Abstract
Staphylococcus aureus is a major cause of skin and soft tissue infections, such as furuncles, carbuncles, and abscesses, but it also frequently colonizes the human skin and mucosa without causing clinical symptoms. Panton-Valentine leukocidin (PVL) is a pore-forming toxin that has been associated with soft tissue infections and necrotizing pneumonia. We have compared the genotypes, virulence gene repertoires, and phage patterns of 74 furunculosis isolates with those of 108 control strains from healthy nasal carriers. The large majority of furunculosis strains were methicillin sensitive. Clonal cluster (CC) 121 (CC121) and CC22 accounted for 70% of the furunculosis strains but for only 8% of the nasal isolates. The PVL-encoding genes luk-PV were detected in 85% of furunculosis strains, while their prevalence among colonizing S. aureus strains was below 1%. luk-PV genes were distributed over several lineages (CCs 5, 8, 22, 30, and 121 and sequence type 59). Even within the same lineages, luk-PV-positive phages characterized furunculosis strains, while their luk-PV-negative variants were frequent among nasal strains. The very tight epidemiological linkage between luk-PV and furunculosis, which could be separated from the genetic background of the S. aureus strain as well as from the gene makeup of the luk-PV-transducing phage, lends support to the notion of an important role for PVL in human furunculosis. These results make a case for the determination of luk-PV in recurrent soft tissue infections with methicillin-sensitive as well as methicillin-resistant S. aureus.
Skin and soft tissue infections (SSTIs) are the most frequent type of disease caused by Staphylococcus aureus outside the hospital setting. SSTIs comprise a diverse range of clinical pictures, such as furuncles, carbuncles, subcutaneous abscesses, folliculitis, bullous impetigo, and staphylococcal scalded skin syndrome (23). Furunculosis is a very common disease characterized by infection of the hair follicles and the local accumulation of pus and necrotic tissue. Even mild lesions are painful and unsightly and often leave a scar after they heal (20). Antibiotic treatment is frequently not effective, and many furunculosis patients suffer from recurrent episodes or develop chronic symptoms over months and years without a period free from outbreaks (20).
Apart from being a major human pathogen, S. aureus is also a frequent colonizer of the human skin and mucosa (53, 57). The bacteria find their primary ecological niche in the human nose; but they are also able to colonize the skin, throat, and intestines, sometimes exclusively (1, 35). About 20% of the healthy population are persistent nasal S. aureus carriers (53, 57). Patients suffering from chronic furunculosis are usually S. aureus carriers, and skin and nose isolates from a given patient commonly have identical characteristics (10, 48, 51).
The species S. aureus displays extensive genetic variability. Genotyping analyses, such as multilocus sequence typing (MLST) and protein A (spa) sequence typing, demonstrated that the S. aureus population structure is highly clonal with 10 major and many minor clonal clusters (CCs) (14, 22, 34, 38). Mobile genetic elements comprise 15% of the S. aureus genome (32). They contain plasmids, phages, and pathogenicity islands and carry a variety of virulence and resistance genes which can strongly enhance the virulence of staphylococci (31, 33). For example, staphylococcal prophages, which are classified into seven types (Sa1int to Sa7int), can harbor the genes for exfoliative toxin A (Sa1int), the pore-forming toxin Panton-Valentine leukocidin (PVL; Sa2int), and superantigens (SAgs; Sa3int) (17). Most mobile genetic elements can readily spread horizontally among S. aureus strains of the same clonal cluster, while transfer between clusters is limited (17, 22, 33, 56).
Despite intensive research efforts, it still remains elusive how staphylococcal virulence is determined on a molecular level. Numerous studies compared the core genome and virulence gene repertoire of blood culture and colonizing isolates but failed to identify factors clearly related to virulence (14, 22, 33). This suggests that invasion into the bloodstream does not require special bacterial virulence traits but mainly depends on host factors, e.g., barrier breakage, the presence of indwelling catheters, or a compromised immune system. In contrast, the causative virulence factors for a number of toxin-mediated diseases are well known. Toxic shock syndrome and food poisoning are caused by SAgs (15), while staphylococcal scalded skin syndrome and bullous impetigo are associated with exfoliative toxins (3, 18, 30).
PVL is a pore-forming toxin which is composed of two protein components (LukF and LukS) that very efficiently disrupt the cell membrane of neutrophils (25). PVL has been associated with chronic or recurrent skin and soft tissue infections and with necrotizing pneumonia, which also affect immune-competent persons (7, 16, 31, 54). One PVL-producing S. aureus clone, USA300, a community-acquired methicillin-resistant S. aureus (CA-MRSA) member of CC8, is epidemic in the community in the United States and causes severe SSTIs and necrotizing pneumonia (40).
The aim of the molecular epidemiological study described here was to further elucidate the molecular determinants of virulence in chronic furunculosis, in particular, to assess the contributions of the bacterial genetic background versus those of virulence factors and phages. By applying spa genotyping and PCR-based virulence gene and phage profiling, we observed strong associations of PVL and the genetic background with furunculosis.
MATERIALS AND METHODS
Study population and bacterial isolates. (i) Furunculosis strains.
S. aureus isolates from 74 patients with furunculosis were obtained from a typical mature furuncle (fresh pus) by a physician during the acute phase of skin infection or by a surgeon during abscess incision. In 11 cases, nose swab specimens were taken in parallel. The study was carried out at the Department of Microbiology and Immunology, Pomeranian Medical University, Szczecin, Poland, between 2002 and 2008.
(ii) Nasal strains.
