Abstract
Oligonucleotide chips targeting the bacterial internal transcribed spacer region (ITS) of the 16S-23S rRNA gene, which contains genus- and species-specific regions, were developed and evaluated. Forty-three sequences were designed consisting of 1 universal, 3 Gram stain-specific, 9 genus-specific, and 30 species-specific probes. The specificity of the probes was confirmed using bacterial type strains including 54 of 52 species belonging to 18 genera. The performance of the probes was evaluated using 825 consecutive samples that were positive by blood culture in broth medium. Among the 825 clinical specimens, 708 (85.8%) were identified correctly by the oligonucleotide chip. Most (536 isolates, or 75.7%) were identified as staphylococci, Escherichia coli, or Klebsiella pneumoniae. Thirty-seven isolates (4.5%) did not bind to the corresponding specific probes. Most of these also were staphylococci, E. coli, or K. pneumoniae and accounted for 6.3% of total number of the species. Sixty-two specimens (7.5%) did not bind the genus- or species-specific probes because of lack of corresponding specific probes. Among them, Acinetobacter baumannii was the single most frequent isolate (26/62). The oligonucleotide chip was highly specific and sensitive in detecting the causative agents of bacteremia directly from positive blood cultures.
Sepsis (bacteremia) is a serious medical condition caused by microbial pathogens in blood, with a mortality rate as high as 50% (14). Identification of microorganisms in the blood thus usually is indicative of a serious invasive infection necessitating urgent antimicrobial therapy. Sepsis most often is caused by bacteria, with fungal, parasitic, or viral infections being uncommon (5). Various pathogens have different antimicrobial susceptibilities, and successful treatment is dependent on prompt administration of the correct drug (19). Therefore, the most important early intervention in sepsis is quick diagnosis. However, the identification of pure colonies on agar plates after broth culture of blood takes 1 day or more. Clearly, a rapid and accurate method for identification of pathogens is necessary.
Technical improvement in molecular techniques such as PCR, sequencing, and oligonucleotide chip assay has increased the sensitivity, specificity, and speed of assays (2). Oligonucleotide chips have recently become a powerful tool for microbial genotyping and identification of drug resistance associated with mutations and single-nucleotide polymorphisms (12). This method allows thousands of specific DNA sequences to be detected simultaneously (13). In most bacteria, the internal transcribed spacer region (ITS) of the 16S-23S rRNA gene contains genus- and species-specific regions and therefore is useful as the target for species identification (7, 12). In this study, we aimed to detect and identify important bacterial pathogens associated with sepsis directly from blood culture by using an ITS-targeted oligonucleotide chip.
MATERIALS AND METHODS
Design of universal and specific probes and alignment of probes in the oligonucleotide chip.
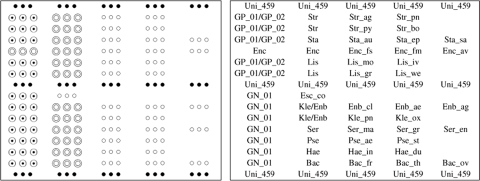
Forty-three sequences were designed consisting of 1 universal, 3 Gram stain-specific, 9 genus-specific, and 30 species-specific probes (Table 1). Genus and species were selected empirically on the basis of isolation frequency in clinical laboratories. The universal probe (Uni-459), which was designed from the sequences of highly conserved regions of the 23S rRNA gene, was used as an internal control to confirm the presence of genomic DNA and the adequacy of PCR. All the other probes were designed from the ITS region between the 16S and 23S rRNA genes using sequences obtained from the GenBank database. Multiple alignment analysis using CLUSTALW was applied to the probe design. The uniqueness of the sequence of each probe was analyzed with a BLAST search. The probes were designed to satisfy the following criteria: the oligonucleotides were between 18 and 27 nucleotides long, and the position of the potential mismatch in similar sequences was close to the center of the probe. The 5′ end of each probe was modified by adding poly(T) sequence and an amino link group to enable covalent immobilizing on the aldehyde-coated glass surface. Figure 1 illustrates the probe array on the oligonucleotide chip. Each of the probes was spotted in triplicate.
TABLE 1.
