus sequence typing (MLST), a method for genotyping and species identification of Burkholderia cepacia complex bacteria, could be applied directly to cystic fibrosis sputum. The redesigned nested-PCR MLST format was successfully used to accurately identify strains in 23 sputum samples, of which 8 were culture negative.
Burkholderia cepacia complex (BCC) organisms have the capacity to spread within populations of patients with cystic fibrosis (CF) and to cause serious epidemic outbreaks (6, 16). Strict infection control measures are therefore in place in many CF centers to prevent person-to-person transmission of the organisms. Correct identification of BCC organisms in clinical specimens is an essential prerequisite for implementing a successful infection control system. The 17 bacterial species that form the BCC taxonomic cluster today (9, 12, 13) can only be accurately distinguished from each other by applying molecular genetic methods. Although several protocols have been developed to discriminate individual species (4, 8, 10), multilocus sequence typing (MLST) has proven to be the only method which has kept pace with the increasing complexity of BCC species. By sequence analysis of seven housekeeping genes, MLST enables unequivocal identification of all established BCC species and also has the advantage of being able to define novel BCC groups (1, 3, 7). In addition, it can serve as a highly standardized genotyping method that provides robust information on strain type, by defining a sequence type (ST).
There is great potential in DNA-based identification methods, as they provide the possibility to detect minute quantities of BCC bacteria directly in clinical specimens. We previously described a diagnostic nested recA PCR with excellent sensitivity (5) that enabled the detection of BCC bacteria in sputum, weeks to months before patients became culture positive. Such direct PCR testing narrowed the diagnostic window between the time of the initial BCC infection and laboratory diagnosis and had a great impact on implementing infection control in advance of what was routine with conventional microbiology.
Using direct PCR analysis, several instances of samples that were BCC positive by PCR, but not by culture, were identified. Follow-up of such cases was rather limited, as the recA PCR we used (5) did not allow strain identification, and with no culture to examine, conventional typing methods were ineffective. Yet obtaining the strain type information for such patients is vital in understanding the epidemiology behind their infection with BCC bacteria. All gel pattern-based genotyping systems such as pulsed-field gel electrophoresis require pure bacterial cultures that were not available in the reproducibly PCR-positive, but culture-negative patients. Although MLST is also a typing method primarily designed to work from pure bacterial cultures, in principle since it is based on DNA and PCR amplification, its applicability to samples where bacteria are present in a small, noncultivable amount was not ruled out.
We hypothesized that typing of the low quantities of BCC strains in such PCR-positive cultures was feasible without the need for obtaining pure cultures, if MLST was directly applied to total DNA extracted from sputum. In order to increase MLST sensitivity and to make it comparable to double-round PCR-based detection of BCC from sputa (5), the original MLST protocol for BCC (1) was modified to a redesigned nested-PCR format. The primers used for sequence analysis in the original MLST method already formed a primer pair that was internal to the allele-specific amplification primers. Therefore, they were employed, in addition to sequencing, for a second round of the nested PCR.
To investigate the applicability of MLST for direct sputum genotyping of BCC bacteria, 23 sputum samples from 17 CF patients were examined (Table 1). The sputum samples were representatives of the following diagnostic eventualities: (i) the extent of BCC positivity, as determined by nested PCR, with 9 samples positive at round 1 and 14 positive only after 2 rounds of this nested PCR; (ii) the differential ability to cultivate BCC organisms, with 8 culture-negative and 15 culture-positive samples; (iii) situations involving multiple BCC species, as identified by the recA species-specific PCRs, with 10 Burkholderia cenocepacia IIIA, 2 B. cenocepacia IIIB, 4 Burkholderia multivorans, and 3 Burkholderia stabilis samples, as well as 4 samples of indeterminate BCC species; and (iv) presence/absence of coinfection with Pseudomonas aeruginosa (10 samples culture positive and 13 culture negative for P. aeruginosa). In addition, for four of the patients studied, sputa were sampled at multiple clinic visits: for 3 patients (patients 10, 14, and 17; Table 1), two samples were obtained 5, 2, and 9 months apart, respectively, and for patient 9, four consecutive samples were received at time intervals of 1, 6, and 7.5 months from the first sampling point.
TABLE 1.