One hundred eight nasal S. aureus isolates were obtained from 362 healthy blood donors at the Department of Microbiology and Immunology, Pomeranian Medical University, in March 2006. Volunteers who reported that they had had skin infections at some time during the previous 2 years were excluded. All participants gave informed consent, and the study was approved by the Ethics Board of Pomeranian Medical University in Szczecin, Poland. The genotypes and virulence genes of a subset of these strains (the 28 CC30 isolates) were previously published by Holtfreter et al. (22).
S. aureus identification and DNA isolation.
S. aureus was identified by standard diagnostic procedures and a gyrase PCR (22). Total DNA of S. aureus was isolated with a DNeasy blood and tissue kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions.
spa genotyping.
PCR for amplification of the S. aureus protein A (spa) repeat region was performed according to the description in the published protocol (2, 19). PCR products were purified with a QIAquick PCR purification kit (Qiagen) and sequenced by a commercial supplier (Agowa, Berlin, Germany) by using both amplification primers. The forward and reverse sequence chromatograms were analyzed with StaphType software (Ridom GmbH, Würzburg, Germany). The spa types were clustered into different groups with the BURP algorithm (Ridom GmbH) and by use of the following setting: the calculated cost between members of a group was less than or equal to five. spa types shorter than five repeats were not clustered, because they do not allow the reliable deduction of ancestries. Since the results of spa typing and MLST are highly concordant (47), the spa typing data could easily be mapped on the MLST types by using the SpaServer database (www.spaserver.ridom.de).
Detection of S. aureus virulence factors and phages by PCR.
PCR was used to screen for a total of 26 genes. Single PCR was applied for the detection of 16S rRNA, gyrase (gyr), methicillin resistance (mecA), PVL (luk-PV), and exfoliative toxin etb. Six sets of multiplex PCRs were applied to amplify (i) sea, seh, sec, and tst; (ii) sed, etd, eta, and sek; (iii) see, seb, sem, sel, and seo; (iv) sen, seg, seq, and sej; (v) sei, ser, seu, and sep; and (vi) agr types 1 to 4, as reported previously (22). Single and multiplex PCRs were performed with the GoTaq Flexi DNA polymerase system (Promega, Mannheim, Germany), as described previously (22). All PCR products were resolved by electrophoresis in 1.5% agarose gels (1× TBE [Tris-borate-EDTA] buffer), stained with ethidium bromide, and visualized under UV light. Positive controls included DNA from SAg gene-positive S. aureus reference strains, while S. aureus strain 8325-4 served as a negative control.
Multiplex PCR for the Sa1int to Sa7int phage integrase genes was performed as reported previously (17).
Statistical analysis.
Categorical variables were assessed by using Pearson's chi-square test. P values of <0.05 were considered statistically significant.
RESULTS
Study cohorts.
To identify virulence determinants in S. aureus furunculosis, we analyzed the genotypes, virulence gene patterns, and phage profiles of 74 S. aureus isolates from furunculosis patients and 108 nasal isolates from healthy carriers (Table 1).
TABLE 1.
Characteristics of the study cohorts
| Characteristic | Furunculosis patients | Colonization patients |
|---|---|---|
| No. of strains | 74 | 108 |
| Mean ± SD age (yr) | 26.6 ± 11.7 | 29.4 ± 9.4 |
| % male | 48.6 | 88.0 |
| Time of sampling | 2002-2008 | March 2006 |
spa-defined clonal lineages.
To clarify the role of the core genome in furunculosis, we performed spa typing of the furunculosis and nasal isolates. This revealed 91 different spa types, which were assigned to 10 CCs and 4 sequence types (STs) by BURP clustering. Singletons, i.e., spa types which could not be assigned to a CC or ST, occurred among nasal strains (9/108) and furunculosis strains (1/74). Nine strains were excluded from BURP clustering because the spa repeats were too short, and two strains were spa negative.
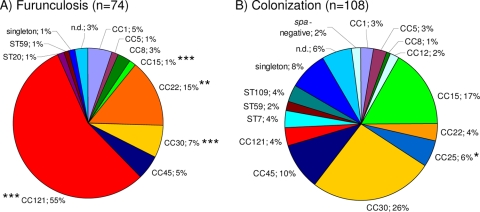
As expected, the nasal strains showed a highly diverse population structure (Fig. 1). The major lineages (i.e., those containing more than 5% of the isolates) included CC30 (26%), CC15 (17%), CC45 (10%), and CC25 (6%), whereas CCs 5, 8, 12, and 121 and STs 7, 59, and 109 were rarely detected.
FIG. 1.
Prevalence of spa-defined CCs among furunculosis strains (A) and colonizing strains (B). CC121 and CC22 together accounted for 70.3% of the furunculosis strains but for only 8% of the colonizing isolates. spa types were clustered into 10 CCs and 4 STs by BURP analysis. The MLST-CC nomenclature was deduced from the spa CCs by using the Ridom SpaServer database. Chi-square test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.d., not determined.
In sharp contrast, 55.4% (41/74; P < 0.001) of all furunculosis strains belonged to the CC121 lineage (Fig. 1). Notably, this lineage was rare among nasal strains (3.7%; 4/108) (Fig. 1). spa types were diverse within this lineage: among the 41 furunculosis-associated CC121 isolates, we observed 14 different spa types, with spa types t159 (13 isolates) and t435 (8 isolates) being the most prevalent. All four commensal CC121 strains belonged to different spa types. Moreover, CC22 was overrepresented among furunculosis strains (14.9% versus 3.7% among colonizing nasal strains; P < 0.01). Together, CC121 and CC22 accounted for 70.3% of all furunculosis isolates, and, accordingly, the prevalence of other lineages such as CC15, CC25, and CC30 was significantly reduced.
Nasal strains were available from 11 of the 74 patients. In all cases, the furunculosis and nasal strains were clonally identical (see Table S1 in the supplemental material). This confirms the findings of an earlier study, which reported the same phage type in the nose and the lesion in the majority of furunculosis patients (57).