Probes included on the chip
| Target bacteria | Probe name | Probe sequence (5′ to 3′) | Accession no. (GenBank) or source |
|---|---|---|---|
| All bacteria | Uni_459 | CCGATAGTGAACCAGTACC | |
| Gram-positive bacteria | |||
| Nonspecific | GP_01 | YTTVTTYAGTTTTGAGAGGTY | |
| GP_02 | TGTATTCAGTTTTGAATGTTY | ||
| Enterococcus spp. | Enc | GAGTTTTTTAATAAGTTCAATTG | |
| E. faecalis | Enc_fs | GCTTATTTATTGATTAACC | AY351321 |
| E. faecium | Enc_fm | AGACTACACAATTTGTTTTT | AY351317 |
| E. avium | Enc_av | GGATACAGAAACAATTTTAA | AY116901 |
| Listeria spp. | Lis | CACAAGTAACCGAGAATCATCTG | |
| L. monocytogenes | Lis_mo | CATAGATAATTTATTATTTATGAC | AF514302 |
| L. ivanovii | Lis_iv | CTGTATAACCTATTTAAGGG | U57913 |
| L. grayi | Lis_gr | GAAACTTTCCGCTTTGGAAG | U57918 |
| L. welshimeri | Lis_we | AGAAAACAAAATATTATTTCC | U57917 |
| Staphylococcus spp. | Sta | AGATTTTACCAAGCAAAACCG | |
| S. aureus | Sta_au | AAACGCGTTATTAATCTTGTGA | U11787 |
| S. epidermidis | Sta_ep | TTGAATTCVTAAATAATCGC | U90018 |
| S. saprophyticus | Sta_sa | CTTACGAAGATGCAGGAAT | EU430992 |
| Streptococcus spp. | Str | CATTGAAAATTGAATAWCKA | |
| S. agalactiae | Str_ag | AGGAAACCTGCCATTTGCGTC | AF489592 |
| S. pneumoniae | Str_pn | ATCACCAAGTAATGCACATTG | AY347556 |
| S. pyogenes | Str_py | ACACGTTTATCGTCTTATTTAG | AY347560 |
| S. bovis | Str_bo | GTTTAAGGTCAACAGAACCAA | AY347547 |
| Gram-negative bacteria | |||
| Nonspecific | GN_01 | CTCAGTTGGTTAGAGCGCSMCM | |
| Bacteroides spp. | Bac | GTAGAGGTCGGCAGTTCAAC | |
| B. fragilis | Bac_fr | GAAAAGGAGATGAATCTGGC | AF172709 |
| B. thetaiotaomicron | Bac_th | GGTTAATACCTGATACTT | AF172710 |
| B. ovatus | Bac_ov | CCAGTATGAGAATAAAACGTT | AF176691 |
| Escherichia coli | Esc_co | AAAAAAGAAGCGTWCTTTGMAGTGCTC | AY684796 |
| Enterobacter/Klebsiella spp. | Kle/Enb | TAATGTGTGTTCGAGTCTCT | |
| E. cloacae | Enb_cl | CGAAGGGGACGTACAGTCTCA | AY116915 |
| E. aerogenes | Enb_ae | AAGTAGAGAAGCAAGGGGTC | AF047426 |
| E. agglomerans | Enb_ag | GATACCTTCCCGCGCAGTGTCC | AF041584 |
| K. pneumoniae | Kle_pn | GACGCGTGCCGATGTATC | AF047425 |
| K. oxytoca | Kle_ox | GCTGCGAAGTCGCGACACCT | AY116899 |
| Haemophilus spp. | Hae | CGAATCCCCGTGGGGACGCCA | |
| H. influenzae | Hae_in | GAGAGAAAGTCTGAGTAGGCA | Our laboratory |
| H. ducreyi | Hae_du | AAGTAGAAAGTCTGAGTAATC | Our laboratory |
| Pseudomonas spp. | Pse | ACAGTATAACCAGATTGCTT | |
| P. aeruginosa | Pse_ae | CCATCTAAAACAATCGTCG | AY684792 |
| P. stutzeri | Pse_st | CACGTTATAGACAGTAACC | AJ635307 |
| Serratia spp. | Ser | CCTCCTTACCTAASGATATT | |
| S. marcescens | Ser_ma | GCCACTCGAACTAATATCTT | Our laboratory |
| S. grimesii | Ser_gr | GATATTGATTGCGTGAAGTGC | Our laboratory |
| S. entomophila | Ser_en | CACTTCACTCGAATCAATATC | Our laboratory |
FIG. 1.
Probe layout on oligonucleotide chip. For explanation of the probe names, see Table 1.
QC probes for the oligonucleotide chip.
