Abstract
Upregulated in infectious sporozoites gene 4 (UIS4) encodes a parasitophorous vacuole membrane protein expressed in the sporozoite and liver stages of rodent malaria parasites. Parasites that lack UIS4 arrest in early liver-stage development, and vaccination of mice with uis4− sporozoites confers sterile protection against challenge with infectious sporozoites. Currently, it remains unclear whether an ortholog of UIS4 is carried in the human malaria parasite Plasmodium falciparum, although the gene PF10_0164 has been identified as a candidate ortholog for UIS4 on the basis of synteny and structural similarity of the encoded protein. We show that PF10_0164 is expressed in sporozoites and blood stages of P. falciparum, where it localizes to the parasitophorous vacuole, and is also exported to the host erythrocyte. PF10_0164 is refractory to disruption in asexual blood stages. Functional complementation was tested in Plasmodium yoelii by replacing the endogenous copy of UIS4 with PF10_0164. PF10_0164 localized to the parasitophorous vacuole membrane of liver stages, but transgenic parasites did not complete liver-stage development in mice. We conclude that PF10_0164 is a parasitophorous vacuole protein that is essential in asexual blood stages and that does not complement P. yoelii UIS4, and it is thus likely not a functional ortholog of UIS4.
Malaria is caused by Plasmodium parasites, which are transmitted by deposition of sporozoites into the dermis of a mammalian host via the bite of infected Anopheles mosquitoes (2). Sporozoites travel through the bloodstream to the liver, where they infect hepatocytes (31). Productive hepatocyte infection requires formation of a parasitophorous vacuole (PV). Within the PV, the parasite transforms into a trophic phase and undergoes growth and schizogony to produce tens of thousands of merozoites, which are released into the circulation to initiate the asexual blood stage of the infection (19). The PV membrane (PVM) forms a barrier between the parasite and hepatocyte and prevents free access of secreted liver-stage proteins to the hepatocyte. Parasite proteins are targeted to the PVM, where they are likely to play a role in the parasite's interaction with its host cell (31).
In the rodent malaria parasites Plasmodium berghei and Plasmodium yoelii, upregulated in infectious sporozoite (UIS) genes are first transcribed in sporozoites that accumulate within the salivary glands of the mosquito vector (14). UIS genes were predicted to be important for infection of the mammalian host. The fourth gene in this group, UIS4, encodes a protein estimated to be 23 kDa in mass. UIS4 contains a single transmembrane domain and localizes to secretory organelles of sporozoites (8) and to the PVM of liver stages (18). UIS4 is not expressed in blood stages or early sporozoites that are produced in oocysts. Targeted deletion of UIS4 by homologous recombination in P. berghei and P. yoelii did not affect asexual blood-stage replication, parasite development in the mosquito, or sporozoite invasion of the salivary glands. uis4− sporozoites also invade hepatocytes within a PV and transform into liver stages normally. However, they show a severe deficiency in causing blood-stage infection (patency) in susceptible mice following infection by either mosquito bite or intravenous injection of sporozoites (18, 28). In vivo uis4− liver stages appear similar in size and morphology to wild-type (WT) parasites until 12 h postinfection, but they arrest in development after this point. Furthermore, mice immunized with uis4− sporozoites are protected against subsequent infection with WT sporozoites (18, 28), and thus uis4− parasites are a potent live-attenuated vaccine.
These results have raised the interest in identifying an ortholog of UIS4 in Plasmodium falciparum, the most virulent human malaria parasite, which is responsible for the majority of malaria mortality. Chromosomal synteny searches of the P. falciparum genome database have previously identified PF10_0164, a candidate UIS4 ortholog located on chromosome 10 (18). PF10_0164 is predicted to be a single-exon gene encoding a protein with a signal peptide and single transmembrane domain, similar to UIS4 in P. yoelii and P. berghei. Also similar to the rodent malaria parasites, PF10_0164 is transcribed in P. falciparum sporozoites (16).
Here, we studied PF10_0164 expression and examined its ability to complement P. yoelii UIS4 (PyUIS4) function. Unlike the rodent parasite UIS4, PF10_0164 was expressed in blood stages, where it localized to the PV and the host erythrocyte. We generated P. yoelii parasites in which the endogenous copy of UIS4 was replaced with a copy of PF10_0164 under the control of the endogenous PyUIS4 promoter. Our results indicate that although the transgene was expressed and its product localized to the PVM of liver stages, it did not complement PyUIS4 function.
MATERIALS AND METHODS
Bioinformatics tools.
Synteny mapping was adapted from PlasmoDB v5.5 (http://plasmodb.org). Protein alignment was adapted from results produced with the Multalin tool (4). Topology prediction was performed with the SOSUI tool (7).
Experimental animals, parasites, and cell lines.
Female Swiss Webster (SW) mice and female BALB/c mice (6 to 8 weeks old) were purchased from Harlan Laboratories (Indianapolis, IN). SW mice were used for routine parasite life cycle maintenance; BALB/c mice were used to obtain all experimental data shown. Animal handling was conducted in accordance with protocols approved by the Seattle Biomedical Research Institute Institutional Animal Care and Use Committee. P. yoelii strain 17XNL (a nonlethal strain) were used for transfections and as controls for all experiments. Parasites were used to infect Anopheles stephensi mosquitoes to produce sporozoites for infections. HepG2-CD81 cells were used for in vitro infections (24).
Generation of transgenic P. yoelii parasites.
To create the PF10_0164myc construct, the PF10_0164 open reading frame (ORF) was amplified by PCR from a 3D7 strain genomic DNA template with the primers SP/PfUIS4-B3D and ASP/PfUIS4-B3D. All primers used in this study are listed in Table 1. The promoter from PyUIS4 was amplified from 17XNL strain genomic DNA with the primers SP/PyUIS4Prom-B3D and ASP/PyUIS4Prom-B3D. The amplified PF10_0164 ORF and the PyUIS4 promoter were combined, and PCR was performed to fuse the products into a new amplicon using the primers SP/PyUIS4Prom-B3D and ASP/PfUIS4-B3D. The resulting product was ligated into the B3D vector, which encodes a 4× tandem myc epitope tag (32), with the SacII and SpeI restriction endonucleases. To create the PF10_0164 construct, the PF10_0164myc construct was digested with SpeI and treated with T4 DNA polymerase to fill in both ends of the digested plasmid, and the blunt ends were ligated back together. This introduced a stop codon between the PF10_0164 ORF and the sequence coding for the myc tag, as was confirmed by DNA sequencing. To create the PyUIS4myc construct, the PyUIS4 promoter and ORF were amplified as one product using the primers SP/PyUIS4Prom-B3D and ASP/PyUIS4-B3D and inserted into the B3D-myc vector with the SacII and SpeI enzymes. The PyUIS4 3′ untranslated region (UTR) was amplified from 17XNL genomic DNA using the primers SP/PyUIS4-B3D/3′UTR and ASP/PyUIS4-B3D/3′UTR, and this was inserted into the B3D-myc vectors containing PyUIS4 or PF10_0164 with the HindIII and KpnI enzymes. These constructs were digested with SacII and KpnI and transfected into P. yoelii 17XNL schizonts as previously described (11). Parasites were selected for transfectants with pyrimethamine, and genomic DNA was extracted from mouse blood using the DNeasy kit (Qiagen). The parasites were genotyped with the following primers: SP/UIS4/T1 and ASP/UIS4/T1 to test for integration at the 5′ terminus of the construct, SP/UIS4/T2 and ASP/UIS4/T2 to test for integration at the 3′ terminus of the construct, SP/UIS4/ORF and ASP/UIS4/ORF to test for the presence of the PyUIS4 ORF, and SP/PfUIS4 and ASP/PfUIS4/1 to test for the presence of the PF10_0164 ORF. Transgenic parasites were cloned in SW mice by limiting dilution.