Virulence gene repertoire.
To address the contribution of virulence factors to furunculosis, we next determined the genes encoding the methicillin resistance (mecA); PVL toxin (luk-PV); SAgs (sea to seu, tst); exfoliative toxins A, B, and D (eta, etb, and etd, respectively); and agr types 1 to 4.
Methicillin resistance.
Except for three isolates, all furunculosis and nasal isolates were methicillin sensitive. Among the nasal strains, we detected one MRSA isolate (isolate SZ148) which belonged to CC45, a known MRSA lineage. Moreover, one furunculosis isolate and one nasal isolate (isolates H5391 and SZ179, respectively) belonged to ST59 and were mecA and luk-PV positive, which is characteristic for CA-MRSA.
PVL.
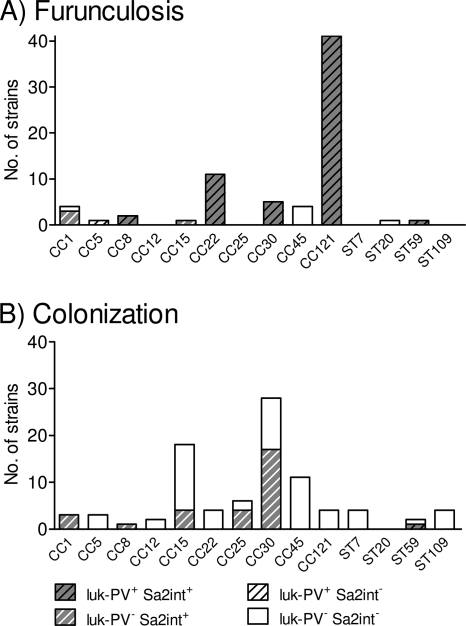
The genes encoding the PVL toxin were a characterizing feature of the furunculosis strains but were almost absent from the nasal isolates. In total, 85.1% (64/74) of the furunculosis strains but only one nasal isolate were luk-PV positive (P < 0.001; Fig. 2 and 3). The phage-encoded luk-PV genes were widely distributed among the different lineages. All CC5, CC8, CC22, CC30, CC121, and ST59 isolates were luk-PV positive, whereas strains belonging to CC1, CC15, CC45, and ST20 lacked the luk-PV genes (Fig. 3A).
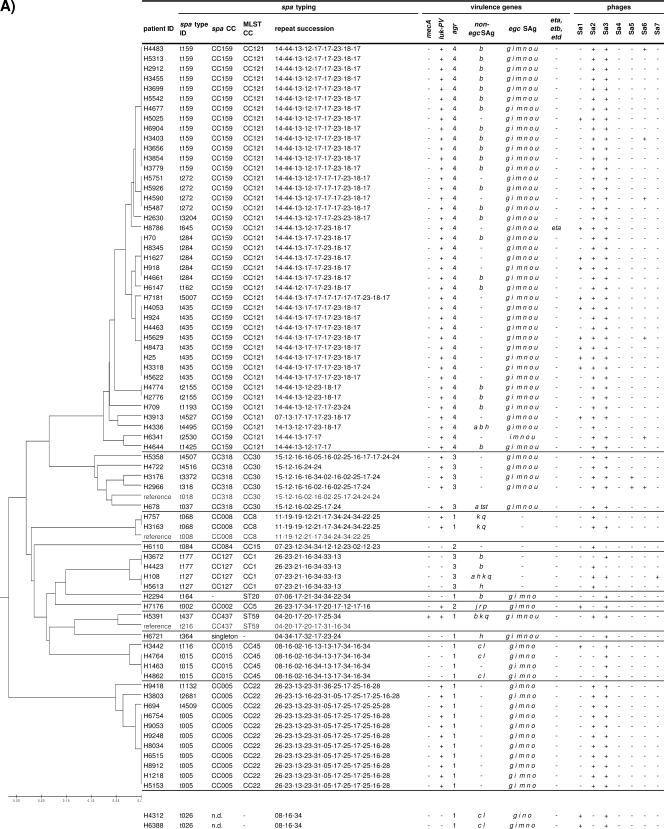
FIG. 2.
Distribution of virulence genes and phages within spa-defined CCs among furunculosis strains (A) and colonizing strains (B). luk-PV genes were detected in 85% of furunculosis strains, while their prevalence among nasal strains was below 1%. Furunculosis and colonizing strains did not differ in their SAg gene patterns. For a reliable construction of the consensus tree, some reference spa types were included in the BURP clustering (gray lettering). Virulence genes (SAg genes, agr, eta, etb, etd, luk-PV, and mecA) and phage types were determined by PCR. Staphylococcal enterotoxins (SEs) are indicated by single letters (a sea, etc.). ID, identifier; n.d., not determined.
FIG. 3.
Distribution of luk-PV genes and PVL-encoding phage Sa2int within the spa-defined CCs among furunculosis strains (A) and colonizing strains (B). luk-PV-positive (luk-PV+) phages characterized the furunculosis strains, whereas their luk-PV-negative (luk-PV−) counterparts were frequent among the colonizing strains. The total number of strains per CC is represented by the overall height of the bar, whereas the numbers of strains positive for luk-PV and phage Sa2int (Sa2int+) are represented by stripes and gray shading, respectively. Sa2int−, phage Sa2int negative.
SAg.
SAg genes were more or less tightly linked to staphylococcal lineages, which is in agreement with the findings of previous studies (Fig. 2) (22, 36, 39). For example, egc SAgs, which are encoded on the vSAβ genomic island, were strictly linked to CC5, ST20, CC22, CC30, CC45, and CC121. Other SAgs with strong CC linkages were tst (CC30), sea (CC30), sec and sel (CC45), sep (ST7), and seb (CC121) (Fig. 2). However, within certain CCs and even within the same spa type there was remarkable variation in the SAg gene patterns. This suggests that the horizontal transmission of SAg-encoding mobile genetic elements occurs frequently within lineages but might be limited between lineages.