Quality control (QC) probes were used to confirm the uniformity of the spots. The 6-carboxytetramethylrhodamine (TAMRA)-labeled QC probe [15-mer poly(T)] was modified by an amino link group at the 5′ end. We spotted mixtures of each specific probe and QC probe. All probes were stored in 50-μl volumes at 100 pM/μl concentrations at −20°C for less than 6 months before use. Before hybridization, proper spotting was confirmed by scanning at 532 nm.
Oligonucleotide chip fabrication.
For printing, the probes were dispensed into a 384-well microplate. Each well of the plate contained a mixture of a specific probe and a QC probe (9:1 ratio) in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0). The oligonucleotide probes were spotted on silylated glass slides (CEL Associates, Houston, TX) using a PixSys nQUAD 4500 Microarrayer (Cartesian Technologies, Inc., Irvine, CA). The spotted slides were left for at least 5 h at 50°C to fix the probes to the surface. Slides were then washed once with 0.2% SDS and distilled H2O (dH2O) for 1 min each. To reduce Schiff base formation between the amino groups of the oligonucleotide probes and the aldehyde groups of the glass slide, the slides were treated for 5 min with a freshly prepared 0.25% NaBH4 solution (1.0 g of NaBH4, 300 ml of phosphate-buffered saline [PBS], and 100 ml of absolute ethanol). Slides were washed once in dH2O at 95°C for 2 min and once with 0.2% SDS and dH2O for 1 min each and dried.
Type strains of bacteria and blood culture isolates.
The bacterial strains listed in Table 2 were used to hybridize the oligonucleotide chip, which included 54 strains of 52 species belonging to 18 genera. The type strains were obtained from the American Type Culture Collection (ATCC) or the Korean Collection for Type Cultures (KCTC). All the type strains were used for evaluating the specificity of the probe binding. Additionally, 825 consecutive samples that were positive by blood culture broth medium assay were included, resulting in 825 bacterial species; these were collected from Pusan National University Hospital, Kosin University Gospel Hospital, Inje University Busan Paik Hospital, Ulsan University Hospital, and Gyeongsang National University Hospital (Table 3) by the method described below. Blood culture assays were performed according to each hospital's own protocol using the BacT/Alert 3D system (Organon Teknika, Durham, NC) or the BACTEC 9240 blood culture system (Becton Dickinson Diagnostic Instrument Systems, Sparks, MD). When positive signals appeared, 1 ml of broth medium was drawn from the culture for the oligonucleotide chip assay in addition to routine identification and susceptibility testing. Identification of bacteria was performed by biochemical reactions, the Vitek 2 System (bioMérieux, Durham, NC), or both. Analysis of oligonucleotide chips was performed by persons blinded to the identification results of the blood culture.
TABLE 2.
Organisms tested
| Organism and strain no. | Probe types hybridizeda |
|---|---|
| Acinetobacter baumanii ATCC 19606 | U, N |
| Bacillus cereus KTCC 3711 | U, P |
| Bacteroides fragilis ATCC 25285 | U, N, G, S |
| Bacteroides ovatus ATCC 8483 | U, N, G, S |
| Bacteroides thetaiotaomicron ATCC 29741 | U, N, G, S |
| Bacteroides vulgatus ATCC 29327 | U, N, G |
| Citrobacter freundii ATCC 33940 | U, N |
| Enterobacter aerogenes ATCC 13048 | U, N, G, S |
| Enterobacter agglomerans ATCC 27987 | U, N, G, S |
| Enterobacter cloacae ATCC 13047 | U, N, G, S |
| Enterobacter gergoviae ATCC 33426 | U, N, G |
| Enterobacter sakazakii ATCC 29544 | U, N, G |
| Enterococcus avium ATCC 14025 | U, P, G, S |
| Enterococcus faecalis ATCC 19433 | U, P, G, S |
| Enterococcus faecium ATCC 8043 | U, P, G, S |
| Enterococcus flavescens ATCC 49996 | U, P, G |
| Enterococcus solitarius ATCC 49428 | U, P, G |
