Abstract
The Roco family consists of multidomain Ras-GTPases that include LRRK2, a protein mutated in familial Parkinson's disease. The genome of the cellular slime mold Dictyostelium discoideum encodes 11 Roco proteins. To study the functions of these proteins, we systematically knocked out the roco genes. Previously described functions for GbpC, Pats1, and QkgA (Roco1 to Roco3) were confirmed, while novel developmental defects were identified in roco4- and roco11-null cells. Cells lacking Roco11 form larger fruiting bodies than wild-type cells, while roco4-null cells show strong developmental defects during the transition from mound to fruiting body; prestalk cells produce reduced levels of cellulose, leading to unstable stalks that are unable to properly lift the spore head. Detailed phylogenetic analysis of four slime mold species reveals that QkgA and Roco11 evolved relatively late by duplication of an ancestor roco4 gene (later than ∼300 million years ago), contrary to the situation with other roco genes, which were already present before the split of the common ancestor of D. discoideum and Polysphondylium pallidum (before ∼600 million years ago). Together, our data show that the Dictyostelium Roco proteins serve a surprisingly diverse set of functions and highlight Roco4 as a key protein for proper stalk cell formation.
The Roco protein family is characterized by sharing a conserved core, consisting of a Ras-like GTPase called Roc (Ras of complex proteins) and a COR (C-terminal Of Roc) domain, often with a C-terminal kinase domain and several N-terminal leucine-rich repeats (LRR) (5, 14). The Dictyostelium cyclic GMP (cGMP)-binding protein GbpC was the seed of the family, in combination with 10 other genes encoding Roco proteins in Dictyostelium. Although the family did not draw much attention in the first years after its discovery, this rapidly changed when mutations in the human Roco protein LRRK2 were linked to the development of Parkinson's disease (PD) (3, 15, 17, 22, 28). Since then, most work on Roco proteins has focused on the biological and biochemical characterization of LRRK2 and GbpC. Phosphorylation studies have revealed that pathogenic mutations in LRRK2 lead to an increase in kinase activity and neuronal toxicity (26, 27). Currently, it is not well understood how mutations in LRRK2 exactly lead to the loss of dopaminergic neurons and formation of so-called Lewis bodies, which are characteristic for the development of PD, but recent evidence hints at a role for LRRK2 in the activation of programmed cell death, through activation of caspase-8 (10).
Dictyostelium cells that lack the cGMP-binding protein GbpC show abnormal phosphorylation and assembly of myosin II, which is needed to control the back of the cell during chemotaxis (7). The role of cGMP, and thus GbpC, in chemotaxis becomes even more evident when two other signaling pathways for chemotaxis (PLA2 and PI3K) are inhibited: under these circumstances, cells become solely dependent on the cGMP pathway for chemotaxis toward the chemoattractant cAMP (25). The biochemical properties of GbpC show similarities with those of LRRK2: the Roc domain is also activated upon GDP/GTP exchange, which likely increases kinase activity. Roc activation occurs intramolecularly in GbpC: cGMP binding to the cGMP binding domains causes the RasGEF domain to stimulate activation at the Roc domain (23). It is currently unknown how the Roc domain of LRRK2 is activated. GbpC contains additional domains (GRAM and DEP) that are involved in membrane translocation of the protein.
As highlighted by the work on GbpC, the slime mold Dictyostelium discoideum provides an excellent model system to study the other 10 Roco proteins. All proteins contain the characteristic Roc, COR, and kinase domains, and most also have LRR. In addition, various types of domains are fused to these conserved domains, such as RhoGAP, WD40, PH, DEP, RGS, and Kelch repeats (5). Phylogenetic analysis suggests that all these genes arose from gene duplications after the common ancestor of mammals and Dictyostelium diverged, about 1 billion years ago. (12, 13). To characterize the Roco family in Dictyostelium, we have knocked out all roco genes in this organism. Here, we report on phenotypes of several of these null cells. We show that mutants with deletion of roco genes are defective in cytokinesis and development; most strikingly, a severe defect in stalk stability is found in cells that lack Roco4.
MATERIALS AND METHODS
Strains, cell culture, aggregation assays, and fluorescence microscopy.
Wild-type AX2 cells and all mutant cells were grown at 21°C on petri dishes or in shaking culture to a maximum density of 5 × 106 cells/ml in HL5-C medium including glucose (Formedium). The gbpC-null and qkgA-null cell lines were described before (1, 23). Selection of transformed cells was accomplished using 10 μg/ml blasticidin, whereas cells expressing proteins from extrachromosomal plasmids were grown in the presence of the appropriate selection marker (10 μg/ml G418 or 50 μg/ml hygromycin). To monitor development, cells were harvested and washed twice with phosphate buffer (PB) (11 mM KH2PO4 and 2.8 mM Na2HPO4) and plated at a density of 1 × 106 cells/cm2 on nonnutrient plates containing phosphate buffer and 15 g/liter agar, and pictures were taken at various time points. To visualize localization in slugs, cells expressing green fluorescent protein (GFP) from the roco4 promoter were allowed to aggregate on NN agar and when slugs were formed, pieces of agar were excised and examined using a Zeiss LSM510 confocal fluorescence microscope. For mixing experiments, AX2 cells were labeled with the GFP-expressing plasmid MB74GFP, while roco4-null cells were labeled with the red fluorescent protein (RFP)-expressing plasmid pDM318 (24).
Phylogenetic analyses.
The Dictyostelium discoideum Roco sequences were used to search for the presence of Roco proteins from D. fasciculatum and Polysphondylium pallidum (http://sacgb.fli-leibniz.de/cgi/index.pl) and D. purpureum (http://www.jgi.doe.gov). Some open reading frames from D. fasciculatum and P. pallidum were not yet correctly assembled and annotated, which was detected and corrected by making other selections of intron/exon boundaries, resulting in much improved alignments with the corresponding sequences from the other two dictyostelia. Domain analyses were done using SMART and PFAM, and the alignment of the Roc-COR-kinase supradomains was made using ClustalW. Low-complexity sequences, such as poly-Q or poly-N stretches, were removed from the alignment. The final tree was constructed with Mega 4.1 software by using the maximum parsimony model.