To avoid a bias caused by the uneven distribution of CCs among furunculosis and nasal strains, we next compared the SAg gene patterns for each CC separately. The seb SAg gene was significantly more frequent among furunculosis-associated strains than among nasal CC121 strains (23/41 and 0/4, respectively; P < 0.05). Except for seb, the furunculosis and nasal strains did not differ in their SAg gene patterns. Earlier studies also found no particular association of enterotoxin genes with impetigo or furunculosis (10, 18, 24).
ETs.
Exfoliative toxins (ETs) ETA and ETB but probably not ETD are strongly associated with bullous impetigo and staphylococcal scalded skin syndrome but are absent from furunculosis strains (18, 59). In line with this finding, eta, etb, and etd were rare among our furunculosis and nasal strains. The etd gene was strictly linked to CC25 (Fig. 2), a lineage which contained only nasal strains. This confirms the microarray data of Monecke et al., who detected the pathogenicity island comprising edinB and etd exclusively in CC25 strains (38, 39).
Accessory gene regulator (agr).
agr is a global regulator of virulence gene expression; and four different agr types, agr-1 to agr-4, are known. The agr locus belongs to the core variable genome and is thus strictly linked to CCs (33). In agreement with the findings of other studies (22, 36, 39), we observed that agr-1 was linked to CC8, CC22, CC45, ST7, and ST59; agr-2 was present in CC5, CC12, CC15, and ST109; agr-3 was associated with CC1 and CC30; and agr-4 was detected in CC121.
On the basis of the results of our PCR analyses, we could define the virulence gene signature of furunculosis CC121 isolates as follows: mecA negative, luk-PV positive, egc positive, frequently seb positive, and agr-4. CC22 furunculosis strains were characterized as mecA negative, luk-PV positive, egc positive, and agr-1.
Phages.
Several S. aureus virulence factors, including PVL; ETA; and SAgs SEA, SEP, SEK, and SEQ, are encoded by staphylococcal phages. To correlate the observed virulence gene profile with the prevalence of phages, we applied a multiplex PCR for the phage-specific integrase genes Sa1int to Sa7int which was recently described by Goerke et al. (17). Almost all strains (96.7%) carried phages, usually between one and three. Phage Sa3int was by far the most prevalent, followed by Sa2int, Sa1int, Sa6int, Sa5int, and Sa7int (Table 2). Except for Sa1int and Sa6int, the phage frequency profiles of the 108 nasal strains were very similar to the frequencies reported by Goerke and coworkers for nasal isolates from Germany; Sa1int and Sa6int, however, were more abundant in the Polish strain collection (17).
TABLE 2.
Prevalence of phages among furunculosis and colonizing strains
| Phage | No. (%) of positive isolates |
P valuea | |
|---|---|---|---|
| Furunculosis (n = 74) | Colonization (n = 108) | ||
| Sa1int | 15 (20.3) | 19 (17.6) | |
| Sa2int | 64 (86.5) | 36 (33.3) | *** |
| Sa3int | 69 (93.2) | 80 (74.1) | *** |
| Sa4int | 0 (0.0) | 0 (0.0) | |
| Sa5int | 2 (2.7) | 12 (11.1) | * |
| Sa6int | 6 (8.1) | 25 (23.1) | ** |
| Sa7int | 1 (1.4) | 10 (9.3) | * |
| None | 0 (0) | 6 (5.5) | * |
Chi-square test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The prophage prevalence was linked with the spa-defined clonal background. For example, the Sa2int phage was highly abundant among CC25 and CC30 isolates but absent from all CC45 and CC5 isolates (Table 2). Similarly, the highly prevalent Sa3int phage was present in all CC5, CC22, CC25, and CC45 strains but was rare among CC15 isolates. This linkage between phage groups and clonal lineages was previously reported by Goerke at al. and indicates that the spread of phages in the S. aureus population is at least partially restricted (17).
The phage patterns of the furunculosis and nasal strains were remarkably different, which can be partially explained by a bias in the prevalence rates of the CCs. Furunculosis strains carried, on average, more phages per strain (2.12 versus 1.69). The Sa2int and Sa3int phages were more frequent among furunculosis strains, while phages Sa5int, Sa6int, and Sa7int were more often detected among nasal strains (Table 2).
However, the most striking difference concerned phage Sa2int. In our strain collection, we observed luk-PV-positive and luk-PV-negative variants of this phage group. While luk-PV-positive Sa2int phages were found in 81% of the furunculosis strains but in only one nasal isolate (P < 0.001), its luk-PV-negative counterpart was present in one-third of the nasal strains but in only 5% of the isolates associated with furunculosis (P < 0.001) (Fig. 3). Remarkably, CC30 (and CC8) strains from furunculosis patients harbored luk-PV-positive Sa2int phage, whereas commensal strains belonging to the same clonal lineage contained its luk-PV-negative variant. All furunculosis-associated isolates from the typical lineages CC121 and CC22 harbored luk-PV-positive phage Sa2int, while the commensal CC121 and CC22 isolates did not contain this phage at all and were therefore luk-PV negative. This very strong association of luk-PV genes with furunculosis indicates an important pathogenic role for the PVL toxin.