| Escherichia coli ATCC 10799 | U, N, S |
| Escherichia coli ATCC 33572 | U, N, S |
| Escherichia coli ATCC 39403 | U, N, S |
| Haemophilus ducreyi ATCC 33940 | U, N, G, S |
| Haemophilus influenzae ATCC 19418 | U, N, G, S |
| Klebsiella oxytoca ATCC 13182 | U, N, G, S |
| Klebsiella planticola ATCC 15380 | U, N, G |
| Klebsiella pneumoniae ATCC 15380 | U, N, G, S |
| Listeria grayi ATCC 19120 | U, P, G, S |
| Listeria innocua ATCC 33090 | U, P, G |
| Listeria ivanovii subsp. ivanovii ATCC 19119 | U, P, G, S |
| Listeria monocytogenes ATCC 19111 | U, P, G, S |
| Listeria welshimeri ATCC 35897 | U, P, G, S |
| Neisseria meningitidis ATCC 13077 | U, N |
| Pseudomonas aeruginosa ATCC 10145 | U, N, G, S |
| Pseudomonas reptilivora ATCC 14878 | U, N, G |
| Pseudomonas resinovorans ATCC 14235 | U, N, G |
| Pseudomonas stutzeri ATCC 17588 | U, N, G, S |
| Salmonella bongori ATCC 43975 | U, N |
| Serratia entomophila ATCC 43705 | U, N, G, S |
| Serratia ficaria ATCC 33105 | U, N, G |
| Serratia grimesii ATCC 14460 | U, N, G, S |
| Serratia marcescens ATCC 13880 | U, N, G, S |
| Serratia odorifera ATCC 33077 | U, N, G |
| Shigella boydii ATCC 8700 | U, N |
| Staphylococcus aureus ATCC 25923 | U, P, G, S |
| Staphylococcus epidermidis ATCC 12228 | U, P, G, S |
| Staphylococcus homminis ATCC 27844 | U, P, G |
| Staphylococcus saprophyticus ATCC 15305 | U, P, G, S |
| Staphylococcus warneri ATCC 35897 | U, P, G |
| Streptococcus agalactiae ATCC 13813 | U, P, G, S |
| Streptococcus bovis ATCC 33317 | U, P, G, S |
| Streptococcus intermedius KCTC 3268 | U, P, G |
| Streptococcus mutans ATCC 25175 | U, P, G |
| Streptococcus pneumoniae ATCC 33400 | U, P, G, S |
| Streptococcus pyogenes ATCC 19615 | U, P, G, S |
| Vibrio parahaemolyticus ATCC 17802 | U, N |
Expected and tested results were the same. U, universal probe; N, Gram-negative probe; P, Gram-positive probe; G, genus-specific probe; S, species-specific probe.
TABLE 3.
Classification of results
| Probe group and organism | Total no. of specimens | No. of specimens by categorya |
|||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Bacteria targeted by specific probe | |||||
| Bacteroides spp. | 2 | 1 | 1 | ||
| Escherichia coli | 164 | 154 | 8 | 2 | |
| Enterobacter aerogenes | 7 | 6 | 1 | ||
| Enterobacter intermedius | 1 | 1 | |||
| Enterobacter cloacae | 19 | 19 | |||
| Enterococcus avium | 1 | 1 | |||
| Enterococcus faecalis | 25 | 25 | |||
| Enterococcus faecium | 33 | 33 | |||
| Enterococcus, other species | 7 | 7 | |||
| Haemophilus influenzae | 1 | 1 | |||
| Klebsiella oxytoca | 3 | 3 | |||
| Klebsiella ozaenae | 1 | 1 | |||
| Klebsiella pneumoniae | 72 | 66 | 5 | 1 | |
| Pseudomonas aeruginosa | 27 | 25 | 2 | ||
| Serratia marcescens | 4 | 4 | |||
| Serratia, other species | 1 | 1 | |||
| Staphylococcus aureus | 77 | 64 | 13 | ||
| Staphylococcus epidermidis | 136 | 136 | |||
| Staphylococcus, other species | 87 | 79 | 8 | ||
| Streptococcus agalactiae | 9 | 9 | |||
| Streptococcus pneumoniae | 31 | 31 | |||
| Streptococcus, other species | 45 | 43 | 2 | ||
| Subtotal for group | 753 | 708 | 37 | 2 | 6 |
| Bacteria not targeted by specific probe | |||||
| Acinetobacter baumannii | 27 | 26 | 1 | ||
| Acinetobacter junii | 1 | 1 | |||
| Aeromonas hydrophila | 5 | 5 | |||
| Aeromonas sobria | 7 | 7 | |||
| Alcaligenes xylosoxidans | 1 | 1 | |||
| Bacillus spp. | 8 | 8 | |||
| Burkhoderia cepacia | 3 | 2 | 1 | ||
| Citrobacter freundii | 6 | 1 | 5 | ||
| Corynebacterium xerosis | 2 | 2 | |||
| Flavobacterium meningosepticum | 3 | 2 | 1 | ||
| Gemella morbillorum | 1 | 1 | |||
| Morganella morganii | 2 | 2 | |||
| Pasteurella hemolytica | 1 | 1 | |||
| Stenotrophomonas maltophilia | 5 | 5 | |||
| Subtotal for group | 72 | 62 | 10 | ||
| Total both groups | 825 | 708 | 99 | 12 | 6 |
For a description of the categories, see Materials and Methods.