Expression of kinase domains.
The kinase domains of all Roco proteins (except Roco9) were amplified from cDNA using primer sets A (see Table S1 in the supplemental material). Each forward and reverse primer contained an introduced restriction site (for cloning in expression vectors), and the forward primers additionally contained a Kozak sequence and a start codon.
Gene disruptions.
All roco genes (except gbpC, qkgA, and roco9) were disrupted using the DNA fragments encoding the kinase domains. Because no kinase PCR product could be amplified for roco9, the first kilobase of coding sequence was amplified instead for this gene. DNA fragments were cloned in the EcoRV site of pBluescript, digested at unique restriction sites located approximately in the center of the insert, and made blunt using Klenow polymerase, if necessary (see Fig. S1A and Table S2 in the supplemental material). Next, the HincII/SmaI-digested bsr selection cassette (21) was inserted and the resulting plasmid was used as a template to amplify at least 5 μg of an approximately 2-kb knockout construct, using the primer sets A (see Table S1 in the supplemental material). DNA was transfected to AX2 wild-type cells, and after selection with blasticidin, single colonies were analyzed for correct integration sites by PCR (for primer sequences, see Fig. S1C and Table S3 in the supplemental material).
Reverse transcriptase PCR (RT-PCR).
RNA was isolated as described before (18). cDNA was generated using reverse transcriptase (Fermentas) following the manufacturer's protocol. For all roco genes, small fragments in the 5′ region were amplified using forward and reverse primers that anneal on both sides of an intron, to detect possible genomic DNA (gDNA) contamination (for primer sequences, see Table S4 in the supplemental material); only roco6 does not contain introns. IG7 was used as a control, yielding a 370-bp fragment.
DAPI staining.
Cells were grown on coverslips, washed once with phosphate-buffered saline (PBS), and fixed for 15 min in PBS plus 2% paraformaldehyde. After two washes in PBS plus 0.1% Triton X-100 (10 min each), cells were incubated in PBS with DAPI (4′,6-diamidino-2-phenylindole) (1.5 μg/ml, 15 min) and washed once more with PBS. Images were taken using a Zeiss Axiophot microscope with a Plan-NEOFLUAR 40×/0.70 lens.
Cloning of Roco4.
The roco4 open reading frame (ORF) was amplified in three steps from cDNA; the first part (bp 1 to 960) was amplified using the primers Roco4fwA and Roco4rv1; the second part (bp 847 to 3128) was amplified using Roco4fw2 and Roco4rv2; the third part (bp 2942 to 5178) was amplified using Roco4fw3 and Roco4rvA (for primer sequences, see Table S5 in the supplemental material). These fragments were first cloned separately in pBluescript and sequenced to ensure mutation-free DNA. Subsequently, the second part was fused to the first part, using a unique EcoRV site in the roco4 gene and in the multiple cloning site (MCS) from pBluescript. Next, the third part was constructed in this resulting plasmid, using the unique StyI site in roco4 and the MCS, resulting in the complete roco4 open reading frame (plasmid Roco4-pBluescript). This plasmid was subsequently digested with BamHI, and the 5,187-bp fragment was ligated in the BglII site from plasmids MB74GFP (to obtain plasmid Roco4-MB74GFP) and pDM363 (to obtain plasmid Roco4-pDM363). pDM363 was made by replacing the XhoI/BamHI-flanked G418 cassette from pDM323 with a XhoI/BamHI-flanked hygromycin cassette (24). No difference in expression levels or ability to rescue the roco4-null phenotype was found between both plasmids.
Cloning and activity of roco4 promoter.
A 959-bp PCR fragment containing the roco4 promoter was amplified from gDNA using the forward primer CTCGAGACCGGTCAAATAGTGTGGTGCCTGTAAAAC and the reverse primer ACTAGTTTGTGATGAATCCATTTTTTTA (XhoI and BcuI sites, respectively, in bold). After cloning in pBluescript, the promoter was excised with XhoI/BcuI and exchanged with the XhoI/BcuI-flanked actin15 promoter in pDM363. The resulting plasmid contains the roco4 promoter, the first five codons of the roco4 ORF, followed in frame by the gene coding for GFP. Smaller promoter fragments were made using forward primers starting with an XhoI site, followed by the desired promoter sequence, and the same reverse primer as above (for primer sequences, see Table S6 in the supplemental material). To assay promoter activity, equal numbers of cells in phosphate buffer were allowed to settle and aggregate in parallel on six-well plates, at a density of 1 × 106 cells/cm2. At various time points, cells from one well were harvested in PB and proteins were extracted by adding SDS loading buffer and heating for 3 min at 80°C. Equal amounts of total protein were loaded on a 13% SDS-PAGE gel, and Western blot analysis using anti-green fluorescent protein (GFP) antibodies was performed as described before (23). After exposure, the membranes were stripped in stripping buffer (100 mM mercaptoethanol, 62.5 mM Tris, pH 6.8, 2% SDS) for 45 min at 60°C, washed twice with PBS, and blocked again in 5% milk solution. Next, the same Western blot procedure was repeated using an anti-actin antibody (1:10,000 dilution; Santa Cruz).
Cloning of QkgA.
The complete qkgA ORF was constructed using two PCRs; reaction A (encompassing bp 1 to 830) was done on cDNA using the primers QkgAfwA/QkgArv1, while reaction B (encompassing bp 670 to 4659) was done on gDNA using QkgAfw2/QkgArvA (see Table S5 in the supplemental material). Both PCR products were ligated in the same orientation in the EcoRV site of pBluescript and cut with AdeI (which cuts once in the ORF and once in pBluescript), and the obtained fragments of 1,227 and 6,410 bp were ligated, resulting in plasmid QkgA-pBluescript. As a last step, the complete qkgA ORF was excised with BcuI and inserted in the BcuI site of expression plasmid pDM323, yielding a C-terminal GFP fusion to QkgA.
Cloning of Roco11.