Characteristics of sputum samples examined in this study
| Patient | Sample ID no. | Identification of BCC by nested PCR and culture |
MLST analysis of BCC |
|||
|---|---|---|---|---|---|---|
| PCR round at which detected | Species identified | Culture | ST | Species identified | ||
| 1 | 5884 | 1 | B. cenocepacia IIIA | + | 32 | B. cenocepacia IIIA |
| 2 | 6140 | 1 | B. cenocepacia IIIA | + | 32 | B. cenocepacia IIIA |
| 3 | 5475 | 1 | B. cenocepacia IIIA | + | 32 | B. cenocepacia IIIA |
| 4 | 5938 | 2 | B. cenocepacia IIIA | − | 32 | B. cenocepacia IIIA |
| 5 | 5883 | 2 | B. cenocepacia IIIA | + | 211 | B. cenocepacia IIIA |
| 6 | 5780 | 2 | B. cenocepacia IIIA | − | 234 | B. cenocepacia IIIA |
| 7 | 6144 | 1 | B. cenocepacia IIIA | + | 234 | B. cenocepacia IIIA |
| 8 | 5911 | 1 | B. cenocepacia IIIA | + | 234 | B. cenocepacia IIIA |
| 9 | 4871 | 2 | Indeterminate | + | 102, 482 | B. contaminans |
| 4941 | 1 | B. cenocepacia IIIA | + | 482 | B. contaminans | |
| 5315 | 1 | B. cenocepacia IIIA | + | 102, 482 | B. contaminans | |
| 5445 | 2 | Indeterminate | − | 482 | B. contaminans | |
| 10 | 5662 | 2 | Indeterminate | + | 102 | B. contaminans |
| 6070 | 2 | Indeterminate | − | 102 | B. contaminans | |
| 11 | 5983 | 1 | B. cenocepacia IIIB | + | 184 | B. cenocepacia IIIB |
| 12 | 6072 | 2 | B. cenocepacia IIIB | + | 184 | B. cenocepacia IIIB |
| 13 | 5699 | 1 | B. multivorans | + | 180 | B. multivorans |
| 14 | 5781 | 2 | B. multivorans | − | 564 | B. multivorans |
| 5984 | 2 | B. multivorans | − | 564 | B. multivorans | |
| 15 | 5747 | 2 | B. multivorans | − | 562 | B. multivorans |
| 16 | 4894 | 2 | B. stabilis | − | 51 | B. stabilis |
| 17 | 4934 | 2 | B. stabilis | + | 563 | B. stabilis |
| 5702 | 2 | B. stabilis | + | 563 | B. stabilis | |
The PCR conditions for MLST round 1 amplification were as follows. A PCR for each MLST allele was carried out in a 20-μl volume with 2.0 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 10 pmol “amplification primers” (1), 1× Q solution, and 0.75 U Taq polymerase (all PCR reagents were obtained from Qiagen, Hilden, Germany) and 1 μl bacterial DNA extracted from clinical samples with an Amplicor respiratory specimen preparation kit (Roche, Indianapolis, IN). The PCR program was run on a TGradient thermocycler (Biometra, Goettingen, Germany) with initial denaturation for 5 min at 96°C and a subsequent 30 cycles of 30 s at 96°C, 30 s at 55°C, and 2 min at 72°C. An internal DNA fragment was then amplified in a second PCR with reaction chemistry identical to round 1, except that the sequencing primers (1) and 1 μl PCR product from round 1 were incorporated. The thermal cycling profile for round 2 was as follows. Initial denaturation was at 96°C for 2 min followed by a subsequent run of 35 cycles, each comprising 15 s at 96°C, 30 s of annealing (see below), and 45 s at 72°C. The annealing temperature was set to decrease from 60°C in the first cycle by 0.5°C per cycle over the next 10 cycles to reach the final annealing temperature of 55°C, which was then kept for the last 25 cycles of PCR. The detection limit of the nested PCR was determined by serial decimal dilutions of DNA from a B. cenocepacia culture of known viability that had been extracted in the same way as the sputum samples.
Successful MLST data were obtained for every sample examined, including those samples which were culture negative and those which were amplification negative after 1 round of the recA diagnostic PCR (summarized in Table 1). If a single MLST allele was not detected after completion of the first MLST-PCR round, all seven genes of the sample were subjected to the round 2 PCR. The threshold of this setup was found for each of the seven MLST genes at ∼104 CFU/ml (10 CFU per reaction). Out of 161 sequencing PCRs performed (23 samples × 7 MLST genes), only one reaction failed repeatedly to give a readable sequence (the phaC allele for sample 5445); however a complete ST profile for this strain was obtained from another sample from the same patient (Table 1). Thus, we did not have to exploit alternative primers which were recently redesigned in order to achieve more reliable amplification results for all BCC groups and other Burkholderia species (11).