DNA extraction and PCR amplification.
DNA was extracted using an InstaGene Matrix Kit (Bio-Rad Laboratories, Richmond, CA) according to the manufacturer's protocol. The DNA of reference strains was extracted from colonies on plated medium, and DNA of clinical isolates was from 1 ml of cultivated broth. For PCR, the universal primers 5′-biotin-GCCTTGTACACWCCGCCC-3′ (forward primer) within the bacterial 16S rRNA gene and 5′-biotin-NAGAACCTGAAACCGTGTGC-3′ (reverse primer) within the bacterial 23S rRNA gene region were used. The PCR amplifications were performed in 25-μl volumes containing 1 U of Taq DNA polymerase (Qiagen Inc., Valencia, CA), 1× Qiagen PCR buffer with 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate (dNTP) mix, 0.02% bovine serum albumin (BSA), and 10 pmol of each primer. Four microliters of sample (extracted DNA) was added to the PCR mixture, and PCR was performed in a PCT 100 thermal cycler (MJ Research, Waltham, MA) as follows: 4 min of denaturation at 94°C, followed by 35 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 50°C, and 1 min of extension at 72°C, with a final extension step of 10 min at 72°C.
Hybridization and scanning.
Biotin-labeled PCR products were denatured for 5 min at 95°C and then chilled on ice for 5 min. Two microliters of single-stranded biotin-labeled target DNA was mixed with 8 μl of hybridization buffer containing 1× SSC, 3 M formamide, salmon sperm DNA, BSA, dH2O, and Cy5-streptavidin (Amersham Pharmacia Biotech, Piscataway, NJ). The oligonucleotide chip was incubated for 30 min at 40°C. After hybridization, the slides were washed for 4 min with 2× SSC and 0.2× SSC at room temperature and dried by centrifugation to remove any remaining solution and reduce the background signal. The fluorescent image of the specific probes was obtained at 635 nm (Cy5), and that of the QC probes was obtained at 532 nm (TAMRA) by a nonconfocal fluorescent scanner (GenePix 4000A; Axon Instruments, Foster City, CA). The signal intensity (SI) of hybridization was analyzed using an intelligent analysis program (CombiView software; GeneIn, Inc., Busan, South Korea).
Sequencing of the bacterial 16S rRNA gene.
After probe hybridization, the result was compared with the blood culture identification result. If the two were the same, the result was classified as category 1 (for definitions, see the next paragraph), and no further study was performed. For all other specimens, the PCR products were subjected to DNA sequencing. For 16S rRNA gene sequencing, the primers 16S-27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 16S-518R (5′-ATTACCGCGGCTGCTGG-3′) were used to obtain approximately 491-bp nucleotides. The amplicons were purified using a QIAquick PCR kit (Qiagen). The sequences of the PCR products were determined using an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA) in the BaseStation-1 DNA fragment analyzer (MJ Research).
Data comparison of blood culture, oligonucleotide chip, and 16S rRNA gene sequencing.
The oligonucleotide chip or sequencing data were compared with the conventional identifications of blood culture isolates. The results were divided into four categories. In category 1, the chip results were the same as the blood culture results. These specimens were not subjected to DNA sequence analysis. In category 2 the results for conventional identification and sequence analysis were the same, but in the chip analysis there was an absence of binding to the genus- or species-specific probes. In category 3 the results for conventional identification and sequence analysis were the same, but in the chip analysis there was nonspecific probe binding to either the genus- or the species-specific probe. In category 4 the results for oligonucleotide chip reaction patterns and sequence analysis were the same, but these differed from the conventional identification.
RESULTS
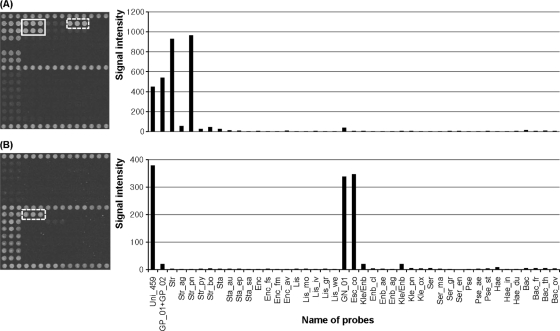
For all 54 type strains used for specific probe binding, the amplified products reacted with the universal probes, the corresponding Gram-positive or -negative probes, and the corresponding genus- and/or species-specific probes with unique hybridization patterns, as expected. For example, Streptococcus pneumoniae was expected to hybridize with the probes Uni_459, GP_01, GP_02, and Str and Str_pn, and the result was as expected. Hybridization signal and intensity results of S. pneumoniae and Escherichia coli are presented in Fig. 2 as examples.
