Abstract
A molecular survey of 16,057 mosquitoes captured in Southwest Germany during the summer of 2009 demonstrated the presence of Sindbis virus (SINV) in Culex spp. and Anopheles maculipennis sensu lato. Phylogenetic analysis of the German SINV strains linked them with Swedish SINV strains, the causative agent of Ockelbo disease in humans.
Sindbis virus (SINV), the prototype virus of the genus Alphavirus within the family Togaviridae, is an arthropod-borne (arbo), single-stranded RNA virus and the most widely distributed of all known arboviruses (16). SINV was first isolated from a pool of 63 Culex pipiens and Culex univittatus mosquitoes collected in the Sindbis health district, near Cairo, in 1952 (15) and was further demonstrated to be the causative agent of a febrile illness associated with maculopapular rash and joint pain in humans in Africa, Eurasia (Ockelbo disease, Pogosta disease, and Karelian fever), and Australia (8, 9, 13). In Europe, the vectors are ornithophilic mosquitoes of the species Culex torrentium, C. pipiens, and Culiseta morsitans, as well as Ochlerotatus spp. and Aedes spp. (5). Birds of the orders Pas-seriformes and Anseriformes are counted among the main hosts of the virus and have been shown to be responsible for the geographic distribution of SINV and the possible introduction of SINV to so-far-uninfested areas (5). Phylogenetic analysis of the nucleotide sequence data of several SINV strains demonstrated two distinct genetic lineages (paleoarctic/Ethiopian and oriental/Australian) (7, 14). It was not known if SINV circulates in Germany, and therefore, the natural vectors of SINV and the geographic distribution of SINV in its vectors have remained unknown as well.
Mosquitoes were trapped from July to September 2009 at three sites in Southwest Germany (Fig. 1). The Kühkopf and Waghäusel trapping sites are in the wildlife sanctuaries at these locations on the Rhine river; they get flooded regularly and were chosen as trapping sites because of the great abundance of mosquitoes and high occurrence of migratory birds. At these two sites, the mosquitoes were trapped with CO2-baited EVS (encephalitis vector surveillance) traps (BioQuip, Compton, CA). The traps were set in the afternoon and retrieved the following morning. A total of 12 traps were used at each site, and they were placed in shaded positions that were sheltered from wind and rain at a distance of about 1.5 m above ground. The Weinheim trapping site is a small garden within the city of Weinheim, which is in one of the warmest parts of Germany. The average temperature during the summer is 19.6°C. The mosquitoes were trapped with gravid traps designed according to the CDC gravid trap model 1712 (John W. Hook Company, Gainesville, FL). The oviposition attractant was a hay infusion (11).
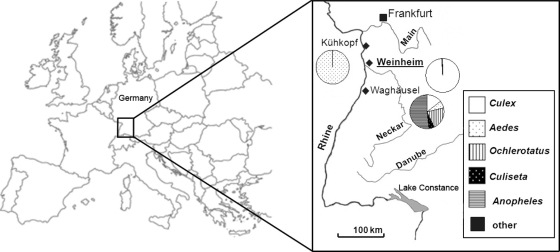
FIG. 1.
Location of the study area (box) in Europe and locations of the study sites, Kühkopf (49°49′N, 8°24′E), Waghäusel (49°15′N, 8°31′E), and Weinheim (49°33′N, 8°40′E). For each trapping site, the proportions of collected mosquitoes are given as a pie chart. Mosquitoes that assayed positive for Sindbis virus were exclusively trapped at Weinheim (in bold and underlined).