At the level of species identification, the direct MLST analysis outperformed the original recA-based diagnostic PCR (5). MLST provided correct species identification in 4 samples that were left undetermined by recA species-specific PCR (Table 1; patients 9 and 10). All of these samples were assigned to the recently described species Burkholderia contaminans, a relevant pathogen in CF (7, 12), for which no species-specific recA PCR has been described. In addition, MLST corrected previous species misidentification of two B. contaminans isolates from patient 9 (Table 1) which had been misclassified as B. cenocepacia IIIA with recA species-specific PCRs. This finding is in agreement with recent studies (3, 4) that highlight potential problems in BCC species identification performed solely with first-generation molecular identification approaches, such as recA restriction fragment length polymorphism (RFLP) or recA species-specific PCRs; sequence analysis of the recA gene as part of the MLST is far more accurate than these indirect ways of examining DNA polymorphisms.
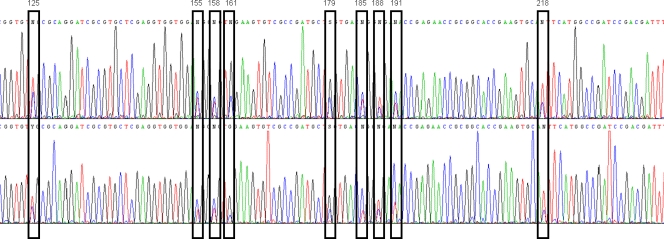
Analysis of two sputum samples from patient 9 (ID 4871 and 5315; Table 1) revealed the coexistence of two distinct strains of B. contaminans (ST102 and ST482). These two strains differ from each other by all seven MLST alleles, out of which the gyrB gene carries the most pronounced sequence polymorphisms (18-nucleotide difference). Nucleotide sequence differences for each allele variant were clearly apparent in the sequence traces obtained (Fig. 1). Clearly discernible composite nucleotide peaks were observed at each nucleotide polymorphism site that could be readily separated into the two discrete allele sequences. Consequently, the presence of both strains was tracked down and confirmed by more thorough analysis of the culture that was isolated from sputum ID 4941 and that alone showed an MLST result corresponding only to one of the two strains. By screening the sequences of the gyrB allele of 50 individual colonies which were isolated from the parental culture ID 4941, we detected gyrB 76 (i.e., ST102) in five colonies. While this may have indicated that the ST482 strain was the more predominant strain from the pair, neither strain persisted and caused chronic infection, as 5 subsequent samples collected over the period of 1 year were BCC free (data not shown). Although coinfection and strain replacement with different BCC species have been previously demonstrated in CF (2, 14, 15), such studies required the cultivation and purification of each strain present, which may not always be possible if the colonial morphology of each isolate is not discriminatory. Direct MLST as described here enabled the detection of mixed BCC populations in an analysis without the need for strain separation. Nevertheless, this ST identification approach brings a potential limitation in that we selected a combination of 7 out of 14 obtained alleles (7 alleles × 2 strains) based on known profiles submitted to the public MLST database (http://pubmlst.org/bcc/); without an option to validate a not-yet-described combination of alleles, this method would not allow identification of a completely new ST. In addition, definition of the correct 7-allele combination would have been impossible if we had detected multiple shared alleles that occur in multiple STs, including the two in question. The ST assembly derived for the two B. contaminans strains (Table 1) was simplified by the fact that every allele was either unique for one strain or did not occur in combination with other alleles for the other strain.
FIG. 1.
Identification of two B. contaminans strains within the sputum of patient 9. The sequence trace file of the gyrB allele resulting from MLST analysis of clinical sample ID 5315 is shown. Two distinct strains (ST102 and ST482) were present in the sample, and nucleotide polymorphisms representing each allele of the two STs (i.e., gyrB 76 and gyrB 245) are highlighted; positions of nucleotide differences in allele sequence are indicated above the bars.
Potential cross-reactivity with DNA of another origin was one of our major concerns about the applicability of MLST directly to sputum. However, we obtained unambiguous sequencing results from both PCR rounds, which indicated that neither human DNA nor non-BCC bacterial DNAs (such as P. aeruginosa) that were present in inflammatory CF sputum did interfere with the MLST protocol.