DISCUSSION
SSTIs are the most common diseases caused by S. aureus in the community, and the recent spread of PVL-positive CA-MRSA has spurred scientific and public interest in this neglected disease. In the United States, CA-MRSA clone USA300 causes about 60% of the cases of severe SSTIs among patients presenting to U.S. emergency departments (7, 40). However, in Europe, the large majority of SSTIs are caused by methicillin-susceptible S. aureus (MSSA) strains and PVL-positive CA-MRSA strains are still uncommon, accounting for less than 1% of all MRSA isolates (45). The prevalence of CA-MRSA was also low in our study: only one CA-MRSA (ST59) strain was isolated from a furunculosis patient.
A major finding of our study is that only two lineages, CC121 and CC22, accounted for 70% of the furunculosis MSSA strains. Notably, all were luk-PV positive, while their colonizing counterparts never harbored luk-PV genes. The pronounced genetic diversity of the CC121 isolates (16 spa types) and CC22 isolates (6 spa types) from patients and controls cannot be explained by clonal outbreaks but suggests a long-term endemic persistence and diversification of these lineages. CC121 and CC22 differed in agr types but had similar virulence gene signatures: egc SAg genes and frequently in CC121 isolates seb genes but no other SAg genes and no exfoliative toxin genes.
SSTIs caused by luk-PV-positive MSSA CC121 strains have been reported from Saxony and Brandenburg in Germany as well as from hospitals in Russia, showing that this CC has a worldwide distribution (4, 39, 55, 58). Similar strains also caused highly lethal community-acquired pneumonia and severe sepsis with progressive metastatic soft tissue infection (4, 46), demonstrating the virulence potential of this lineage.
A different subclass of CC121 isolates harbors the eta and/or etb gene but lacks the luk-PV locus. These strains commonly cause staphylococcal scalded skin syndrome and bullous impetigo (28, 43), toxin-mediated staphylococcal diseases which are associated with eta or etb in 65 to 100% of the cases (10, 18, 26, 30, 60). One such CC121 clone, characterized by ST123 and spa type t171, is notorious as the “epidemic European fusidic acid-resistant impetigo clone (EEFIC)” (27-29, 42, 44, 52). This suggests that within a given clonal background, i.e., CC121, the virulence gene repertoire can shape the clinical symptoms.
Despite the high degree of selection pressure exerted by the frequent antibiotic treatment of patients with severe skin infections, CC121 strains apparently have not acquired the staphylococcal chromosome cassette mec (SCCmec) (37) (U. Nübel, Robert Koch Institute, Wernigerode, Germany, personal communication), suggesting restraints on resistance gene transfer. However, considering the wide distribution of SCCmec among diverse S. aureus clonal lineages, this might change in the future and CC121 might emerge as highly virulent CA-MRSA.
In our molecular epidemiological study, we further consolidated the role of PVL in furunculosis. Generally, the potent PVL toxin is epidemiologically associated with furunculosis, abscesses, and skin lesions but is absent from isolates causing impetigo, blisters, or staphylococcal scalded skin syndrome (21, 31). In our cohort, 85% of MSSA isolates from patients with furunculosis harbored luk-PV genes. Others have reported luk-PV gene frequencies of from 30 to 97% among isolates causing SSTIs (10, 18, 21, 31, 39). In striking contrast, the prevalence of luk-PV among colonizing S. aureus strains was below 1%, which, again, is in agreement with the findings of previous studies (22, 38). However, it must be emphasized that not all furunculosis-associated strains harbored the PVL gene, implying that additional factors, either host or pathogen derived, affect the development of furuncles.
For example, the SAg gene seb was present significantly more frequently among furunculosis-associated strain than among nasal CC121 strains. However, about half of these CC121 strains were seb negative. Moreover, while PVL was distributed over a broad range of staphylococcal lineages, seb was detected only in CC121, CC1, and ST59 strains. This suggests that SEB might contribute to the disease process in CC121 isolates, but its linkage with furunculosis is much weaker than that of PVL.
The present study highlights the genetic diversity of luk-PV-positive MSSA strains. luk-PV genes were detected in isolates belonging to CC5, CC8, CC22, CC30, CC121, and ST59, confirming and extending the findings from Saxony, Germany, that have been reported by Monecke et al. (39). Similarly, the luk-PV genes were demonstrated in a range of MRSA isolates worldwide, which belonged to 10 different MLST sequence types (STs 1, 5, 8, 22, 30, 59/359, 80/583, 88, 93, and 152) (9, 36, 54). This diversity suggests a dominant role for PVL above that of the clonal background in the pathogenesis of furunculosis. Moreover, it indicates the occurrence of frequent and independent luk-PV gene acquisition events by S. aureus (36, 39). The high degree of genetic mobility of luk-PV can be attributed to its localization on phage of group Sa2int, which transduces PVL genes within the species S. aureus (17, 41). However, there appear to be restrictions in the transfer of phages between lineages, since we and others (almost) never observed phage Sa2int in CC45 strains (17, 39). Phage group Sa2int comprises 11 closely related luk-PV-positive and luk-PV-negative phages (17). In our study, only luk-PV-positive Sa2int phage was closely linked to furunculosis strains, whereas luk-PV-negative phage variants were frequent among colonizing strains. Remarkably, this was also the case when furunculosis-associated and colonizing strains belonged to the same lineage, CC30 and CC8, underlining the very strong association of luk-PV with chronic furunculosis.
The importance of PVL as a virulence factor in S. aureus infections is currently under debate, because studies addressing its role in mouse models of skin and lung infections produced contradictory results (5, 6, 8, 50). This study shows a very tight epidemiological linkage between luk-PV and furunculosis, which can be separated from the genetic background of the S. aureus strain as well as from the gene makeup of the luk-PV-transducing phage. The findings therefore lend support to the notion of a causative role for PVL in human furunculosis.
However, this does not explain the strong overrepresentation of CC121 and CC22 among the furunculosis isolates, which indicates that PVL and the core genome act in synergy, as has been proposed by Fan and coworkers (13). Apart from bacterial virulence factors, the disposition of the host, such as the presence of immune suppression and gene polymorphisms, exposures, and personal hygiene, might play an important role (11, 12).