FIG. 2.
Hybridization pattern of clinical isolates with the oligonucleotide chip. (A) S. pneumoniae. (B) E. coli. Hybridization images (left) and each probe's relative signal intensity after hybridization (right) are shown. Solid and dotted boxes in the left images indicate corresponding genus-specific and species-specific probes, respectively.
Among the 825 clinical specimens that were positive by blood culture broth medium assay, 708 (85.8%) were identified correctly by the oligonucleotide chip (correct answer group) (Table 3, category 1,). Most (536, or 75.7%) were identified as staphylococci, E. coli, or Klebsiella pneumoniae. In category 2, there were 99 specimens that did not bind to any of the genus- or species-specific probes, and all of the amplified products proved to be the same species as detected by the blood cultures. Thirty-seven (4.5%) did not bind to the specific probes even though the corresponding probes were included on the chip, suggesting low sensitivity of hybridization (low-sensitivity group). Most of these also were identified as staphylococci, E. coli, or K. pneumoniae and accounted for 6.3% of the total number of species: staphylococci, 21/300 (7.0%); E. coli, 8/164 (4.9%); and K. pneumoniae, 5/72 (6.9%). The other 62 (7.5%) belonged to a species for which genus- or species-specific probes were not included in the current chip format, so no specific probes were present for binding (no probe group). Among them, Acinetobacter baumannii was the single most frequent isolate (26/62). In category 3, 2 specimens for which there were specific probes and 10 for which there were no specific probes bound to the wrong probes, such that a total 1.5% of isolates yielded incorrect results (wrong answer group). In category 4, six specimens (0.7%) showed different results in the culture and chip identifications and were confirmed to have a DNA sequence of the amplified products concordant with the chip reaction (unexplained answer group).
Among 30 bacterial species for which specific probes were included on the chip, 14 (Bacteroides ovatus, Bacteroides thetaiotaomicron, Enterobacter agglomerans, Haemophilus ducreyi, Listeria grayi, Listeria ivanovii, Listeria monocytogenes, Listeria welshimeri, Pseudomonas stutzeri, Staphylococcus saprophyticus, Serratia entomophila, Serratia grimesii, Streptococcus bovis, and Streptococcus pyogenes) were not found among the 825 blood culture isolates, indicating that probes for low-frequency isolates were included.
DISCUSSION
Rapid and correct identification of microorganisms in blood culture is critical for appropriate antibiotic use, eliminating unnecessary antibiotic administration and facilitating early recovery of the patient. Therefore, many automated identification and susceptibility testing systems are available (3, 11). These can identify microorganisms and determine antibiotic susceptibility in hours; however, they require pure colonies, making overnight subculture of positive blood cultures inevitable. In a previous study, the oligonucleotide chip proved to be the most effective way of detecting bacteria in clinical practice (15). Such chips offer the possibility of rapid and accurate diagnosis of a wide range of microorganisms through hybridization (16). In the current study, we evaluated oligonucleotide chip-based identification of microorganisms directly from culture medium and demonstrated that 94% of species for which probes were included were identified correctly in hours. This chip contains most of the species frequently isolated from blood such as coagulase-negative staphylococci and E. coli, as judged by previous reports (9, 10). However, A. baumannii, one of the frequent isolates, could not be identified because specific probes were inadvertently omitted.
One problem implied by the results in category 2 is the sensitivity of the oligonucleotide chip. Among 99 isolates in that category that did not bind to any specific probes, 62 actually had no appropriate specific probes, so this problem may be solved with additional probes. However, the remaining 37 isolates (5%) did not bind to their specific probes even though such probes were present. We assume that the current version of our oligonucleotide chip was not sensitive enough to detect all isolates because these same strains were detected by sequencing. Another problem of the current chip was that it gave us wrong information about 12 isolates (1.5% of the total). One interesting finding was that the culture results were sometimes not consistent with the oligonucleotide chip and the sequencing results. These results might be attributable to erroneous culture identification or to mixed blood cultures. Unfortunately, we could not determine the explanation by reculturing the medium because the stock specimens were used for molecular work.