The complete roco11 ORF was constructed using a strategy similar to that for qkgA, but for roco11 three PCRs were necessary to obtain the complete ORF; reaction A (encompassing bp 1 to 1654) was done on cDNA using the primers Roco11fwA/Roco11rv1, reaction B (encompassing bp 1483 to 3511) was done on gDNA using Roco11fw2/Roco11rv2, and reaction C (encompassing bp 3302 to 4461) was done on gDNA using Roco11fw3/Roco11rvA (see Table S5 in the supplemental material). After ligation in the same orientation in pBluescript, PCR products A and B were fused using HindIII, which cuts once in the ORF and once in pBluescript. Next, part A/B was fused to part C using SalI/NdeI (which cut in pBluescript and in the ORF, respectively), resulting in the complete roco11 ORF in pBluescript. As a last step, the gene was excised with XbaI and ligated in the compatible BcuI site of pDM323.
Calcofluor staining.
AX2 and roco4-null cells were starved on agar plates as described above. Fruiting bodies of wild-type cells were allowed to gently fall over on the agar surface by carefully tapping the plates. Next, drops of 0.01% (wt/vol) calcofluor white (Sigma) were placed on the fruiting bodies, and the liquid was removed after 10 min. Images were made using a fluorescent microscope (Axiophot; Carl Zeiss MicroImaging, Inc.), and assembly of pictures was done using graphical software.
RESULTS
Phylogenetic analysis of the Dictyostelium Roco family.
The Dictyostelium Roco family was discovered by homology searches with the sequence of GbpC (5). The domain compositions of Roco proteins generally consist of a conserved Roc-COR-kinase core, in addition to several regulatory domains. More extensive searches against the recently updated regular domain databases reveal several additional domains that were not identified before: Roco8 has an additional N-terminal DEP domain, Roco4 contains C-terminal WD40 repeats, and Pats1 contains N-terminal myotubulin-related and catalytic protein tyrosine phosphatase (PTP) domains (Fig. 1A). The structural similarities of the Dictyostelium roco genes with LRRK genes are due to independent acquisitions of distantly related protein kinase domains (12). We also used the conserved Roc-COR-kinase supradomain to search for Roco proteins in Dictyostelium purpureum, Dictyostelium fasciculatum, and Polysphondylium pallidum. Although the nuclear genomes of D. purpureum, D. fasciculatum, and P. pallidum are not completely assembled and annotated yet, we found homologues for all roco genes in all four genomes, not only the homologous Roc-COR-kinase module but also the associated regulatory domains that are typical for a specific gene (data not shown).
Fig. 1.
Overview and phylogeny of the Dictyostelium Roco protein family. (A) Domain organization of the Dictyostelium Roco family. The proteins are characterized by a conserved core that consists of a Roc, COR, and kinase domain, in addition to multiple regulatory domains. Red bars represent places where the genes were disrupted. (B) Phylogenetic tree of the Roc-COR-kinase domain modules of Roco proteins from Dictyostelium discoideum (Dd), Dictyostelium purpureum (Dp), Dictyostelium fasciculatum (Df), and Polysphondylium pallidum (Pp); human (Hs) LRRK2 was used as an outgroup. The tree was constructed with Mega 4.1 software, using an alignment that was created in ClustalW. Bootstrap values of >90 are indicated in the figure. Locus tags for all sequences are listed in Table S7 in the supplemental material. These sequence data were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) and the Jena Centre for Bioinformatics (http://sacgb.fli-leibniz.de/cgi/index.pl), in collaboration with the user community.
To gain more insight into the evolutionary history of the Dictyostelium Roco proteins, we made a detailed phylogenetic analysis of the conserved Roc-COR-kinase supradomain of Roco proteins from Dictyostelium discoideum (Dd), Dictyostelium purpureum (Dp), Dictyostelium fasciculatum (Df), and Polysphondylium pallidum (Pp), using human LRRK2 as an outgroup (Fig. 1B). D. purpureum is closely related to D. discoideum (Dictyostelium taxonomic group 4) (20), while D. fasciculatum (group 1) and P. pallidum (group 2) are more distantly related amoebae. The Roco tree confirms these conclusions, because the closest homologues of all D. discoideum Roco proteins are found in D. purpureum, while the D. fasciculatum and P. pallidum Roco proteins mostly significantly cluster together in a separate branch. The tree also reveals that qkgA, roco4, and roco11 are highly similar. roco4 is present in all four genomes, while qkgA and roco11 are present only in D. discoideum, which suggests that qkgA and roco11 arose from gene duplications of the ancestor roco4 gene. These gene duplications occurred relatively late in evolution, at least after the split of D. discoideum and D. purpureum, which is estimated to have occurred 300 million years ago (20). Not only are the Roc, COR, and kinase domains of these three homologous genes very similar (60 to 80% identity); they also share a large amount of conservation in their entire N termini, including the LRR. Moreover, these N termini do not show significant homology to any other known protein sequence in the NCBI protein sequence database, supporting the conclusion of the described gene duplications. Interestingly, QkgA and Roco11 do not have the WD40 repeats that are present in all Roco4 proteins, suggesting that during or after this duplication, qkgA and roco11 have lost these repeats. Together, we conclude that qkgA and roco11 are duplications from an ancestor roco4 gene that was duplicated late in evolution (later than ∼300 million years ago), contrary to the findings for other roco genes, which were already present before the split of the common ancestor of D. discoideum and P. pallidum (before ∼600 million years ago).
Developmental expression.
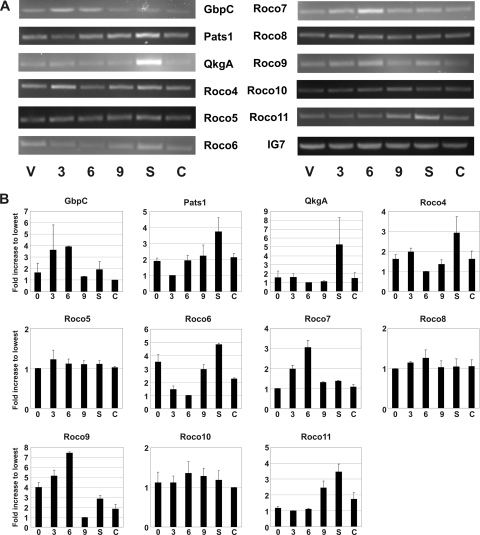
To examine if expression is developmentally regulated, we performed RT-PCR on mRNA for all roco genes (Fig. 2A and 2B); gbpC was shown before to be expressed most highly during aggregation (9), a finding which was confirmed here. No major variations in expression levels were found during development for roco5, roco8, and roco10. In contrast, pats1, qkgA, roco4, roco6, and roco11 show elevated expression levels during the slug phase. roco7 and roco9 are expressed mostly during aggregation, similar to GbpC. Together, the results show that several Dictyostelium roco genes have distinct expression patterns during multicellular development.