Table 1.
Primers used in this study
| Primer name | Primer sequence (5′ → 3′) |
|---|---|
| SP/PfUIS4-B3D | CATCTGAATAAAATGAAGGTTTCTAGGCATACCGTTC |
| ASP/PfUIS4-B3D | ACTAGTTTCATCTTTGTCTTTGTCCTCTTTA |
| SP/PyUIS4Prom-B3D | CCGCGGGCTTCTTTGAGCAAATACTG |
| ASP/PyUIS4Prom-B3D | ACTAGTTTTATTCAGATGTAATTATGTGCTG |
| ASP/PyUIS4-B3D | ACTAGTTATGTATGGGTCAAATGGTT |
| SP/PyUIS4-B3D/3′UTR | AAAAGCTTTTCATTATGAGGGTAATTCAGAAAG |
| ASP/PyUIS4-B3D/3′UTR | AAGGTACCGACTTTTAAAAATAATATATATGAAA |
| SP/UIS4/T1 | TCTTTTATTCAGCGATTTTATGATGTTATATATA |
| ASP/UIS4/T1 | CGGGATCCCTAAGCTGCTAA |
| SP/UIS4/T2 | AAACACCATTTTTGAAAAGA |
| ASP/UIS4/T2 | CAAATGCTACATTATATGCA |
| SP/PfUIS4 | GAAGGTTTCTAGGCATACCGTTCTTTTAAA |
| ASP/PfUIS4/1 | CTAATTCCTCAGAGTCGGATCCATCATTCACC |
| ASP/PfUIS4/2 | GGTTCATTCTTGAAATATGGGTGTTCAATG |
| SP/PfUIS4/C-1 | GAATTCAATGAAGAGCTGGAAAAACAAAAA |
| ASP/PfUIS4/C-1 | GAGCTCTTTTCATCTTTGTCTTTGTCCT |
| SP/PfUIS4/C-2 | GAGCTCAAAGAGCTGGAAAAACAAAAA |
| ASP/PfUIS4/C-2 | AAGCTTTTCATCTTTGTCTTTGTCCT |
| SP/PfUIS4/C-3 | AAGCTTAAGAGCTGGAAAAACAAAAA |
| ASP/PfUIS4/C-3 | GCGGCCGCTTCATCTTTGTCTTTGTCCTCTTTAGAA |
| SP/UIS4/ORF | TATAGGATCCAAAATGAAAACCACATACGTTTCTCTC |
| ASP/UIS4/ORF | TAATGAATTCTATGTATGGGTCAAATGGTTTAT |
| SP/PF_CSP | GAGAAAATTAGCTATTTTATCTGTTTCTTCC |
| ASP/PF_CSP | ATCAGGATTACCATCCGCTGGTTGCTTTAA |
| SP/PF_HSP70 | CCTGACGAAGCTGTCGCTTTAGGTGCTGCT |
| ASP/PF_HSP70 | TACACCTCTTGGTGCTGGTGGAATTCCAAC |
| SP/PfUIS4-KO/5′ | CCATTTATCATAAAATTGTGGTCC |
| ASP/PfUIS4-KO/5′ | GTCGATATCCTTCAACGCTCTCTTATCATCATTTGCAG |
| SP/PfUIS4-KO/3′ | GGATCCGACTCTGAGGAATTAGATAGTTCTAAAGAGG |
| ASP/PfUIS4-KO/3′ | GTAACAAGCACATTACGTACAG |
In vitro and in vivo infections.
Anopheles stephensi mosquitoes were infected with transgenic or WT parasites by feeding on SW mice infected with sexual stages (exflagellation of microgametes was confirmed microscopically for each mouse). On day 14 postfeed, mosquitoes were dissected, and salivary glands were extracted and homogenized. Sporozoites were counted on a hemacytometer. For in vitro liver-stage infections, 105 sporozoites were loaded into wells of microscope coverslips seeded the previous day with 105 HepG2-CD81 cells. Cells were washed 1 h after the addition of sporozoites and grown in complete medium supplemented with 20 μg/ml gentamicin until the time point indicated, when they were fixed in 4% formalin and stained with primary and secondary antibodies. For in vivo infections, sporozoites were injected into the tail veins of BALB/c mice. Mice subsequently were either monitored for patency by thin blood smear or had their livers harvested, fixed in 4% paraformaldehyde, and made into 50-μm sections using a vibratome (Ted Pella, Inc., Redding, CA). The sections were incubated with anti-myc (SC-789; Santa Cruz Biotechnology, Santa Cruz, CA), anti-PyUIS4 (8), anti-Hep17, or anti-circumsporozoite protein (CSP; 2F6) primary antibodies and then with fluorophore-conjugated goat anti-mouse and goat anti-rabbit secondary antibodies, as well as 4′,6-diamidino-2-phenylindole (DAPI). P. falciparum blood stages were cultured (30) and fixed and stained (29) as previously described. Parasites were incubated with anti-P. falciparum HSP70 (anti-PfHSP70) and anti-PF10_0164 primary antibodies and then with fluorophore-conjugated secondary antibodies, as well as DAPI.
P. falciparum in vitro liver stages.
Severe combined immunodeficiency (SCID) mice homozygous for the urokinase type plasminogen activator transgene (SCID Alb-uPA) received transplanted human primary hepatocytes and were screened 6 weeks later for successful engraftment (21). Chimeric mice were infected with 1 × 106 to 1.5 × 106 P. falciparum sporozoites, and livers were harvested at 7 days postinfection. Liver sections were prepared and imaged as described previously (21).
Custom antisera.
Three copies of the PF10_0164 C-terminal domain-coding sequence were amplified with primers SP/PfUIS4/C-1 through SP/PfUIS4/C-3 and the respective antisense primers ASP/PfUIS4/C-1 through ASP/PfUIS4/C-3. These were digested with EcoRI, SacI, HindIII, and NotI and ligated in tandem into the pET-28B(+) vector (Novagen, EMD Chemicals, Gibbstown, NJ). This construct was used to express three tandem copies of the C-terminal domain of PF10_0164 (three tandem copies of the peptide KSWKNKNKSKDKVNDGSDSEELDSSKEDKDKDE) fused to a 6×His tag. The protein was purified and used to immunize rabbits (Pocono Rabbit Farm and Laboratories, Inc., Canadensis, PA). The resulting antisera were affinity purified on a column containing the recombinant antigen. Specificity of the antisera was confirmed by immunofluorescence assay (IFA) on in vitro liver-stage Pyuis4(−)PF10_0164myc or PyUIS4myc parasites (data not shown).