Mosquitoes collected at the study sites were frozen at −70°C, transported to the laboratory, and identified on chill tables according to species and sex using morphological characteristics (1). Morphological species determination of pools that tested SINV positive was confirmed by sequence analysis of the internal transcribed spacer 2 region ribosomal DNA according to a previously published protocol (4). After identification, the mosquitoes were pooled (25 specimens) according to species, placed in sterile 2-ml cryovials, and then maintained at −70°C until being assayed for virus. Each mosquito pool was triturated in 500 μl of cell culture medium (high-glucose Dulbecco's modified Eagle's medium [DMEM; Sigma-Aldrich, St. Louis, MO] with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml amphotericin B) with two stainless steel beads (5 mm; Qiagen, Hilden, Germany) in a TissueLyser (Qiagen) for 2 min at 50 oscillation/s. The suspensions were clarified by centrifugation (5,000 × g for 1 min), and the supernatant was used for RNA extraction with a QIAamp viral RNA mini kit according to the manufacturer's protocol (Qiagen). For virus isolation, the supernatant was centrifuged again (10,000 × g for 8 min). Twenty microliters of the clarified supernatant were inoculated into a well of a 48-well plate containing a confluent African green monkey kidney (Vero) cell monolayer (17). Cells were incubated for 1 h at 37°C, and after this, 200 μl of cell culture medium was added and the cells were incubated at 37°C in the presence of 5% CO2 for 1 week and observed daily for evidence of cytopathology. Cell cultures showing a cytopathic effect (CPE) were frozen at −70°C for later virus identification studies. The extracted RNA was analyzed by a newly designed SINV-specific real-time reverse transcription (RT)-PCR using the primers SIND F (5′-CACWCCAAATGACCATGC-3′; nucleotide [nt] position 161 to 178 [the nt positions are given according to the numbering in the SINV reference strain Edsbyn, GenBank accession number M69205]) and SIND R (5′-KGTGCTCGGAAWACATTC-3′; nt position 277 to 294) and probe SIND P (5′-FAM-CAGAGCATTTTCGCATCTGGC-BHQ-1-3′; nt position 185 to 205 [FAM, 6-carboxyfluorescein; BHQ-1, black hole quencher 1]) to target a 134-nucleotide region of the nonstructural protein 1. Real-time RT-PCR was performed with a QuantiTect probe RT-PCR kit according to the manufacturer's protocol (Qiagen).
A total of 16,057 female mosquitoes were collected from July to September 2009. With the CO2-baited EVS traps, 4,111 (25.6%) mosquitoes were trapped at Kühkopf and 7,889 (49.1%) were trapped at Waghäusel. With the gravid traps, 4,057 (25.3%) mosquitoes were trapped in Weinheim. The mosquitoes represented 10 of the 46 known mosquito species in Germany (2). The floodwater mosquito Aedes vexans was the most abundant mosquito (99%) trapped at Kühkopf (Fig. 1). In contrast, the species diversity was greatest at Waghäusel, and the most common mosquitoes were Anopheles claviger (46%) and Ochlerotatus sticticus (14%) (Fig. 1). The mosquitoes most frequently trapped with the gravid traps in Weinheim were Culex spp. (99%) (Fig. 1).
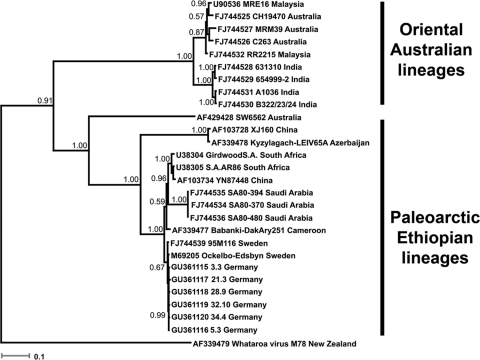
Overall, 643 pools were assayed (cell culture and real-time RT-PCR) for the presence of SINV. Ten SINV RNA-positive pools, all originating from Weinheim, were detected by real-time RT-PCR. Eight pools included mosquitoes of the species Cx. torrentium (pools 3.3, 3.4, 4.2, 4.5, 21.3, 28.9, 32.10, and 34.4), one included mosquitoes of the species Cx. pipiens (pool 3.5), and another one included mosquitoes of the species complex Anopheles maculipennis sensu lato (pool 5.3). Maximum likelihood estimates of mosquito infection rates were calculated using the software program PooledInfRate, version 3.0 (3) and revealed that the highest infection rate (4.9) in the Culex mosquitoes trapped in Weinheim was in the beginning of July (Table 1). Three (pools 3.3, 5.3, and 28.9) of 643 pools caused virus-like CPE in Vero cells after 48 h. All three pools previously tested positive for SINV-specific RNA by real-time RT-PCR, and SINV-specific RNA was also detected by real-time RT-PCR in the supernatants of the infected cell cultures after 5 passages. Moreover, electron microscopy of the infected cell cultures demonstrated enveloped viral particles measuring 65 nm in diameter (data not shown). For extensive phylogenetic analysis, the partial SINV structural polyprotein coding sequences (nt position 8122 to 10696; refers to SINV strain Edsbyn, GenBank accession number M69205) from 6 pools (Fig. 2) were amplified by RT-PCR in 4 overlapping fragments using the primers listed in Table 2. The overall nucleotide identities between the German SINV strains and the Swedish SINV strains were 99.4 to 99.6%. Phylogenetic analysis by Bayesian inference revealed a close relationship of the newly described SINV strains from Germany with the SINV strains (Ockelbo-Edsbyn and 95M116) circulating in Sweden (Fig. 2).