Fig. 2.
Developmental expression of roco genes. (A) Wild-type cells were allowed to starve on nutrient-free agar plates, RNA was extracted at various time points during starvation, and RT-PCR was performed on isolated RNA using roco-specific primers (see Table S4 in the supplemental material). IG7 is expressed continuously during development and was therefore used as a positive control. Abbreviations: V, vegetative state; S, slug phase; C, culmination phase. Numbers refer to hours of starvation. (B) Quantification of the data from panel A. The results shown are the means ± SD of results from two or three experiments. Data were quantified using ImageJ software and normalized against the lowest value for each gene.
Gene disruptions.
In an initial study to attribute functions to members of the Dictyostelium Roco family, all Roco kinase domains (except Roco9) were overexpressed in wild-type AX2 cells. No obvious defects were found in cell proliferation, cell division, or development. Also, expression of these domains fused to GFP revealed solely cytosolic distributions (data not shown). We also expressed GFP fusions of several regulatory domains from various Roco proteins in wild-type cells, such as the WD40 repeats of Roco4 and Roco7, the RhoGEF-PH module of Roco5, the WD40-PH-WD40 module of Roco6, the DEP-DEP module of Roco8, the Kelch-RGS-Kelch module of Roco10, and the RhoGAP domain of Roco9. Also here, no distinct phenotypes were found and all proteins showed a cytosolic distribution (data not shown). To extend the search for phenotypes, we disrupted all roco genes that are encoded in the Dictyostelium genome by taking advantage of the previously amplified kinase domains and an N-terminal sequence of roco9. A bsr cassette was inserted in all genes, and clones containing correct integration sites were identified by PCR; primers flanking the knockout constructs were designed to distinguish between clones containing a correct integration in the gene and integration elsewhere in the genome (see Fig. S1A to C in the supplemental material). Because qkgA was already disrupted before in the AX2 background (1), while gbpC was disrupted in our lab before (23), these genes were not knocked out again during this study. The only other previously described roco knockout is pats1 (2), but because this gene was disrupted in the distant DH1 wild-type cell line, we made this knockout again in AX2 for better comparison.
Development of roco-null cells.
To study functions during development, wild-type AX2 and all roco-null cells were subjected to starvation on nutrient-free agar plates, and development was followed over time. All cell lines with disrupted roco genes were able to aggregate and form mounds like wild-type cells (data not shown). The only found aggregation defect was that qkgA-null cells were consistently delayed (about 2 to 3 h) in the initiation of stream formation. However, this delay was caught up with wild-type cells in the later stages of development, and mature fruiting bodies were formed with approximately the same timing as AX2 cells. All knockout cell lines (except roco4-null; see below) were able to form slugs and fruiting bodies with timing similar to that of AX2. Morphology defects were detected in roco11-null cells by the end of the differentiation process, as these cells develop significantly larger fruiting bodies; in particular, the final structures have longer stalks than those of wild-type cells, while the sizes of the spore heads appeared similar. Expression of Roco11 in roco11-null cells from an extrachromosomal plasmid rescues this defect (Fig. 3A). Together, the results show that disruption of roco genes leads to various developmental defects, although these defects are (except for roco4; see below) mostly mild.
Fig. 3.
Phenotypes of qkgA-null, roco11-null, and pats1-null cells. (A) roco11-null cells produce fruiting bodies with longer stalks than those of wild-type cells. AX2, roco11-null cells, and roco11-null cells expressing Roco11 from an extrachromosomal plasmid were starved on nutrient-free agar and allowed to develop. After culmination, pieces of agar were excised and fruiting bodies were photographed from the side. (B) QkgA expression regulates cell proliferation. Doubling times were calculated from three independent growth curves; average values ± SD of results for exponentially growing cells are presented. Data were analyzed with paired Student's t test; *, significantly less than AX2 at P < 0.05; **, significantly more than AX2 at P < 0.05. (C) pats1-null cells have a cytokinesis defect on a solid support. AX2 and pats1-null cells were grown on plates and in shaking culture and photographed. (D) Quantification of the number of nuclei per cell. Cells were stained with DAPI, and the amount of nuclei per cell was counted. Data shown are from 616 AX2 and 335 pats1-null cells.
Role of QkgA and Pats1.
All roco-null cell lines were examined for possible growth defects. Doubling times in shaking cultures were not significantly affected compared to those of wild-type cells, except for those of qkgA-null cells. These cells were reported before to grow slightly faster in shaking culture (1), which was reproduced here; in shaking conditions, wild-type AX2 cells had an average doubling time (Td) of 10.0 ± 0.8 h (mean ± standard deviation [SD]) (n = 3), while qkgA-null cells grew consistently faster (Td = 9.5 ± 0.9 h; Fig. 3B). To confirm this role for QkgA in cell proliferation, we also overexpressed QkgA-GFP in qkgA-null cells and AX2 cells and found doubling times of these cell lines to be 10.9 ± 0.5 h and 11.7 ± 0.6 h, respectively (Fig. 3B). These data suggest that larger amounts of QkgA lead to slower cell proliferation, thus confirming a role for QkgA in this process.