Differential permeabilization.
HepG2-CD81 cells were fixed in 4% paraformaldehyde in the absence of methanol at 10 h after infection with sporozoites. Cells were then permeabilized with the indicated concentrations of the mild detergent digitonin and incubated with anti-CSP and anti-myc primary antibodies and then with goat anti-mouse and goat anti-rabbit secondary antibodies, as well as DAPI.
Image acquisition and image processing.
IFA images were captured on a Deltavision Spectris RT microscope (Applied Precision, Issaquah, WA) using a 100× oil objective and subjected to deconvolution with the softWoRx software package.
Reverse transcription-PCR (RT-PCR).
Total RNA was extracted from P. falciparum midgut and salivary gland sporozoites as well as mixed blood stages in Trizol reagent (Invitrogen). RNA was treated with DNase I (Invitrogen), purified on an RNeasy column (Qiagen), and used to synthesize cDNA with the Superscript III first-strand synthesis supermix (Invitrogen), containing oligo(dT) and random hexamer primers. The conditions of the PCR were as described previously (1). PF10_0164 was amplified with SP/PfUIS4/1 and ASP/PfUIS4/1. P. falciparum HSP70 was amplified with SP/PF_HSP70 and ASP/PF_HSP70. P. falciparum CSP was amplified with SP/PF_CSP and ASP/PF_CSP.
Gene disruption in P. falciparum parasites.
The 5′ and 3′ regions of PF10_0164 were amplified using PCR from NF54 strain P. falciparum genomic DNA (supplied by the Walter Reed Army Institute for Research). The fragments were cloned into the transfection vector pCC-1 (13). The 5′ region of the gene spanned nucleotides 682385 to 683047 of chromosome 10 and was amplified using the primers SP/PfUIS4-KO/5′ and ASP/PfUIS4-KO/5′. The 3′ region spanned nucleotides 681722 to 682237 of chromosome 10 and was amplified using the primers SP/PfUIS4-KO/3′ and ASP/PfUIS4-KO/3′. The 5′ and 3′ regions of the target gene, PF10_0164, were cloned on either side of the human dihydrofolate reductase (hDHFR) cassette of pCC-1 to act as targets for homologous recombination (13). The 3D7 P. falciparum parasites were synchronized at ring stage with sorbitol prior to transfection. Transfection of ring stages with 100 μg of DNA was performed by electroporation at 0.31 kV and 950 μF with a Bio-Rad Gene Pulser (Bio-Rad, La Jolla, CA). Cultures were placed on positive selection using WR99210 (Jacobus Pharmaceuticals, Princeton, NJ) at 6 h posttransfection and maintained as described previously (5, 30). This was followed by negative selection against the cytosine deaminase/uracil phosphoribosyl transferase (CDUP) gene product with 5′-fluorocytosine (12). Parasites resistant to positive and negative selection underwent Southern blot analysis. Genomic DNA was prepared with a DNeasy tissue kit (Qiagen), and Southern blot analysis was performed using the digoxigenin (DIG) system (Roche) according to the manufacturer's instructions.
RESULTS
Identification of a putative UIS4 ortholog in P. falciparum.
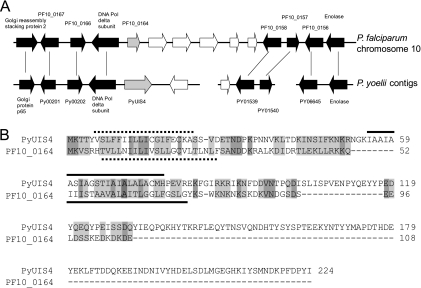
BLAST searches of the P. falciparum genome database did not identify genes with significant similarity to P. yoelii UIS4. However, several conserved genes immediately upstream of PF10_0164 are syntenous with the genes upstream of PyUIS4 (Fig. 1A). The genes downstream of PF10_0164 are syntenous with genes on a different P. yoelii contig, and one study has proposed that a synteny breakpoint exists in this region (10). The intervening genes (indicated by white arrows in Fig. 1A) have no recognized ortholog in rodent malaria parasites, and their products are all less similar to UIS4 than is PF10_0164, on the basis of sequence similarity and the number of predicted transmembrane domains. We confirmed that PF10_0164 is encoded by a single-exon gene by using RT-PCR and by sequencing the products of 3′ rapid amplification of cDNA ends (data not shown). The amino acid sequence identity between P. falciparum PF10_0164 and P. yoelii UIS4 is 9% (17% conserved residues), and the predicted PF10_0164 protein is considerably shorter (Fig. 1B). Topology prediction tools suggest that the transmembrane domain is located at a position in the protein that makes the lengths of the N termini comparable in the two proteins, whereas the C terminus of PF10_0164 is considerably shorter. Both, however, possess a high density of charged residues in their C termini (51% and 64% of residues in PF10_0164 and PyUIS4, respectively), with a net negative charge for each.
Fig. 1.
PF10_0164 is a candidate ortholog of PyUIS4. (A) Synteny map of P. falciparum chromosome 10 and unassembled P. yoelii contigs. Genes upstream of PF10_0164 on P. falciparum chromosome 10 are homologous to genes upstream of PyUIS4 on the P. yoelii contig MALPY00055. Genes in black are connected to their homologs with vertical lines; genes in white have no recognized homolog. Homologous genes are labeled with their predicted functions where appropriate and with gene identifier numbers where no predicted function is available. (B) Alignment of the predicted protein sequences of PyUIS4 and PF10_0164. Identical residues are shaded dark gray; similar residues are shaded light gray. The regions representing the predicted signal peptide and transmembrane domain are indicated by a dotted line and a solid line, respectively, for each protein.