Our results have consequences for the diagnosis and therapy of S. aureus infections. luk-PV genes are recognized worldwide as markers for epidemic CA-MRSA (36, 49, 54). This study and work by others in recent years have demonstrated that in Europe, SSTIs with luk-PV-positive MSSA strains are common (36, 45). As shown in this study, SSTI-associated strains will, in most cases, be characterized by PVL and/or a permissive genetic background and will exhibit a particular virulence. Thus, PVL-associated symptoms might frequently be encountered, even in settings where CA-MRSA is rare. The implementation of a luk-PV test for the diagnosis of S. aureus infection would therefore provide important information for the treating clinician. Our findings are also relevant for therapy, because we and others have shown that the vast majority of patients with chronic furunculosis (87 to 100%) are nasal carriers of their infecting S. aureus strain (10, 48, 51). To prevent repeated reinfection, the elimination of this S. aureus strain from the whole body as well as the patient's environment should be the aim of therapy for chronic furunculosis (58, 61).
Acknowledgments
This work was supported by grants from the DFG (grants SFB-TR34 and GRK-840) and the BMBF.
We thank Ulrich Nübel for helpful comments on the manuscript.
Footnotes
Published ahead of print on 3 March 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Acton, D. S., M. J. Plat-Sinnige, W. van Wamel, N. de Groot, and A. van Belkum. 2009. Intestinal carriage of Staphylococcus aureus: how does its frequency compare with that of nasal carriage and what is its clinical impact? Eur. J. Clin. Microbiol. Infect. Dis. 28:115-127. [DOI] [PubMed] [Google Scholar]
- 2.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amagai, M., N. Matsuyoshi, Z. H. Wang, C. Andl, and J. R. Stanley. 2000. Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat. Med. 6:1275-1277. [DOI] [PubMed] [Google Scholar]
- 4.Baranovich, T., H. Zaraket, I. I. Shabana, V. Nevzorova, V. Turcutyuicov, and H. Suzuki. 4 August 2009. Molecular characterization and susceptibility of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from hospitals and the community in Vladivostok, Russia. Clin. Microbiol. Infect. [Epub ahead of print.] doi: 10.1111/j.1469-0691.2009.02891.x. [DOI] [PubMed]
- 5.Brown, E. L., O. Dumitrescu, D. Thomas, C. Badiou, E. M. Koers, P. Choudhury, V. Vazquez, J. Etienne, G. Lina, F. Vandenesch, and M. G. Bowden. 2009. The Panton-Valentine leukocidin vaccine protects mice against lung and skin infections caused by Staphylococcus aureus USA300. Clin. Microbiol. Infect. 15:156-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bubeck Wardenburg, J., A. M. Palazzolo-Ballance, M. Otto, O. Schneewind, and F. R. DeLeo. 2008. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J. Infect. Dis. 198:1166-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S. J., H. S. Deshmukh, C. L. Nelson, I. G. Bae, M. E. Stryjewski, J. J. Federspiel, G. T. Tonthat, T. H. Rude, S. L. Barriere, R. Corey, and V. G. Fowler, Jr. 2008. Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J. Clin. Microbiol. 46:678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diep, B. A., A. M. Palazzolo-Ballance, P. Tattevin, L. Basuino, K. R. Braughton, A. R. Whitney, L. Chen, B. N. Kreiswirth, M. Otto, F. R. DeLeo, and H. F. Chambers. 2008. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS One 3:e3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 10.Durupt, F., L. Mayor, M. Bes, M. E. Reverdy, F. Vandenesch, L. Thomas, and J. Etienne. 2007. Prevalence of Staphylococcus aureus toxins and nasal carriage in furuncles and impetigo. Br. J. Dermatol. 157:1161-1167. [DOI] [PubMed] [Google Scholar]
- 11.El-Gilany, A. H., and H. Fathy. 2009. Risk factors of recurrent furunculosis. Dermatol. Online J. 15:16. [PubMed] [Google Scholar]
- 12.Emonts, M., A. G. Uitterlinden, J. L. Nouwen, I. Kardys, M. P. Maat, D. C. Melles, J. Witteman, P. T. Jong, H. A. Verbrugh, A. Hofman, P. W. Hermans, and A. Belkum. 2008. Host polymorphisms in interleukin 4, complement factor H, and C-reactive protein associated with nasal carriage of Staphylococcus aureus and occurrence of boils. J. Infect. Dis. 197:1244-1253. [DOI] [PubMed] [Google Scholar]
- 13.Fan, J., M. Shu, G. Zhang, W. Zhou, Y. Jiang, Y. Zhu, G. Chen, S. J. Peacock, C. Wan, W. Pan, and E. J. Feil. 2009. Biogeography and virulence of Staphylococcus aureus. PLoS One 4:e6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. J. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, J. D., and T. Proft. 2008. The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225:226-243. [DOI] [PubMed] [Google Scholar]
- 16.Gillet, Y., B. Issartel, P. Vanhems, J. C. Fournet, G. Lina, M. Bes, F. Vandenesch, Y. Piemont, N. Brousse, D. Floret, and J. Etienne. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753-759. [DOI] [PubMed] [Google Scholar]
- 17.Goerke, C., R. Pantucek, S. Holtfreter, B. Schulte, M. Zink, D. Grumann, B. M. Broker, J. Doskar, and C. Wolz. 2009. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 191:3462-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gravet, A., P. Couppie, O. Meunier, E. Clyti, B. Moreau, R. Pradinaud, H. Monteil, and G. Prevost. 2001. Staphylococcus aureus isolated in cases of impetigo produces both epidermolysin A or B and LukE-LukD in 78% of 131 retrospective and prospective cases. J. Clin. Microbiol. 39:4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen, D., H. Claus, W. Witte, J. Rothganger, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hay, R. J., and B. Adriaans. 2004. Bacterial infections, p. 1-85. In T. Burn, S. Breathnach, N. Cox, and C. E. Griffiths (ed.), Rook's textbook of dermatology. Blackwell Publishing Company, London, United Kingdom.