In a previous study, pats1 was disrupted in DH1 cells, resulting in large multinuclear cells in shaking culture, but these cells divided normally when grown on plates (2). We observed all roco knockout cell lines and found only pats1-null cells to have cytokinesis defects. Remarkably, these cells showed large multinuclear cells when grown on plates but not in shaking culture (Fig. 3C), a result which is opposite from that for pats1-null cells that were created in a DH1 background. To further compare both cell lines, we expressed the kinase domain of Pats1. In pats1/DH1 cells, this is reported to rescue the phenotype of pats1-null cells, whereas overexpression in DH1 resulted in large multinucleated cells again (2). In our cells, expression of the kinase domain alone was insufficient to rescue the pats1-null phenotype, and it did not result in a cytokinesis defect in wild-type cells (data not shown). Cell nuclei were visualized using DAPI staining (see Fig. S2 in the supplemental material), and we determined the number of nuclei in these cells (Fig. 3D). AX2 appeared mostly as mononucleated cells (87%), and a small fraction of the cells had two (12%) or three (1%) nuclei. In contrast, only 50% of the pats1-null cells were mononucleated and 10% of the cells contained five or even more nuclei (Fig. 3D). Together, the results suggest that Pats1 has an important role in cytokinesis, but the division mechanism involved might vary among different wild-type strains.
Phenotype of roco4-null cells.
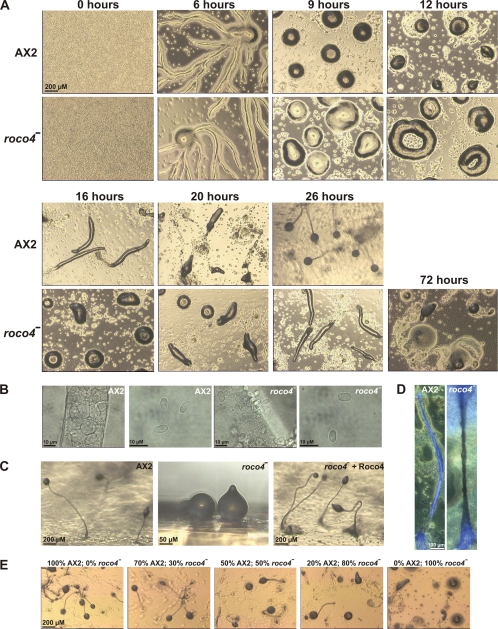
During development, both roco4-null and AX2 cells start to aggregate and form characteristic streams after 6 h of starvation. After 9 h, aggregation is complete and both cell lines have formed mounds, although the mounds of roco4-null cells are somewhat more “loose” than those of AX2. From here on, major phenotypical differences appear (Fig. 4A): after 12 h, wild-type cells are at the onset of forming slugs and first fingers, while roco4-null mounds have mostly transformed into circular, doughnut-shaped structures that last for about 1 to 4 h. When starved on bacterial plates, these circular forms sometimes appear for up to 10 h. After 16 h of starvation, some of the roco4-null mounds slowly form first fingers, to develop into slugs, while most mounds have transformed to slugs only after 26 h. These slugs migrate for many hours before making multiple attempts to culminate, a process that sometimes takes up to 72 h after the onset of starvation. Eventually, this aberrant culmination results in fruiting bodies consisting of spore heads that are located on the agar surface, because a proper stalk is not present to lift the spore head into the air (Fig. 4A to C). When starved at high cell densities, these spore heads often break open, thus spreading the spores on the surface; these spores have normal morphology and are viable (Fig. 4B). We observed improved development when the plates were incubated with the lid down; the fruiting bodies collapse when such plates are carefully rotated lid up, suggesting that the formation and stability of a hanging fruiting body are better than those of an erecting fruiting body. A delicate stalk of roco4-null cells that can better cope with pulling stress than with compressive stress may explain this unusual effect of gravity. Close inspection of the stalks revealed that the roco4-null stalk cells, but not the spores, have a different morphology than wild-type cells (Fig. 4B): roco4-null stalk cells are smaller and pile up in an unordered structure. Stalk cell induction by differentiation-inducing factor 1 (DIF) in vitro (11) appeared to be indistinguishable from that of wild-type cells (see Fig. S3 in the supplemental material). To determine the cause of the inability of roco4-null stalks to raise the spore head in the air, we stained stalks with calcofluor (Fig. 4D). This compound stains cellulose, which is present in large amounts in stalks to provide stability (4, 11). Usually, wild-type stalks stain well over the entire length, but they may contain a small area just above the basal disc with reduced staining. The stalks of roco4-null cells exhibit good staining of the basal disk and the part of the stalk that is located inside the spore head. However, the entire region of the stalk in between basal disk and spore head is not stained. The absence of cellulose, together with the observation that stalk cells in this region of the stalk are arranged in an irregular pattern, may explain why these stalks are not firm enough to keep up a spore head in the air.
Fig. 4.
Phenotypes of roco4-null cells. (A) Development of roco4-null. Wild-type AX2 and roco4-null cells were plated on nutrient-free agar and allowed to develop. Photographs were taken at various time points. At the end of the mound phase, roco4-null cells transform into circular structures that take hours to form slugs. These slugs finally form spores and aberrant stalks, resulting in spore heads that are located at the surface instead of in the air. (B) Close-up pictures of stalk and spore cells, respectively, from wild-type and roco4-null cells. (C) Rescue of aberrant roco4-null fruiting body morphology by reexpression of Roco4. Cells were allowed to develop for 48 h (plates lid up), and pieces of agar were excised and photographed from the side. (D) roco4-null stalks produce little cellulose. Wild-type and roco4-null cells were allowed to develop on nutrient-free agar upside down (plates lid down). After fruiting body formation, plates were turned, causing roco4-null fruiting bodies to spontaneously fall over on the agar; wild-type fruiting bodies were allowed to fall over by gentle tapping of the plate. Fruiting bodies were stained with 0.01% calcofluor to visualize cellulose production. Fluorescent and bright-light pictures were taken simultaneously and assembled afterwards using assembly software. (E) Fruiting body formation of wild-type and roco4-null chimeras. AX2 and roco4-null cells were mixed in various compositions and allowed to develop on nutrient-free agar. Photographs are shown of final structures of these chimeras at 48 h after the start of starvation. The number of fruiting bodies with wild-type-like morphology gradually diminishes at higher percentages of roco4-null cells, suggesting that the developmental defect in these cells is cell autonomous.