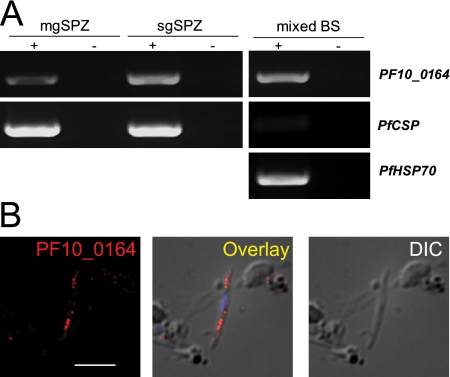
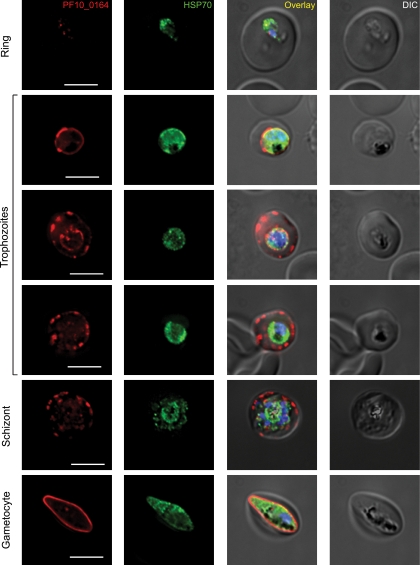
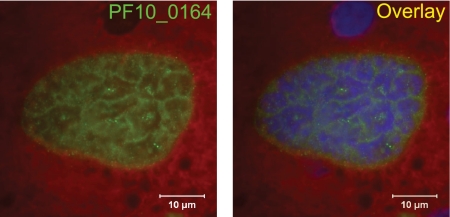
We next assayed for expression of PF10_0164 in different life cycle stages by RT-PCR (Fig. 2A). Transcripts were present in all stages tested, including midgut sporozoites and blood stages. To determine the localization of the protein in different stages of P. falciparum parasites, we raised antisera against the C-terminal domain of PF10_0164 and performed immunofluorescence assays (IFAs). In sporozoites, granular, internal staining that is consistent with potential localization to secretory organelles was present (Fig. 2B), reminiscent of that seen for PyUIS4 in P. yoelii sporozoites (see Fig 7A). Expression of PF10_0164 in asexual blood stages is presented in Fig. 3. Staining was weak or absent in ring stages. In larger uninucleate trophozoite stages, the protein could be seen to localize in a circumferential manner around the parasite, which suggests association with the PV, and/or in vesicular structures at the periphery of the host erythrocyte. These parasites are likely trophozoites, although it is possible that some are immature gametocytes. In schizonts, PF10_0164 is found predominantly in the host erythrocyte, in a pattern suggestive of localization to the Maurer's clefts. Expression of PF10_0164 was also detected in gametocytes, where it appeared to be associated with the PV. To visualize expression in P. falciparum liver stages, sporozoites were used to infect humanized mice that harbor human hepatocytes and infected livers were analyzed 7 days after infection (21). Staining with anti-PF10_0164 antiserum shows expression of the protein throughout the developing parasite and excluded from nuclei (Fig. 4). Expression of PF10_0164 was not detected at earlier time points.
Fig. 2.
P. falciparum PF10_0164 is expressed in all life cycle stages examined. (A) RT-PCR data showing expression of PF10_0164 in P. falciparum midgut sporozoites (mgSpz), salivary gland sporozoites (sgSpz), and mixed blood stages (mixed BS). Thirty-five cycles of amplification were performed from RNA treated with (+) or without (−) reverse transcriptase. Controls include HSP70 for blood stages and CSP for blood stages and sporozoites. (B) P. falciparum salivary gland sporozoite labeled with antibodies recognizing PF10_0164. The protein shows internal, granular staining similar to that seen for PyUIS4 in P. yoelii. Overlay image includes DAPI for labeling nuclei. Scale bar, 5 μm.
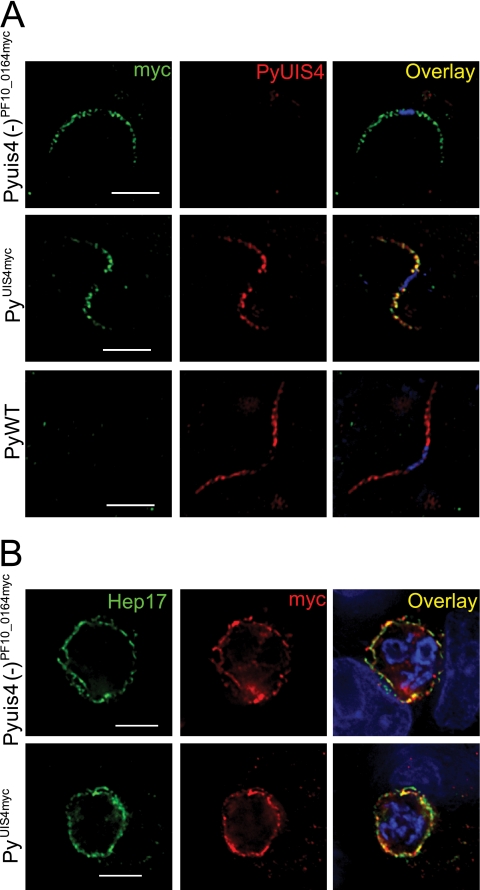
Fig. 7.
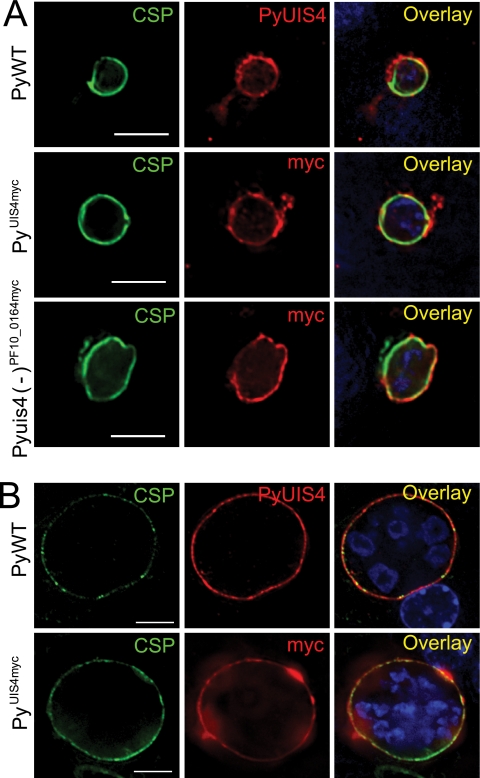
Expression of PF10_0164myc or PyUIS4myc in transgenic P. yoelii parasites. (A) PF10_0164myc and PyUIS4myc are expressed in salivary gland sporozoites and localize to secretory organelles, similar to the localization of PyUIS4 in PyWT sporozoites. Overlay images include DAPI for staining nuclei. Scale bars, 5 μm. (B) PF10_0164myc is expressed in Pyuis4(−)PF10_0164myc liver stages and PyUIS4myc is expressed in PyUIS4myc liver stages in vitro. Both epitope-tagged proteins colocalize with the integral PVM protein Hep17. Parasites are shown at 24 h postinfection of HepG2-CD81 cells. Scale bars, 5 μm.
Fig. 3.
P. falciparum in vitro blood stages labeled with antibodies recognizing PF10_0164. Parasites were from synchronized cultures fixed at the indicated developmental stage. The protein appears to be absent in rings. It localizes to the PV in trophozoites and gametocytes and is exported to the host erythrocyte in schizonts and some trophozoites. Overlay images in all figures include DAPI for labeling nuclei. Scale bars, 5 μm.
Fig. 4.
PF10_0164 is expressed in late P. falciparum liver stages formed in chimeric mouse livers. Liver sections stained with anti-PF10_0164 antiserum, from SCID/Albu-uPA mice which were infected with P. falciparum sporozoites following engraftment with human primary hepatocytes, are shown. Sections were made at 7 days postinfection and were counterstained with Evans blue dye. Overlay image includes DAPI for labeling nuclei.