In conclusion, we have demonstrated that BCC strain identification is feasible directly from sputum samples without a need to work with pure cultures. We found this method useful particularly in situations where culture results remain negative due to a low bacterial load during infection. In such cases, only direct MLST on sputum can provide information on strain type and facilitate the complete analysis of a local epidemiological situation, a vital prerequisite for effective BCC surveillance and infection control in every CF center.
Acknowledgments
This work was supported by grants from the Czech Ministry of Health (NS 10543-3) and the Czech Ministry of Education (MSM0021620812).
We thank Helena Reitzova for excellent technical assistance.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Baldwin, A., E. Mahenthiralingam, K. M. Thickett, D. Honeybourne, M. C. Maiden, J. R. Govan, D. P. Speert, J. J. LiPuma, P. Vandamme, and C. G. Dowson. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhardt, S. A., T. Spilker, T. Coffey, and J. J. LiPuma. 2003. Burkholderia cepacia complex in cystic fibrosis: frequency of strain replacement during chronic infection. Clin. Infect. Dis. 37:780-785. [DOI] [PubMed] [Google Scholar]
- 3.Cesarini, S., A. Bevivino, S. Tabacchioni, L. Chiarini, and C. Dalmastri. 2009. recA gene sequence and multilocus sequence typing for species-level resolution of Burkholderia cepacia complex isolates. Lett. Appl. Microbiol. 49:580-588. [DOI] [PubMed] [Google Scholar]
- 4.Drevinek, P., A. Baldwin, C. G. Dowson, and E. Mahenthiralingam. 2008. Diversity of the parB and repA genes of the Burkholderia cepacia complex and their utility for rapid identification of Burkholderia cenocepacia. BMC Microbiol. 8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drevinek, P., H. Hrbackova, O. Cinek, J. Bartosova, O. Nyc, A. Nemec, and P. Pohunek. 2002. Direct PCR detection of Burkholderia cepacia complex and identification of its genomovars by using sputum as source of DNA. J. Clin. Microbiol. 40:3485-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones, A. M., M. E. Dodd, J. R. Govan, V. Barcus, C. J. Doherty, J. Morris, and A. K. Webb. 2004. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59:948-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahenthiralingam, E., A. Baldwin, P. Drevinek, E. Vanlaere, P. Vandamme, J. J. LiPuma, and C. G. Dowson. 2006. Multilocus sequence typing breathes life into a microbial metagenome. PLoS One 1:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 10.Moore, J. E., J. Xu, B. C. Millar, M. Crowe, and J. S. Elborn. 2002. Improved molecular detection of Burkholderia cepacia genomovar III and Burkholderia multivorans directly from sputum of patients with cystic fibrosis. J. Microbiol. Methods 49:183-191. [DOI] [PubMed] [Google Scholar]
- 11.Spilker, T., A. Baldwin, A. Bumford, C. G. Dowson, E. Mahenthiralingam, and J. J. LiPuma. 2009. Expanded multilocus sequence typing for Burkholderia species. J. Clin. Microbiol. 47:2607-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanlaere, E., A. Baldwin, D. Gevers, D. Henry, E. De Brandt, J. J. LiPuma, E. Mahenthiralingam, D. P. Speert, C. Dowson, and P. Vandamme. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102-111. [DOI] [PubMed] [Google Scholar]
- 13.Vanlaere, E., J. J. Lipuma, A. Baldwin, D. Henry, E. De Brandt, E. Mahenthiralingam, D. Speert, C. Dowson, and P. Vandamme. 2008. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evol. Microbiol. 58:1580-1590. [DOI] [PubMed] [Google Scholar]
- 14.Waine, D. J., D. Honeybourne, E. G. Smith, J. L. Whitehouse, and C. G. Dowson. 2009. Cross-sectional and longitudinal multilocus sequence typing studies of Pseudomonas aeruginosa in cystic fibrosis sputum. J. Clin. Microbiol. 47:3444-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wellinghausen, N., and J. Kothe. 2006. Evidence of coinfection with distinct strains of Burkholderia multivorans in a cystic fibrosis patient. Infection 34:289-291. [DOI] [PubMed] [Google Scholar]
- 16.Whiteford, M. L., J. D. Wilkinson, J. H. McColl, F. M. Conlon, J. R. Michie, T. J. Evans, and J. Y. Paton. 1995. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax 50:1194-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]