Reexpression of Roco4 from a constitutive actin15 promoter in roco4-null cells rescues the roco4-null phenotype, although some of the resulting fruiting bodies have slightly smaller stalks (Fig. 4C). This difference might be due to the constitutive expression of Roco4 in all cells instead of their being expressed after aggregation and in the proper cell type. Roco4 (fused to GFP) showed a uniform cytosolic distribution in the cell, and overexpression of Roco4 in wild-type cells did not lead to a recognizable phenotype (data not shown). To determine whether the developmental defect of roco4-null cells is cell autonomous, roco4-null cells were mixed with wild-type cells and fruiting body formation was monitored (Fig. 4E). The chimeric slugs had the phenotype of roco4-null cells with up to 20% wild-type cells in the mixture and had problems forming fruiting bodies up to a proportion of 50% wild-type cells, while normal fruiting bodies were formed at more than 70% wild-type cells in the mixture.
Expression and cell-type-specific localization of Roco4.
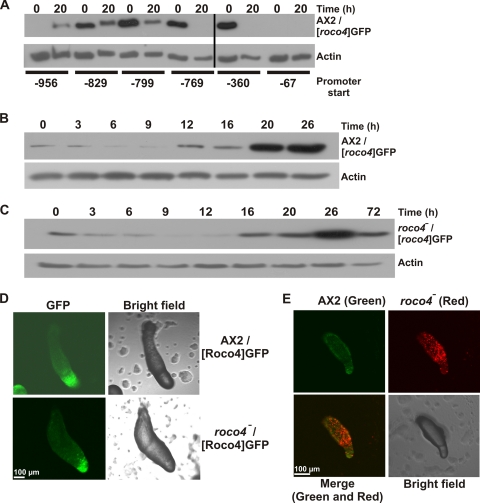
RT-PCR analysis suggested that expression of Roco4 is upregulated during the later stages of development, consistent with the observed phenotype of roco4-null cells (Fig. 2). To confirm this upregulation, protein expression was also analyzed by developmentally regulated expression of GFP from the roco4 promoter. Because most regulatory promoter elements in Dictyostelium are within the first ∼1,000 bp upstream of the start codon, we replaced the actin15 promoter from a GFP expression plasmid with 956-bp promoter sequences upstream of the roco4 ORF (see Fig. S4 in the supplemental material) and examined the expression of GFP from this promoter sequence in AX2 cells by extracting proteins from cells at the onset of starvation and after 20 h, representing the late slug phase (Fig. 5A). Computational analysis from the Dictybase center predicted a possible regulatory element (TCATTCACTCA) at position −783. Therefore, the promoter analysis included this hypothetical element by testing sequences that start just before and just after this sequence. A promoter starting at −67 yielded no detectable expression of GFP. Promoters starting at −360 and −769 showed expression in the vegetative state, but this expression was lost during the developmental cycle. Promoter sequences that included the predicted regulatory element (start at −799 or further upstream), however, were also active during development, suggesting that the regulatory element could serve as an activation sequence during development. The promoter sequence starting at −956 yielded somewhat lower expression levels but showed the expected increase during later development. Apparently the region between −956 and −799 contains an element that inhibits expression during early development. The promoter sequence starting at −956 yielded the expression profile that was also observed during RT-PCR experiments for the endogenous gene. Therefore, we expected this sequence to represent the complete roco4 promoter, and it was used for more extensive analysis to examine expression of Roco4 during development. Similar experiments with higher degrees of time resolution revealed an increase in expression around 12 h after starvation (representing the late mound stage), which further increased during the late slug phase (Fig. 5B). When the experiments were done using roco4-null cells, a similar expression profile was observed but with a 4- to 6-h delay, consistent with the roco4-null phenotype (Fig. 5C).
Fig. 5.
Expression of GFP from the roco4 promoter. (A) Promoter activity analysis for roco4. Expression of GFP from the putative roco4 promoter with increasing size was analyzed in developing wild-type cells. Proteins were extracted at the start and after 20 h starvation and subjected to Western blotting, using anti-GFP antibody. (B and C) Analysis of roco4 promoter activity during development in wild-type AX2 and roco4-null cells, respectively. GFP was expressed from the roco4 promoter starting at −956 bp upstream of the ATG start site. Cells were allowed to develop on nutrient-free agar, harvested at various time points, and analyzed for GFP expression using Western blotting with an anti-GFP antibody. Expression in AX2 increases after 12 h and peaks at the late-slug phase. Expression is delayed in roco4-null cells but also peaks in the later stages of development. (D) Localization of GFP, expressed from the roco4 promoter, in slugs from AX2 and roco4-null cells. For both cell lines, localization is enriched in the anterior part of the slugs, representing the prestalk cell fraction. (E) Cell sorting in AX2/roco4-null chimeras. A 95% portion of GFP-labeled wild-type cells were mixed with 5% RFP-labeled roco4-null cells and allowed to develop. Confocal pictures were taken using both colors and show that roco4-null cells sort out almost exclusively to the posterior part of the slug, representing the prespore fraction.
To examine cell-type-specific expression of Roco4, we analyzed slugs expressing GFP from the roco4 promoter (Fig. 5D). Expression was highly enriched in the anterior of slugs of both AX2 and roco4-null cells, representing the prestalk cell fraction. These observations suggest a specific role for Roco4 in prestalk cells and also that cell sorting occurs correctly in roco4-null cells. To further confirm a role for Roco4 in prestalk cells, 95% green-labeled wild-type cells were mixed with 5% red-labeled roco4-null cells and the distribution of both cell lines in slugs was analyzed (Fig. 5E). The results show that roco4-null cells are almost completely excluded from the prestalk cell zone, suggesting that these cells are not able to develop into prestalk cells in the amount of time that wild-type cells need for this process.
DISCUSSION
The Dictyostelium genome encodes 11 roco genes (that are all being expressed, according to our RT-PCR results), many more than any other sequenced genome to date, suggesting that this slime mold may be a suitable model system to study Roco proteins. This was previously highlighted in biochemical and functional studies involving the cGMP-binding protein GbpC, Pats1, and QkgA, although these latter two proteins were not yet recognized as Roco proteins at the time of publication (5; reviewed in reference 14). To further extend our knowledge about Dictyostelium Roco proteins, we systematically disrupted all remaining roco genes and searched for recognizable phenotypes. Cells that lack Pats1 have a cytokinesis defect when grown on a solid plate, but these cells divide without problems in shaking conditions. Previously, a pats1 knockout was made in a different wild-type strain (DH1), and these cells showed a cytokinesis defect during shaking conditions but not on a solid surface (2). Although the exact reason for this apparent discrepancy remains unknown, it is obvious that Pats1 has an important role during cell division in Dictyostelium.