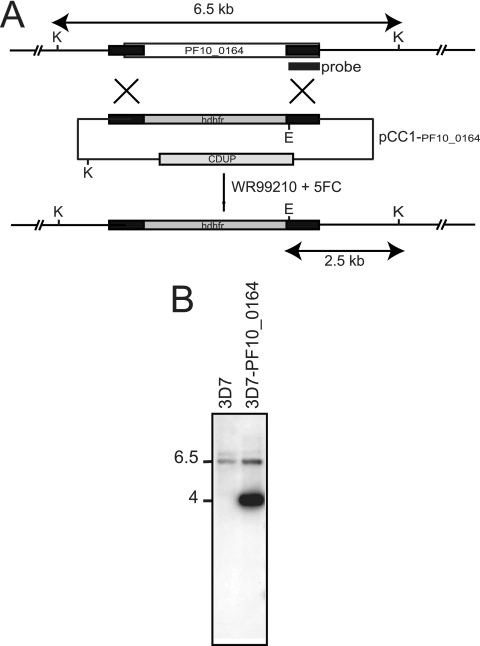
Because PF10_0164 expression was evident in blood stages, we tested whether PF10_0164 is important during the parasites' asexual replication. Repeated attempts to disrupt PF10_0164 by homologous recombination in transfected P. falciparum blood stages were unsuccessful, suggesting that the gene might be essential for intraerythrocytic development (Fig. 5).
Fig. 5.
The PF10_0164 gene in P. falciparum is refractory to deletion. (A) Schematic of the expected products for insertion of the hDHFR (human dihydrofolate reductase) gene into PF10_0164 by double homologous recombination using the vector pCC1-PF10_0164. The areas in black represent the sequences cloned into pCC1 in order to target the PF10_0164 locus for homologous recombination. The location of the hybridization probe is indicated below the schematic of the gene. CDUP refers to the cytosine deaminase/uracil phosphoribosyl transferase gene used for negative selection with 5′-fluorocytosine (5FC). The expected restriction fragments for digestion of the genomic DNA from WT or transgenic parasites with KpnI (K) and EcoRI (E) enzymes are indicated. (B) Southern blot analysis of genomic DNA from parasites transfected with pCC-PF_0164 and selected by cycles of WR99210 and 5′-fluorocytosine (5FC). The presence of a 6.5-kb band in both 3D7 and 3D7-PF10_0164 indicates retention of the WT gene. The 4-kb band in 3D7-PF10_0164 represents the KpnI and EcoRI fragment released from the episomal pCC1-PF10_0164.
Transgenic P. yoelii parasites expressing PF10_0164myc or PyUIS4myc.
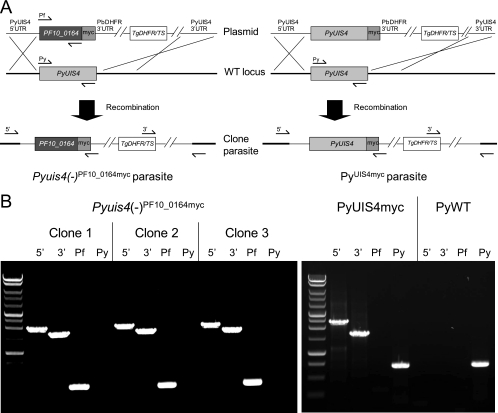
In order to evaluate PF10_0164 as an ortholog of PyUIS4, we tested its ability to complement the function of UIS4 in P. yoelii liver stages. We hypothesized that the protein would be expressed and targeted to the PVM and, if orthologous to UIS4, would allow transgenic parasites to complete liver-stage development and cause blood-stage patency. In order to examine expression and localization of the transgene, we introduced a C-terminal myc epitope tag to PF10_0164. As a control, the same tag was fused to the C terminus of PyUIS4 to ensure that the tag does not interfere with the essential function of the protein. P. yoelii parasites were transfected with constructs encoding PF10_0164 or PyUIS4 with four tandem copies of the c-myc epitope fused to the C terminus of each protein. Both transgenes were under the control of 1.4 kb of the endogenous PyUIS4 promoter and 1kb of the PbDHFR 3′ UTR. Recombination mediated by homology between the 5′ and 3′ UTRs of PyUIS4, contained within the constructs, and their respective sequences in the genomic locus led to replacement of endogenous PyUIS4 with PF10_0164myc or PyUIS4myc (Fig. 6A) [the transgenic parasites are hereafter referred to as Pyuis4(−)PF10_0164myc or PyUIS4myc, respectively]. Transgenic parasites were cloned by limiting dilution and genotyped to confirm replacement of the endogenous locus (Fig. 6B). The clones developed normally as asexual blood stages and produced numbers of salivary gland sporozoites that were comparable to those produced by the WT in Anopheles stephensi mosquitoes (data not shown).
Fig. 6.
Construction of transgenic P. yoelii parasites to test for functional complementation of UIS4. (A) Strategy for replacing PyUIS4 with PF10_0164myc or PyUIS4myc in P. yoelii. The transgenes are under the control of 1.4 kb of the endogenous PyUIS4 promoter and the PbDHFR 3′ UTR. The PyUIS4 5′ UTR and 3′ UTR target the constructs to the endogenous PyUIS4 locus. The mutant TgDHFR/TS enzyme is used in conjunction with pyrimethamine to select for transgenic parasites. Genotyping primers anneal where indicated to test for the presence of the PF10_0164 ORF (Pf) or the PyUIS4 ORF (Py) or for integration of the construct into the genomic locus from the 5′ or 3′ end. The boundary between the thick and thin black bars in the “clone parasite” line indicates the transition from chromosomal DNA to the integrated construct. (B) PCR-based genotyping of Pyuis4(−)PF10_0164myc, PyUIS4myc, and PyWT parasite clones. Amplicons represent the presence of the PyUIS4 ORF (Py) and the PF10_0164 ORF (Pf) and integration of the relevant construct into the PyUIS4 genomic locus from the 5′ or 3′ end.
We confirmed the expression of PF10_0164myc or PyUIS4myc in salivary gland sporozoites by IFA using anti-myc antibodies (Fig. 7A). Each myc-tagged protein exhibited a granular localization pattern in the sporozoite interior but was excluded from the nucleus, similar to the localization for PyUIS4 in WT sporozoites (8). We next evaluated expression in liver stages in vitro and determined that PF10_0164myc and PyUIS4myc localize in a circumferential pattern around the developing parasite (Fig. 7B) Both tagged proteins overlapped extensively with HEP17, a protein known to localize to the PVM. The PVM and parasite plasma membrane (PPM), however, are closely apposed in the liver stage, and therefore an exact distinction between localization to either membrane required additional investigation.