TABLE 1.
Maximum likelihood estimates of infection rates (per 1,000 Culex mosquitoes) of Sindbis virus, found exclusively at the Weinheim trapping site
| Collection time | Infection ratea | 95% CI |
|---|---|---|
| July 1-15 | 4.9 | 1.8-10.8 |
| July 16-31 | 1.4 | 0.1-6.7 |
| August 1-15 | 2.1 | 0.5-5.8 |
| August 16-31 | 0 | 0 |
| September 1-10 | 0 | 0 |
| Total | 2.3 | 1.1-4.2 |
Values were calculated using the software program PooledInfRate per the instructions of the software author (3).
FIG. 2.
Bayesian phylogenetic tree based on partial structural polyprotein nucleotide sequences (length, 2,116 nucleotides) of Sindbis virus strains. For each sequence used, the GenBank accession number, strain designation, and strain origin are shown. Phylogenetic analysis was performed using MrBayes 3.0 (12) with a general time-reversible (GTR) substitution model. Posterior probabilities are shown on each node. Scale bar indicates number of nucleotide substitutions per site. One partial structural polyprotein nucleotide sequence of Whataroa virus is used as an outgroup.
TABLE 2.
Primers used to amplify and sequence the partial Sindbis virus structural polyprotein gene
| Primer | Sequence (5′-3′) | Nucleotide positiona | Amplicon size (nt) | Temp (°C) |
|---|---|---|---|---|
| Sind15F | CCAAGTCGTCAGCATACGACATGGAG | 8122-8147 | 764 | 66 |
| Sind15R | GATGTCATCCATGGTGCCTTCTTT | 8862-8885 | 61 | |
| Sind16F | GACTTCCGCCCAGTTTGGATACGA | 8774-8797 | 768 | 64 |
| Sind16R | GCCCCTAGTCTCCTGGTGGTGAGCA | 9517-9541 | 69 | |
| Sind17F | ACGGCTTTAAACACATCAGCCTCCA | 9469-9493 | 606 | 63 |
| Sind17R | CATTTGGAACAGTGGTCGCATGTTCG | 10073-10098 | 65 | |
| Sind18F | TTAGTGGTTGCCGGCGCCTACCTG | 10032-10055 | 665 | 68 |
| Sind18R | GTCAAGGAGGTAGCTTGAATGTCTCC | 10671-10696 | 65 |
The nucleotide positions are given according to the numbering in the Sindbis virus reference strain Edsbyn (GenBank accession number M69205).
In conclusion, the first molecular survey of SINV in Germany demonstrated the presence of SINV in three different mosquito species (Cx. torrentium, Cx. pipiens, and An. maculipennis sensu lato). The ornithophilic C. torrentium and C. pipiens mosquitoes are the principal vectors for SINV in Sweden (5) and are responsible for hundreds of human SINV infections in Fennoscandia (6, 13). In contrast, the zoophilic An. maculipennis sensu lato was previously not identified as a vector for SINV. In line with the results of a previous study (10), it was demonstrated that the floodwater mosquito Ae. vexans is not a vector for SINV in Germany. The close phylogenetic relationship of German SINV strains with Swedish SINV strains suggests that migratory birds serve as a host for the virus, allowing the dissemination of SINV over large areas within a short period of time. Future studies will investigate the risk of human SINV infections in Germany in areas with high SINV infection rates in mosquitoes.
Footnotes
Published ahead of print on 24 March 2010.
REFERENCES
- 1.Becker, N., D. Petric, M. Zgomba, C. Boase, C. Dahl, J. Lane, and A. Kaiser. 2003. Mosquitoes and their control. 1st ed. Kluwer, New York, NY.
- 2.Becker, N., and A. Kaiser. 1995. Die Culicidenvorkommen in den Rheinauen des Oberrhein-gebiets mit besonderer Berücksichtigung von Uranotaenia (Culicidae, Diptera)—einer neuen Stechmückengattung für Deutschland. Mitt. dtsch. Ges. Allg. Angew. Ent. 10:407-413. (In German.) [Google Scholar]
- 3.Biggerstaff, B. J. 2006. PooledInfRate, version 3.0: a Microsoft Excel add-in to compute prevalence estimates from pooled samples, 1st ed. Centers for Disease Control and Prevention, Fort Collins, CO.