GbpC was shown before to function in chemotaxis and aggregation of Dictyostelium cells (6, 7, 25). In our present work, we studied the role of the remaining Roco proteins during development, which resulted in the recognition of mild developmental phenotypes in qkgA-null and roco11-null cells and a strong phenotype in roco4-null cells. Cells that lack Roco11 form larger fruiting bodies without initially forming larger aggregation centers. A defect during late differentiation is consistent with RT-PCR data that show an increase in Roco11 expression during the multicellular stages of development. qkgA-null cells have a 2- to 3-h delay in the initiation of aggregation, which hints at a function of QkgA in early cell sensing or regulation of other proteins that are involved in this process. QkgA also has a nondevelopmental function: qkgA-null cells proliferate faster than wild-type cells, while overexpression leads to a growth delay. Together, these two functions could indicate a general role for QkgA in cell sensing: absence of this protein causes hyperactive growth because the cell does not sense the presence of other cells that secrete growth factors to reduce proliferation speed. Similar factors are not sensed during the initial stages of development, thus causing a delay in developmentally regulated gene expression.
The expression of several Roco proteins increases during development. However, apart from Roco4, no strong phenotypes could be recognized in cells that lack or overexpress (part of) these proteins, suggesting less pronounced roles during development. Cells that lack Roco4 have a strong developmental phenotype; these cells aggregate normally, but defects occur during the transition from mounds to slugs. First, the mounds form doughnut-shaped structures before entering the delayed slug phase. Doughnut-shaped structures were also observed in cells that express constitutively activated Gα1-G45V (8). Later, it was demonstrated that ring formation in these Gα1-G45V cells is a result from spiral cAMP waves that do not evolve to a scroll-organizing center in the tip but instead transform into a circularly closed scroll ring wave. During further development, the doughnut increases in diameter and the twisted scroll wave converts into a train of planar waves, resulting in periodic rotational cell movement (19). Temporarily increasing ring diameters of the doughnut were also observed in roco4-null cells (in particular during development after growth on bacterial plates), and it could well be that the underlying mechanism of ring formation is similar to that in the Gα1-G45V mutant. However, it seems unlikely that Roco4 is a direct target of active Gα1: cells expressing constitutively active Gα1 are able to form small, thick-based fruiting bodies, a finding which is different from findings for roco4-null cells. Moreover, the Gα1-G45V mutant was reported to have reduced cAMP-stimulated activation of guanylyl cyclases and severely reduced activation of adenylyl cyclases. Similar experiments with roco4-null cells revealed wild-type-like activation of guanylyl cyclase and overactivation of adenylyl cyclase, a result which is opposite from that found for the Gα1-G45V mutant (our unpublished results). The strongest defect in roco4-null cells is observed during the transition from slug to fruiting body; badly differentiated stalks are formed, resulting in fruiting bodies that have their spore heads located on the surface. This function for Roco4 in stalk cells was further confirmed by the observation that the protein is expressed maximally during late development, as judged from RT-PCR analysis and GFP expression from the roco4 promoter. Furthermore, promoter activity analysis shows that the protein is expressed mainly in prestalk cells. In roco4-null cells, the roco4 promoter shows normal spatial and nearly normal temporal regulation in developing slugs, indicating that Roco4 is not required for induction of its own expression during development or for correct cell sorting. DIF induces terminal stalk cell differentiation in vitro, consistent with the notion that cells are vacuolized in the stalk of roco4-null fruiting bodies. Cellulose is known as a compound to give stability to the stalk in Dictyostelium. Using calcofluor staining, we found that roco4-null stalks have severely reduced cellulose levels, especially in the aereal part of the stalk between the basal disk and the sorocarp. In addition, the cells in this region of the stalk are arranged in an irregular pattern. The resulting stalk is very fragile and difficult to lift from the agar. In wild-type cells, gravitation has little effect on fruiting body formation (16). In roco4-null cells, we observed by putting the agar plates upside down that morphogenesis of a hanging fruiting body is very much improved, even though stalks are defective in cellulose. However, when these plates are very gently turned, the roco4-null fruiting bodies collapse en masse, suggesting that the stalks lack the cement that is necessary for stability. We believe that these observations explain the roco4-null fruiting body phenotype: Roco4 is a prestalk-specific protein involved in proper production of cellulose. roco4-null cells exhibit good cell-type specific differentiation and morphogenetic movement, but they form a stalk with defective mechanical properties that cannot lift the sorocarp.