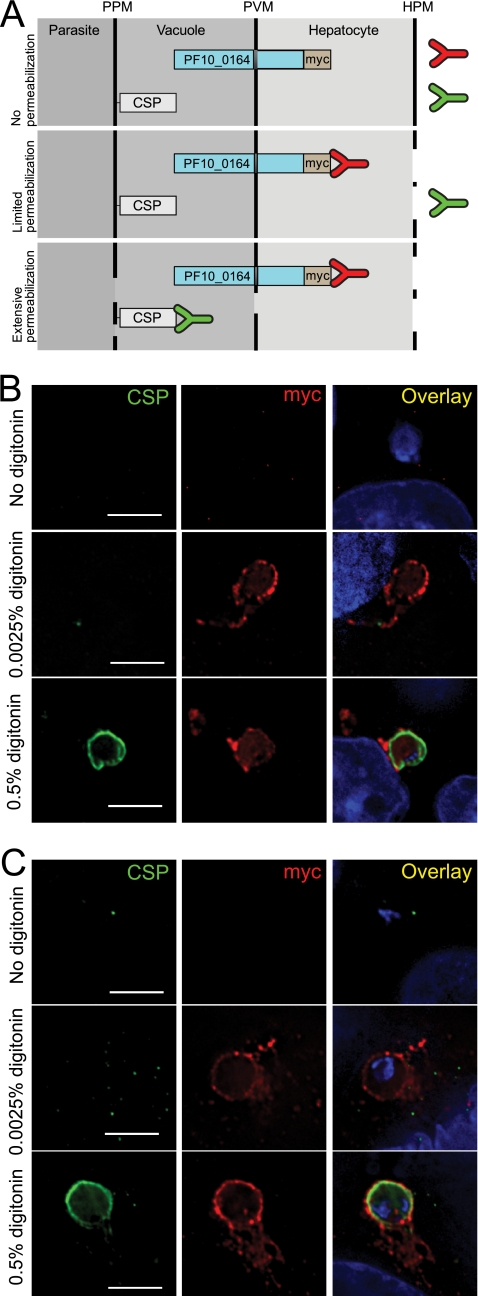
In order to confirm localization of PF10_0164 to the PVM, we performed a differential permeabilization assay. We hypothesized that PF10_0164myc and PyUIS4 are oriented in the PVM with their C-terminal domains facing the host cell cytoplasm (Fig. 8A). This would predict that at low concentrations of a mild detergent, when permeabilization is limited to the hepatocyte plasma membrane, antibodies will be able to access the C-terminal myc epitope tag on PF10_0164 but will not be able to access CSP, which is associated with the liver-stage plasma membrane and faces the lumen of the PV. We found that in in vitro liver stages fixed at 10 h postinfection and subsequently treated with 0.0025% digitonin, the myc tag could be detected via IFA but CSP could not be detected (Fig. 8B). At 0.5% digitonin, both proteins were accessible to antibodies and were thus detected, and in the absence of digitonin, neither could be detected. The same assay was performed on PyUIS4myc liver stages with similar results (Fig. 8C).
Fig. 8.
PF10_0164myc and PyUIS4myc both localize to the PVM of liver stages with their C-terminal domains facing the host cell cytoplasm. (A) Cartoon depicting topology of liver-stage membranes and predicted orientation of PF10_0164myc (PyUIS4myc was hypothesized to have a similar orientation). The parasite cytoplasm, parasitophorous vacuole, host hepatocyte cytoplasm, and extracellular space are separated by the parasite plasma membrane (PPM), parasitophorous vacuole membrane (PVM), and hepatocyte plasma membrane (HPM). During the differential permeabilization assay, antibodies against CSP or myc will bind their antigens based upon the extent of permeabilization of the various membranes. Bound myc in the absence of bound CSP will occur only if the PF10_0164myc protein is localized to the PVM with its C terminus facing outwards. (B) Differential permeabilization assay performed on HepG2-CD81 cells at 10 h after infection with Pyuis4(−)PF10_0164myc parasites. Cells were permeabilized with the indicated concentrations of the mild detergent digitonin. The results indicate that the myc-tagged C terminus faces the hepatocyte cytoplasm. Overlay images include DAPI for staining nuclei. Scale bars, 5 μm. (C) Differential permeabilization assay performed on HepG2-CD81 cells at 10 h after infection with PyUIS4myc parasites. Scale bars, 5 μm.
Failure of PF10_0164myc to complement the function of PyUIS4 in P. yoelii liver stages.
After confirming that PF10_0164myc is expressed in transgenic liver stages and localizes to the PVM similarly to endogenous PyUIS4, we sought to determine whether Pyuis4(−)PF10_0164myc and PyUIS4myc parasites could complete liver-stage development. We infected mosquitoes with both clones, dissected salivary glands to obtain sporozoites, and injected these intravenously into BALB/c mice. Pyuis4(−)PF10_0164myc parasites did not cause patency following injection of 1,000 or 10,000 sporozoites. All mice injected with PyUIS4myc parasites became patent, demonstrating that the C-terminal fusion of the myc tag did not significantly interfere with any essential function of PyUIS4 (Table 2). PF10_0164myc therefore does not complement the function of PyUIS4 sufficiently to allow rescue from the liver-stage developmental block seen in Pyuis4(−) parasites.
Table 2.
Prepatent periods in mice injected with transgenic P. yoelii parasites
| Parasite strain | No. of: |
Prepatent period (days)a | |
|---|---|---|---|
| Sporozoites injected | Mice patent/mice injected | ||
| Pyuis4(−)PF10_0164myc clone 1 | 1,000 | 0/6 | —b |
| 10,000 | 0/6 | ||
| Pyuis4(−)PF10_0164myc clone 2 | 1,000 | 0/3 | — |
| 10,000 | 0/3 | ||
| Pyuis4(−)PF10_0164myc clone 3 | 1,000 | 0/6 | — |
| 10,000 | 0/6 | ||
| PyUIS4-myc clone 1 | 1,000 | 6/6 | 4 |
| 10,000 | 6/6 | 3.5 | |
| PyWT | 1,000 | 7/7 | 3.5 |
| 10,000 | 7/7 | 2.5 | |
Prepatent periods are the means for each group, rounded to the nearest half day.
—, no value.
We next determined whether Pyuis4(−)PF10_0164myc parasites arrest in liver-stage development at a time point similar to that seen for Pyuis4(−) parasites. To that end, we injected one million sporozoites intravenously into BALB/c mice and then harvested livers and made sections to examine the liver stages in vivo. At 12 h postinfection, Pyuis4(−)PF10_0164myc parasites and PyUIS4myc parasites were comparable in size to PyWT liver stages from the same time point (Fig. 9A), although the numbers of Pyuis4(−)PF10_0164myc parasites were greatly reduced compared to those of the PyUIS4myc and PyWT parasites (data not shown), consistent with what has been observed previously for Pyuis4(−) parasites (28). At 24 h postinfection, PyUIS4myc parasites were comparable in size to PyWT parasites, but Pyuis4(−)PF10_0164myc parasites were not detectable (Fig. 9B).
Fig. 9.
Pyuis4(−)PF10_0164myc parasites develop normally as early liver stages. Pyuis4(−)PF10_0164myc, PyUIS4myc, and PyWT liver-stage parasites are shown at 12 (A) or 24 (B) hours postinfection of BALB/c mice with 106 sporozoites. Liver sections (50 μm) were stained with anti-PyCSP and polyclonal rabbit anti-myc or anti-PyUIS4 antibodies. Pyuis4(−)PF10_0164myc liver stages were comparable in size to PyWT liver stages at 12 h postinfection but could not be identified at 24 h postinfection. Overlay images include DAPI for staining nuclei. Scale bars, 5 μm.