We chose the probes for the oligonucleotide chip empirically, so most of the frequent isolates from blood were included. Accidentally, however, one important species (A. baumannii) was not included, whereas probes for many rare isolates were. For example, we did not find any references documenting isolation of either H. ducreyi or L. welshimeri from blood. The reason for their inclusion on the chip was that at least one species of the same genus is an important blood pathogen, and, therefore, many other species of the same genus were subjected to probe design and fitting. However, to incorporate a rare pathogen's probe consumes chip space and is costly, so these probes will be eliminated in the next chip design. Another concern in selecting probes was fungi, especially Candida spp. Fungal isolates are obtained in as many as 5% of all bacteremia and fungemia cases, and most of fungal isolates are Candida spp. (1, 8). Furthermore, episodes of fungemia have increased significantly over the years (1). Universal amplification of fungal isolates is different from that of bacteria, so one more PCR amplification is necessary to detect fungi isolated by the chip.
Recently, a small-scale study of DNA microarray-based identification was reported for detecting bacteria and fungi in blood cultures (20). The study targeted mainly the 23S rRNA gene and the 16S-23S intergenic spacer region for bacteria and 18S rRNA gene for fungi. In target DNA amplification, two separated tubes for bacteria and one tube for fungi were used. It reported 93% sensitivity for 50 bacterial and seven fungal species in 112 blood cultures. The authors did not clarify the selection criteria for positive blood cultures. On the other hand, the current study targeted only the 16S-23S intergenic spacer region for bacteria, and a single tube was used for target DNA amplification. And we included 825 consecutively sampled positive blood cultures from five tertiary care hospitals so that the general frequencies of and target probes for blood pathogens could be revealed. Evaluation of our method by consecutive samples demonstrated that single-tube amplification is good enough to detect major bacterial pathogens. For fungal amplification, however, another tube using universal fungal primers would be necessary. Another concern about the microarray-based detection methods, including the current study, is their direct application to blood. “Most bacteremias in adults have a low number of CFU per milliliter (ml) of blood” (6). Therefore, direct application of a PCR-based method to a small amount of blood is not easy. In some cases, however, bacteria can be seen in a Wright-stained blood smear. Also, high levels of bacteremia (more than 1,000 CFU/ml of blood) are detected in some infants (6). Very recently, direct applications of molecular detection methods have been reported (4, 17, 18). Direct application of an oligonucleotide chip for detecting blood pathogens, if possible, will greatly improve the detection of fastidious or slow-growing microorganisms such as Haemophilus, Actinobacillus, Cardiobacterium, Eikenella, and Kingella (HACEK group). Therefore, efforts to extend the applicability of our method directly to blood specimens should be encouraged.
In conclusion, the oligonucleotide chip was highly specific and sensitive in detecting the causative agents of bacteremia directly from positive blood cultures. However, the current format failed to detect some isolates despite inclusion of specific probes, so improvement of the probe design is required. Also, the application of more probes for frequent isolates and extension to fungi will help in diagnosing bloodstream infection rapidly and correctly.
Acknowledgments
This work was supported by a PNU-IGB joint research center grant of Pusan National University from the Ministry of Education Science and Technology.
Footnotes
Published ahead of print on 17 March 2010.
REFERENCES
- 1.Abelson, J. A., T. Moore, D. Bruckner, J. Deville, and K. Nielsen. 2005. Frequency of fungemia in hospitalized pediatric inpatients over 11 years at a tertiary care institution. Pediatrics 116:61-67. [DOI] [PubMed] [Google Scholar]
- 2.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung, J. W., H. S. Jeon, H. Sung, and M. N. Kim. 2009. Evaluation of MicroScan and Phoenix system for rapid identification and susceptibility testing using direct inoculation from positive BACTEC blood culture bottles. Korean J. Lab. Med. 29:25-34. [DOI] [PubMed] [Google Scholar]
- 4.Dierkes, C., B. Ehrenstein, S. Siebig, H. J. Linde, U. Reischl, and B. Salzberger. 2009. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect. Dis. 9:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fluit, A. C., F. J. Schmitz, and J. Verhoef. 2001. Frequency of isolation of pathogens from bloodstream, nosocomial pneumonia, skin and soft tissue, and urinary tract infections occurring in European patients. Eur. J. Clin. Microbiol. Infect. Dis. 20:188-191. [DOI] [PubMed] [Google Scholar]
- 6.Forbes, B. A., D. F. Sahm, and A. S. Weissfeld. 2007. Bailey and Scott's diagnostic microbiology, 12th ed. Mosby, St. Louis, MO.