Using extensive phylogenetic analyses in various dictyostelia, we have been able to answer some intriguing questions regarding the evolutionary history of the 11 roco genes in the genome from D. discoideum. We found that nine of these genes were already present in the common ancestor of all dictyostelia (more than 600 million years ago), while qkgA and roco11 are unique to D. discoideum, as these duplicated relatively recently (less than 300 million years ago) from the roco4 gene. Consistent with this, we also found that the extensive incorporation of associated domains at the N- or C-terminal part of the LRR-Roc-COR-kinase supradomain has occurred before the split of the dictyostelia, since almost all domains that are found in the D. discoideum roco genes are also present in the corresponding genes in other dictyostelia. The public release of genomic data from species close to the dictyostelia should lead to even more accurate annotation of the phylogenetic history of the roco genes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tsuyoshi Araki and Jeff Williams for sharing the qkgA-null cell line, Ineke Keizer-Gunnink for assistance with the convocal experiments, and Rob Kay and Pauline Schaap for helpful discussions. We would also like to thank the Jena Centre for Bioinformatics and the Joint Genome Institute for sharing genomic sequences from various dictyostelia with the public community.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Abe T., Langenick J., Williams J. G. 2003. Rapid generation of gene disruption constructs by in vitro transposition and identification of a Dictyostelium protein kinase that regulates its rate of growth and development. Nucleic Acids Res. 31:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abysalh J. C., Kuchnicki L. L., Larochelle D. A. 2003. The identification of pats1, a novel gene locus required for cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell 14:14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg D., Schweitzer K., Leitner P., Zimprich A., Lichtner P., Belcredi P., Brussel T., Schulte C., Maass S., Nagele T. 2005. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson's disease. Brain 128:3000–3011 [DOI] [PubMed] [Google Scholar]
- 4.Blanton R. L., Fuller D., Iranfar N., Grimson M. J., Loomis W. F. 2000. The cellulose synthase gene of Dictyostelium. Proc. Natl. Acad. Sci. U. S. A. 97:2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosgraaf L., van Haastert P. J. 2003. Roc, a Ras/GTPase domain in complex proteins. Biochim. Biophys. Acta 1643:5–10 [DOI] [PubMed] [Google Scholar]
- 6.Bosgraaf L., Waijer A., Engel R., Visser A. J., Wessels D., Soll D., van Haastert P. J. 2005. RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J. Cell Sci. 118:1899–1910 [DOI] [PubMed] [Google Scholar]
- 7.Bosgraaf L., Russcher H., Smith J. L., Wessels D., Soll D. R., van Haastert P. J. 2002. A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 21:4560–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dharmawardhane S., Cubitt A. B., Clark A. M., Firtel R. A. 1994. Regulatory role of the G alpha 1 subunit in controlling cellular morphogenesis in Dictyostelium. Development 120:3549–3561 [DOI] [PubMed] [Google Scholar]
- 9.Goldberg J. M., Bosgraaf L., van Haastert P. J., Smith J. L. 2002. Identification of four candidate cGMP targets in Dictyostelium. Proc. Natl. Acad. Sci. U. S. A. 99:6749–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho C. C., Rideout H. J., Ribe E., Troy C. M., Dauer W. T. 2009. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J. Neurosci. 29:1011–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levraud J. P., Adam M., Luciani M. F., de Chastellier C., Blanton R. L., Golstein P. 2003. Dictyostelium cell death: early emergence and demise of highly polarized paddle cells. J. Cell Biol. 160:1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marín I. 2006. The Parkinson disease gene LRRK2: evolutionary and structural insights. Mol. Biol. Evol. 23:2423–2433 [DOI] [PubMed] [Google Scholar]
- 13.Marín I. 2008. Ancient origin of the Parkinson disease gene LRRK2. J. Mol. Evol. 67:41–50 [DOI] [PubMed] [Google Scholar]
- 14.Marin I., van Egmond W. N., van Haastert P. J. 2008. The Roco protein family: a functional perspective. FASEB J. 22:3103–3110 [DOI] [PubMed] [Google Scholar]
- 15.Mata I. F., Wedemeyer W. J., Farrer M. J., Taylor J. P., Gallo K. A. 2006. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci. 29:286–293 [DOI] [PubMed] [Google Scholar]
- 16.Ohnishi T., Takahashi A., Okaichi K., Ohnishi K., Matsumoto H., Takahashi S., Yamanaka H., Nakano T., Nagaoka S. 1997. Cell growth and morphology of Dictyostelium discoideum in space environment. Biol. Sci. Space 11:29–34 [DOI] [PubMed] [Google Scholar]
- 17.Paisán-Ruíz C., Jain S., Evans E. W., Gilks W. P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A. M., Khan N., Johnson J., Martinez J. R., Nicholl D., Carrera I. M., Pena A. S., de Silva R., Lees A., Martí-Massó J. F., Pérez-Tur J., Wood N. W., Singleton A. B. 2004. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron 44:595–600 [DOI] [PubMed] [Google Scholar]
- 18.Pilcher K. E., Gaudet P., Fey P., Kowal A. S., Chisholm R. L. 2007. A general purpose method for extracting RNA from Dictyostelium cells. Nat. Protoc. 2:1329–1332 [DOI] [PubMed] [Google Scholar]
- 19.Rietdorf J., Siegert F., Dharmawardhane S., Firtel R. A., Weijer C. J. 1997. Analysis of cell movement and signalling during ring formation in an activated G alpha1 mutant of Dictyostelium discoideum that is defective in prestalk zone formation. Dev. Biol. 181:79–90 [DOI] [PubMed] [Google Scholar]
- 20.Schaap P., Winckler T., Nelson M., Alvarez-Curto E., Elgie B., Hagiwara H., Cavender J., Milano-Curto A., Rozen D. E., Dingermann T., Mutzel R., Baldauf S. L. 2006. Molecular phylogeny and evolution of morphology in the social amoebas. Science 314:661–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutoh K. 1993. A transformation vector for dictyostelium discoideum with a new selectable marker bsr. Plasmid 30:150–154 [DOI] [PubMed] [Google Scholar]
- 22.Taylor J. P., Mata I. F., Farrer M. J. 2006. LRRK2: a common pathway for parkinsonism, pathogenesis and prevention? Trends Mol. Med. 12:76–82 [DOI] [PubMed] [Google Scholar]
- 23.van Egmond W. N., Kortholt A., Plak K., Bosgraaf L., Bosgraaf S., Keizer-Gunnink I., van Haastert P. J. 2008. Intramolecular activation mechanism of the Dictyostelium LRRK2- homolog Roco protein GbpC. J. Biol. Chem. 283:30412–30420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veltman D. M., Akar G., Bosgraaf L., van Haastert P. J. 2009. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid 61:110–118 [DOI] [PubMed] [Google Scholar]
- 25.Veltman D. M., Keizer-Gunnink I., van Haastert P. J. 2008. Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J. Cell Biol. 180:747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West A. B., Moore D. J., Choi C., Andrabi S. A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K. L., Dawson V. L., Dawson T. M. 2007. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 16:223–232 [DOI] [PubMed] [Google Scholar]
- 27.West A. B., Moore D. J., Biskup S., Bugayenko A., Smith W. W., Ross C. A., Dawson V. L., Dawson T. M. 2005. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U. S. A. 102:16842–16847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Müller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K., Gasser T. 2004. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44:601–607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.