Since the C-terminal domain of PF10_0164 is significantly shorter than that of PyUIS4, it can be argued that the introduction of a myc epitope tag to the C terminus of either protein may affect their functions differently, thus compromising the use of PyUIS4myc parasites as a control in this study. Therefore, as an additional control to ensure that the failure of PF10_0164 to complement the function of PyUIS4 in the liver stage was not due to the presence of the epitope tag, we generated a third transgenic line of P. yoelii parasites in which the PyUIS4 ORF was replaced with an ORF encoding the full-length PF10_0164 without a tag [referred to here as the Pyuis4(−)PF10_0164 strain]. These parasites formed liver stages in vitro that expressed PF10_0164 without a myc tag, and the protein localized in a manner similar to that seen in the Pyuis4(−)PF10_0164myc parasites (Fig. 10A). The mixed population of these transgenic parasites and WT parasites was fed to mosquitoes, and salivary gland sporozoites were injected into BALB/c mice in the manner described above. The mice were subsequently monitored for patency, and the blood-stage parasites that emerged were genotyped. We found that no transgenic parasites completed liver-stage development (Fig. 10B), suggesting that PF10_0164 does not complement PyUIS4 function in the liver, regardless of the presence or absence of an epitope tag.
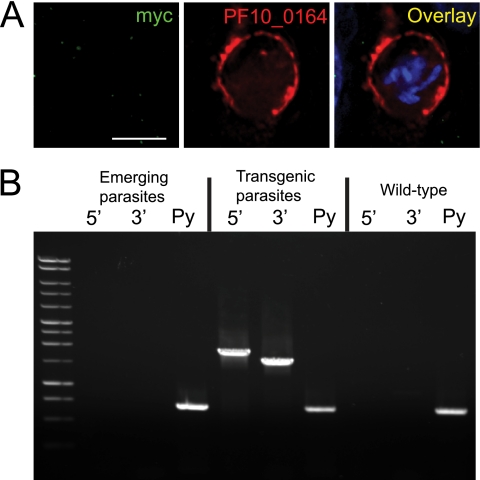
Fig. 10.
PyUIS4(−) parasites expressing the untagged PF10_0164 protein do not complete liver-stage development. (A) IFA of in vitro Pyuis4(−)PF10_0164 strain liver-stage parasites at 40 h after infection of HepG2-CD81 cells. Overlay images include DAPI for staining nuclei. Scale bar, 5 μm. (B) PCR-based genotyping of Pyuis4(−)PF10_0164 strain parasites before (transgenic parasites) and after (emerging parasites) cycling through the mosquito vector and injecting sporozoites into BALB/c mice. The primers and conditions used are the same as in Fig. 5A and B. This experiment was repeated in eight mice with identical results.
DISCUSSION
The PV that surrounds the intracellular stages of Plasmodium parasites remains an enigmatic structure, despite its importance as the primary site of host-parasite interactions. While a number of resident proteins of the PVM in erythrocytic stages are known (9, 27), few have been identified in preerythrocytic stages. The function of these proteins remains largely unknown.
UIS4 has been important to the study of the rodent malaria parasite species P. yoelii and P. berghei for its localization to secretory organelles of sporozoites and the PVM of liver stages and because it is essential for liver-stage development (8, 18, 28). Study of the liver stages of P. falciparum is technically challenging, and therefore the identification of orthologs of liver-stage proteins already partially characterized in rodent models would be greatly advantageous.
In this study we tested whether PF10_0164, a gene that we had suggested to be an ortholog of UIS4 (18), could complement UIS4 function in P. yoelii liver stages. We identified differences in the transcriptional profiles of the two genes, finding that, unlike UIS4, PF10_0164 is also expressed in blood stages. Interestingly, a previous study found that PF10_0164 expression is upregulated in P. falciparum strain 3D7 cultures in which gametocytogenesis is induced by high parasite density, compared to cultures of the F12 clone, which does not produce gametocytes (23). This study named PF10_0164 Pfpeg4, for P. falciparum protein of early gametocytes. Our antisera, however, detect the specific expression of PF10_0164 in asexual blood stages as well. Another study defined PF10_0164 as etramp10.3, a member of the family of early-transcribed membrane proteins transcribed in ring stages of P. falciparum (26). In that study, transcription of PF10_0164 was found to peak in the transition from ring to trophozoite stages, which is consistent with our results from IFAs on synchronized blood-stage cultures, where expression in rings is weak but protein expression in trophozoites and schizonts is robust. Importantly, we find that PF10_0164 is refractory to deletion in blood stages, indicating a potential essential function during intraerythrocytic replication. This is in sharp contrast to UIS4 of rodent malaria parasites, where deletion of the gene does not affect blood-stage replication (18, 28). While the function of PF10_0164 is not apparent, it can be seen to surround the parasite in erythrocytic stages and to localize to vesicular structures in the erythrocyte, suggesting that it might play a role in host-parasite interactions or in remodeling of the infected erythrocyte. In a previous study, ETRAMP10.1 and ETRAMP10.2 were found to localize to the PVM of erythrocytic stages, with their C-terminal domains facing the host cell cytoplasm (26), and this orientation was postulated as a general feature of the ETRAMP gene family. PF10_0164 showed the same orientation when expressed in P. yoelii liver stages and mirrors the orientation of PyUIS4myc in the PVM of liver stages, thus lending credence to a role in host-parasite interactions. We ascertained that the expression of PF10_0164myc in P. yoelii leads to localization to the same cellular compartments as PyUIS4, namely, secretory organelles in sporozoites and the PVM of liver stages. Additionally, we determined that the C-terminal fusion of a myc tag to the protein does not interfere with any essential function of PyUIS4. Despite the satisfaction of these criteria, however, we found that PF10_0164 is unable to complement the function of PyUIS4 in liver stages.
This failure to complement function suggests that the genes are not functional orthologs. The two genes might have arisen from a common ancestor but diverged sufficiently that they no longer perform the same functions. The reduced length of the PF10_0164 C-terminal domain relative to that of PyUIS4 is consistent with this idea. It should also be noted that the other ETRAMPs were initially identified on the basis of shared ring-stage transcription and protein structure, including a single transmembrane domain and highly charged C terminus, yet show limited sequence identity (25, 26). A lack of obvious homology between proteins on the basis of sequence alone does not preclude common ancestry. Another possibility is that PF10_0164 and UIS4 perform analogous functions in their respective species but that incompatible parasite proteins or host proteins are unable to interact appropriately with them. It was recently shown that PyUIS3, which is a predicted ortholog of etramp13 and is similar to UIS4 in terms of gene structure, expression profile, and essentiality for liver-stage development, binds to host hepatocyte liver fatty acid binding protein and that such interactions could be important to parasite growth (15, 22). Additionally, etramp5 was recently found to interact specifically with the human apolipoproteins ApoA1, ApoB, and ApoE in a yeast two-hybrid assay (33). These studies suggest that PVM proteins can be important in direct host-parasite interactions, but little is known about how such specific binding interactions are conserved across Plasmodium species. Specialized functions of parasite proteins could be dependent upon their molecular context, and transplantation into another species would fail to elucidate common function.