- 21.Holmes, A., M. Ganner, S. McGuane, T. L. Pitt, B. D. Cookson, and A. M. Kearns. 2005. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 43:2384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtfreter, S., D. Grumann, M. Schmudde, H. T. Nguyen, P. Eichler, B. Strommenger, K. Kopron, J. Kolata, S. Giedrys-Kalemba, I. Steinmetz, W. Witte, and B. M. Broker. 2007. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 45:2669-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwatsuki, K., O. Yamasaki, S. Morizane, and T. Oono. 2006. Staphylococcal cutaneous infections: invasion, evasion and aggression. J. Dermatol. Sci. 42:203-214. [DOI] [PubMed] [Google Scholar]
- 24.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaneko, J., and Y. Kamio. 2004. Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: structures, pore-forming mechanism, and organization of the genes. Biosci. Biotechnol. Biochem. 68:981-1003. [DOI] [PubMed] [Google Scholar]
- 26.Kanzaki, H., M. Ueda, Y. Morishita, H. Akiyama, J. Arata, and S. Kanzaki. 1997. Producibility of exfoliative toxin and staphylococcal coagulase types of Staphylococcus aureus strains isolated from skin infections and atopic dermatitis. Dermatology 195:6-9. [DOI] [PubMed] [Google Scholar]
- 27.Koning, S., A. van Belkum, S. Snijders, W. van Leeuwen, H. Verbrugh, J. Nouwen, M. Op't Veld, L. W. van Suijlekom-Smit, J. C. van der Wouden, and C. Verduin. 2003. Severity of nonbullous Staphylococcus aureus impetigo in children is associated with strains harboring genetic markers for exfoliative toxin B, Panton-Valentine leukocidin, and the multidrug resistance plasmid pSK41. J. Clin. Microbiol. 41:3017-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen, A. R., R. L. Skov, V. Jarlier, and A. S. Henriksen. 2008. Epidemiological differences between the UK and Ireland versus France in Staphylococcus aureus isolates resistant to fusidic acid from community-acquired skin and soft tissue infections. J. Antimicrob. Chemother. 61:589-594. [DOI] [PubMed] [Google Scholar]
- 29.Laurent, F., A. Tristan, M. Croze, M. Bes, H. Meugnier, G. Lina, F. Vandenesch, and J. Etienne. 2009. Presence of the epidemic European fusidic acid-resistant impetigo clone (EEFIC) of Staphylococcus aureus in France. J. Antimicrob. Chemother. 63:420-421. [DOI] [PubMed] [Google Scholar]
- 30.Lina, G., Y. Gillet, F. Vandenesch, M. E. Jones, D. Floret, and J. Etienne. 1997. Toxin involvement in staphylococcal scalded skin syndrome. Clin. Infect. Dis. 25:1369-1373. [DOI] [PubMed] [Google Scholar]
- 31.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 32.Lindsay, J. A., and M. T. Holden. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12:378-385. [DOI] [PubMed] [Google Scholar]
- 33.Lindsay, J. A., and M. T. Holden. 2006. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics 6:186-201. [DOI] [PubMed] [Google Scholar]
- 34.Melles, D. C., R. F. J. Gorkink, H. A. M. Boelens, S. V. Snijders, J. K. Peeters, M. J. Moorhouse, P. J. van der Spek, W. B. van Leeuwen, G. Simons, H. A. Verbrugh, and A. van Belkum. 2004. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Invest. 114:1732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mertz, D., R. Frei, B. Jaussi, A. Tietz, C. Stebler, U. Fluckiger, and A. F. Widmer. 2007. Throat swabs are necessary to reliably detect carriers of Staphylococcus aureus. Clin. Infect. Dis. 45:475-477. [DOI] [PubMed] [Google Scholar]
- 36.Monecke, S., B. Berger-Bachi, G. Coombs, A. Holmes, I. Kay, A. Kearns, H. J. Linde, F. O'Brien, P. Slickers, and R. Ehricht. 2007. Comparative genomics and DNA array-based genotyping of pandemic Staphylococcus aureus strains encoding Panton-Valentine leukocidin. Clin. Microbiol. Infect. 13:236-249. [DOI] [PubMed] [Google Scholar]
- 37.Monecke, S., R. Ehricht, P. Slickers, H. L. Tan, and G. Coombs. 2009. The molecular epidemiology and evolution of the Panton-Valentine leukocidin-positive, methicillin-resistant Staphylococcus aureus strain USA300 in Western Australia. Clin. Microbiol. Infect. 15:770-776. [DOI] [PubMed] [Google Scholar]
- 38.Monecke, S., C. Luedicke, P. Slickers, and R. Ehricht. 2009. Molecular epidemiology of Staphylococcus aureus in asymptomatic carriers. Eur. J. Clin. Microbiol. Infect. Dis. 28:1159-1165. [DOI] [PubMed] [Google Scholar]
- 39.Monecke, S., P. Slickers, M. J. Ellington, A. M. Kearns, and R. Ehricht. 2007. High diversity of Panton-Valentine leukocidin-positive, methicillin-susceptible isolates of Staphylococcus aureus and implications for the evolution of community-associated methicillin-resistant S. aureus. Clin. Microbiol. Infect. 13:1157-1164. [DOI] [PubMed] [Google Scholar]
- 40.Moran, G. J., A. Krishnadasan, R. J. Gorwitz, G. E. Fosheim, L. K. McDougal, R. B. Carey, D. A. Talan, and the EMERGEncy ID Net Study Group. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666-674. [DOI] [PubMed] [Google Scholar]