- 7.Gilbert, G. L. 2002. Molecular diagnostics in infectious diseases and public health microbiology: cottage industry to postgenomics. Trends Mol. Med. 8:280-287. [DOI] [PubMed] [Google Scholar]
- 8.Gray, J., S. Gossain, and K. Morris. 2001. Three-year survey of bacteremia and fungemia in a pediatric intensive care unit. Pediatr. Infect. Dis. J. 20:416-421. [DOI] [PubMed] [Google Scholar]
- 9.Karlowsky, J. A., M. E. Jones, D. C. Draghi, C. Thornsberry, D. F. Sahm, and G. A. Volturo. 2004. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann. Clin. Microbiol. Antimicrob. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh, E. M., S. G. Lee, C. K. Kim, M. Kim, D. Yong, K. Lee, J. M. Kim, D. S. Kim, and Y. Chong. 2007. Microorganisms isolated from blood cultures and their antimicrobial susceptibility patterns at a university hospital during 1994-2003. Korean J. Lab. Med. 27:265-275. [DOI] [PubMed] [Google Scholar]
- 11.Lee, K. K., S. T. Kim, K. S. Hong, H. J. Huh, and S. L. Chae. 2008. Evaluation of the Phoenix automated microbiology system for detecting extended-spectrum beta-lactamase in Escherichia coli, Klebsiella species and Proteus mirabilis. Korean J. Lab. Med. 28:185-190. [DOI] [PubMed] [Google Scholar]
- 12.Park, H., H. Jang, E. Song, C. L. Chang, M. Lee, S. Jeong, J. Park, B. Kang, and C. Kim. 2005. Detection and genotyping of Mycobacterium species from clinical isolates and specimens by oligonucleotide array. J. Clin. Microbiol. 43:1782-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, H., E. J. Song, E. S. Song, E. Y. Lee, C. M. Kim, S. H. Jeong, J. H. Shin, J. Jeong, S. Kim, Y. K. Park, G. H. Bai, and C. L. Chang. 2006. Comparison of a conventional antimicrobial susceptibility assay to an oligonucleotide chip system for detection of drug resistance in Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 44:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reimer, L. G., M. L. Wilson, and M. P. Weinstein. 1997. Update on detection of bacteremia and fungemia. Clin. Microbiol. Rev. 10:444-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang, S., G. Chen, Y. Wu, L. Du, and Z. Zhao. 2005. Rapid diagnosis of bacterial sepsis with PCR amplification and microarray hybridization in 16S rRNA gene. Pediatr. Res. 58:143-148. [DOI] [PubMed] [Google Scholar]
- 16.Soini, H., and J. M. Musser. 2001. Molecular diagnosis of mycobacteria. Clin. Chem. 47:809-814. [PubMed] [Google Scholar]
- 17.von Lilienfeld-Toal, M., L. E. Lehmann, A. D. Raadts, C. Hahn-Ast, K. S. Orlopp, G. Marklein, I. Purr, G. Cook, A. Hoeft, A. Glasmacher, and F. Stuber. 2009. Utility of a commercially available multiplex real-time PCR assay to detect bacterial and fungal pathogens in febrile neutropenia. J. Clin. Microbiol. 47:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallet, F., S. Nseir, L. Baumann, S. Herwegh, B. Sendid, M. Boulo, M. Roussel-Delvallez, A. V. Durocher, and R. J. Courcol. 18 Aug 2009, posting date. Preliminary clinical study using a multiplex real-time PCR test for the detection of bacterial and fungal DNA directly in blood. Clin. Microbiol. Infect. doi: 10.1111/j.1469-0691.2009.02940.x. [DOI] [PubMed]
- 19.Wiesinger-Mayr, H., K. Vierlinger, R. Pichler, A. Kriegner, A. M. Hirschl, E. Presterl, L. Bodrossy, and C. Noehammer. 2007. Identification of human pathogens isolated from blood using microarray hybridisation and signal pattern recognition. BMC Microbiol. 7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo, S. M., J. Y. Choi, J. K. Yun, J. K. Choi, S. Y. Shin, K. Lee, J. M. Kim, and S. Y. Lee. 2010. DNA microarray-based identification of bacterial and fungal pathogens in bloodstream infections. Mol. Cell Probes 24:44-52. [DOI] [PubMed] [Google Scholar]