- 4.Collins, F. H., M. A. Mendez, M. O. Rasmussen, P. C. Mehaffey, N. J. Besansky, and V. Finnerty. 1987. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am. J. Trop. Med. Hyg. 37:37-41. [DOI] [PubMed] [Google Scholar]
- 5.Hubalek, Z. 2008. Mosquito-borne viruses in Europe. Parasitology Res. 103:S29-S43. [DOI] [PubMed] [Google Scholar]
- 6.Kurkela, S., T. Manni, A. Vaheri, and O. Vapalahti. 2004. Causative agent of Pogosta disease isolated from blood and skin lesions. Emerg. Infect. Dis. 10:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang, G. D., L. Li, G. L. Zhou, S. H. Fu, Q. P. Li, F. S. Li, H. H. He, Q. Jin, Y. He, B. Q. Chen, and Y. D. Hou. 2000. Isolation and complete nucleotide sequence of a Chinese Sindbis-like virus. J. Gen. Virol. 81:1347-1351. [DOI] [PubMed] [Google Scholar]
- 8.Lvov, D. K., T. M. Skvortsova, L. K. Berezina, V. L. Gromashevsky, B. I. Yakovlev, B. V. Gushchin, V. A. Aristova, G. A. Sidorova, E. L. Gushchina, S. M. Klimenko, S. D. Lvov, N. V. Khutoretskaya, I. A. Myasnikova, and T. M. Khizhnyakova. 1984. Isolation of Karelian fever agent from Aedes communis mosquitoes. Lancet ii:399. [DOI] [PubMed] [Google Scholar]
- 9.McIntosh, B. M., P. G. Jupp, I. Dos Santos, and G. M. Meenehan. 1976. Epidemics of West Nile and Sindbis viruses in South Africa with Culex univittatus Theobold as vector. S. Afr. J. Sci. 72:295. [Google Scholar]
- 10.Modlmaier, M., R. Kuhn, O. R. Kaaden, and M. Pfeffer. 2002. Transmission studies of a European Sindbis virus in the floodwater mosquito Aedes vexans (Diptera: Culicidae). Int. J. Med. Microbiol. 291:164-170. [DOI] [PubMed] [Google Scholar]
- 11.Reiter, P. 1983. A portable, battery-powered trap for collecting gravid culex mosquitos. Mosq. News 43:496-498. [Google Scholar]
- 12.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 13.Skogh, M., and A. Espmark. 1982. Ockelbo disease: epidemic arthritis-exanthema syndrome in Sweden caused by Sindbis-virus like agent. Lancet i:795-796. [DOI] [PubMed] [Google Scholar]
- 14.Sammels, L. M., M. D. Lindsay, M. Poidinger, R. J. Coelen, and J. S. Mackenzie. 1999. Geographic distribution and evolution of Sindbis virus in Australia. J. Gen. Virol. 80:739-748. [DOI] [PubMed] [Google Scholar]
- 15.Taylor, R. M., H. S. Hurlbut, T. H. Work, J. R. Kingston, and T. E. Frothingham. 1955. Sindbis virus: a newly recognized arthropod transmitted virus. Am. J. Trop. Med. Hyg. 4:844-862. [DOI] [PubMed] [Google Scholar]
- 16.Tesh, R. B. 1982. Arthritides caused by mosquito-borne viruses. Annu. Rev. Med. 33:31-40. [DOI] [PubMed] [Google Scholar]
- 17.Turell, M. J., M. L. O'Guinn, J. W. Jones, M. R. Sardelis, D. J. Dohm, D. M. Watts, R. Fernandez, A. Travassos da Rosa, H. Guzman, R. Tesh, C. A. Rossi, V. Ludwig, J. A. Mangiafico, J. Kondig, L. P. Wasieloski, Jr., J. Pecor, M. Zyzak, G. Schoeler, C. N. Mores, C. Calampa, J. S. Lee, and T. A. Klein. 2005. Isolation of viruses from mosquitoes (Diptera: Culicidae) collected in the Amazon Basin region of Peru. J. Med. Entomol. 42:891-898. [DOI] [PubMed] [Google Scholar]