- 41.Narita, S., J. Kaneko, J. Chiba, Y. Piemont, S. Jarraud, J. Etienne, and Y. Kamio. 2001. Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, phiSLT. Gene 268:195-206. [DOI] [PubMed] [Google Scholar]
- 42.O'Neill, A. J., A. R. Larsen, A. S. Henriksen, and I. Chopra. 2004. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob. Agents Chemother. 48:3594-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Neill, A. J., A. R. Larsen, R. Skov, A. S. Henriksen, and I. Chopra. 2007. Characterization of the epidemic European fusidic acid-resistant impetigo clone of Staphylococcus aureus. J. Clin. Microbiol. 45:1505-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osterlund, A., T. Eden, B. Olsson-Liljequist, S. Haeggman, and G. Kahlmeter. 2002. Clonal spread among Swedish children of a Staphylococcus aureus strain resistant to fusidic acid. Scand. J. Infect. Dis. 34:729-734. [DOI] [PubMed] [Google Scholar]
- 45.Robert, J., J. Etienne, and X. Bertrand. 2005. Methicillin-resistant Staphylococcus aureus producing Panton-Valentine leukocidin in a retrospective case series from 12 French hospital laboratories, 2000-2003. Clin. Microbiol. Infect. 11:585-587. [DOI] [PubMed] [Google Scholar]
- 46.Schefold, J. C., F. Esposito, C. Storm, D. Heuck, A. Kruger, A. Jorres, W. Witte, and D. Hasper. 2007. Therapy-refractory Panton Valentine leukocidin-positive community-acquired methicillin-sensitive Staphylococcus aureus sepsis with progressive metastatic soft tissue infection: a case report. J. Med. Case Rep. 1:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strommenger, B., C. Kettlitz, T. Weniger, D. Harmsen, A. W. Friedrich, and W. Witte. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toshkova, K., C. Annemuller, O. Akineden, and C. Lammler. 2001. The significance of nasal carriage of Staphylococcus aureus as risk factor for human skin infections. FEMS Microbiol. Lett. 202:17-24. [DOI] [PubMed] [Google Scholar]
- 49.Tristan, A., T. Ferry, G. Durand, O. Dauwalder, M. Bes, G. Lina, F. Vandenesch, and J. Etienne. 2007. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J. Hosp. Infect. 65:105-109. [DOI] [PubMed] [Google Scholar]
- 50.Tseng, C. W., P. Kyme, J. Low, M. A. Rocha, R. Alsabeh, L. G. Miller, M. Otto, M. Arditi, B. A. Diep, V. Nizet, T. M. Doherty, D. O. Beenhouwer, and G. Y. Liu. 2009. Staphylococcus aureus Panton-Valentine leukocidin contributes to inflammation and muscle tissue injury. PLoS One 4:e6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tulloch, L. G. 1954. Nasal carriage in staphylococcal skin infections. Br. Med. J. 2:912-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tveten, Y., A. Jenkins, and B. E. Kristiansen. 2002. A fusidic acid-resistant clone of Staphylococcus aureus associated with impetigo bullosa is spreading in Norway. J. Antimicrob. Chemother. 50:873-876. [DOI] [PubMed] [Google Scholar]
- 53.van Belkum, A., N. J. Verkaik, C. P. de Vogel, H. A. Boelens, J. Verveer, J. L. Nouwen, H. A. Verbrugh, and H. F. Wertheim. 2009. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 199:1820-1826. [DOI] [PubMed] [Google Scholar]
- 54.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vorobieva, V., T. Bazhukova, A. M. Hanssen, D. A. Caugant, N. Semenova, B. C. Haldorsen, G. S. Simonsen, and A. Sundsfjord. 2008. Clinical isolates of Staphylococcus aureus from the Arkhangelsk region, Russia: antimicrobial susceptibility, molecular epidemiology, and distribution of Panton-Valentine leukocidin genes. APMIS 116:877-887. [DOI] [PubMed] [Google Scholar]
- 56.Waldron, D. E., and J. A. Lindsay. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wertheim, H. F., D. C. Melles, M. C. Vos, W. van Leeuwen, A. van Belkum, H. A. Verbrugh, and J. L. Nouwen. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751-762. [DOI] [PubMed] [Google Scholar]
- 58.Wiese-Posselt, M., D. Heuck, A. Draeger, M. Mielke, W. Witte, A. Ammon, and O. Hamouda. 2007. Successful termination of a furunculosis outbreak due to lukS-lukF-positive, methicillin-susceptible Staphylococcus aureus in a German village by stringent decolonization, 2002-2005. Clin. Infect. Dis. 44:e88-e95. [DOI] [PubMed] [Google Scholar]
- 59.Yamaguchi, T., K. Nishifuji, M. Sasaki, Y. Fudaba, M. Aepfelbacher, T. Takata, M. Ohara, H. Komatsuzawa, M. Amagai, and M. Sugai. 2002. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect. Immun. 70:5835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamasaki, O., T. Yamaguchi, M. Sugai, C. Chapuis-Cellier, F. Arnaud, F. Vandenesch, J. Etienne, and G. Lina. 2005. Clinical manifestations of staphylococcal scalded-skin syndrome depend on serotypes of exfoliative toxins. J. Clin. Microbiol. 43:1890-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimakoff, J., V. T. Rosdahl, W. Petersen, and J. Scheibel. 1988. Recurrent staphylococcal furunculosis in families. Scand. J. Infect. Dis. 20:403-405. [DOI] [PubMed] [Google Scholar]