Our success in expressing a P. falciparum protein in P. yoelii and its ability to localize to the expected membrane indicate that this approach may be useful for testing for orthology between other genes of these species. Previously, rodent malaria parasites have been made to express genes from human malaria parasites, primarily for testing the immunogenicities of particular antigens (3, 6, 17, 20). We show that such approaches can also be used to explore the biology of preerythrocytic stages of human malaria parasites, which remain challenging to study. Accordingly, we have determined that PF10_0164, while likely not an ortholog of UIS4, is expressed in P. falciparum, is a bona fide PV protein of blood stages, and likely has an essential function. This protein may therefore warrant additional studies in the future.
ACKNOWLEDGMENTS
This study was funded by grants from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative and the National Institutes of Health (RO1 AI053709-07).
Footnotes
Published ahead of print on 12 March 2010.
REFERENCES
- 1.Aly A. S., Mikolajczak S. A., Rivera H. S., Camargo N., Jacobs-Lorena V., Labaied M., Coppens I., Kappe S. H. 2008. Targeted deletion of SAP1 abolishes the expression of infectivity factors necessary for successful malaria parasite liver infection. Mol. Microbiol. 69:152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aly A. S., Vaughan A. M., Kappe S. H. 2009. Malaria parasite development in the mosquito and infection of the mammalian host. Annu. Rev. Microbiol. 63:195–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao Y., Zhang D., Pan W. 2009. Construction of transgenic plasmodium berghei as a model for evaluation of blood-stage vaccine candidate of Plasmodium falciparum chimeric protein 2.9. PLoS One 4:e6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabb B. S., Rug M., Gilberger T. W., Thompson J. K., Triglia T., Maier A. G., Cowman A. F. 2004. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol. Biol. 270:263–276 [DOI] [PubMed] [Google Scholar]
- 6.de Koning-Ward T. F., O'Donnell R. A., Drew D. R., Thomson R., Speed T. P., Crabb B. S. 2003. A new rodent model to assess blood stage immunity to the Plasmodium falciparum antigen merozoite surface protein 119 reveals a protective role for invasion inhibitory antibodies. J. Exp. Med. 198:869–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirokawa T., Boon-Chieng S., Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 8.Kaiser K., Matuschewski K., Camargo N., Ross J., Kappe S. H. 2004. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol. Microbiol. 51:1221–1232 [DOI] [PubMed] [Google Scholar]
- 9.Kara U. A., Stenzel D. J., Ingram L. T., Bushell G. R., Lopez J. A., Kidson C. 1988. Inhibitory monoclonal antibody against a (myristylated) small-molecular-weight antigen from Plasmodium falciparum associated with the parasitophorous vacuole membrane. Infect. Immun. 56:903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kooij T. W., Carlton J. M., Bidwell S. L., Hall N., Ramesar J., Janse C. J., Waters A. P. 2005. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 1:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labaied M., Camargo N., Kappe S. H. 2007. Depletion of the Plasmodium berghei thrombospondin-related sporozoite protein reveals a role in host cell entry by sporozoites. Mol. Biochem. Parasitol. 153:158–166 [DOI] [PubMed] [Google Scholar]
- 12.Maier A. G., Braks J. A., Waters A. P., Cowman A. F. 2006. Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Mol. Biochem. Parasitol. 150:118–121 [DOI] [PubMed] [Google Scholar]
- 13.Maier A. G., Rug M., O'Neill M. T., Brown M., Chakravorty S., Szestak T., Chesson J., Wu Y., Hughes K., Coppel R. L., Newbold C., Beeson J. G., Craig A., Crabb B. S., Cowman A. F. 2008. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134:48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matuschewski K., Ross J., Brown S. M., Kaiser K., Nussenzweig V., Kappe S. H. 2002. Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J. Biol. Chem. 277:41948–41953 [DOI] [PubMed] [Google Scholar]
- 15.Mikolajczak S. A., Jacobs-Lorena V., MacKellar D. C., Camargo N., Kappe S. H. 2007. L-FABP is a critical host factor for successful malaria liver stage development. Int. J. Parasitol. 37:483–489 [DOI] [PubMed] [Google Scholar]
- 16.Mikolajczak S. A., Silva-Rivera H., Peng X., Tarun A. S., Camargo N., Jacobs-Lorena V., Daly T. M., Bergman L. W., de la Vega P., Williams J., Aly A. S., Kappe S. H. 2008. Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol. Cell. Biol. 28:6196–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mlambo G., Maciel J., Kumar N. 2008. Murine model for assessment of Plasmodium falciparum transmission-blocking vaccine using transgenic Plasmodium berghei parasites expressing the target antigen Pfs25. Infect. Immun. 76:2018–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller A. K., Camargo N., Kaiser K., Andorfer C., Frevert U., Matuschewski K., Kappe S. H. 2005. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. U. S. A. 102:3022–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prudencio M., Rodriguez A., Mota M. M. 2006. The silent path to thousands of merozoites: the Plasmodium liver stage. Nat. Rev. Microbiol. 4:849–856 [DOI] [PubMed] [Google Scholar]
- 20.Ramjanee S., Robertson J. S., Franke-Fayard B., Sinha R., Waters A. P., Janse C. J., Wu Y., Blagborough A. M., Saul A., Sinden R. E. 2007. The use of transgenic Plasmodium berghei expressing the Plasmodium vivax antigen P25 to determine the transmission-blocking activity of sera from malaria vaccine trials. Vaccine 25:886–894 [DOI] [PubMed] [Google Scholar]
- 21.Sacci J. B., Jr., Alam U., Douglas D., Lewis J., Tyrrell D. L., Azad A. F., Kneteman N. M. 2006. Plasmodium falciparum infection and exoerythrocytic development in mice with chimeric human livers. Int. J. Parasitol. 36:353–360 [DOI] [PubMed] [Google Scholar]
- 22.Sharma A., Yogavel M., Akhouri R. R., Gill J. 2008. Crystal structure of soluble domain of malaria sporozoite protein UIS3 in complex with lipid. J. Biol. Chem. 283:24077–24088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silvestrini F., Bozdech Z., Lanfrancotti A., Di Giulio E., Bultrini E., Picci L., Derisi J. L., Pizzi E., Alano P. 2005. Genome-wide identification of genes upregulated at the onset of gametocytogenesis in Plasmodium falciparum. Mol. Biochem. Parasitol. 143:100–110 [DOI] [PubMed] [Google Scholar]
- 24.Silvie O., Greco C., Franetich J. F., Dubart-Kupperschmitt A., Hannoun L., van Gemert G. J., Sauerwein R. W., Levy S., Boucheix C., Rubinstein E., Mazier D. 2006. Expression of human CD81 differently affects host cell susceptibility to malaria sporozoites depending on the Plasmodium species. Cell. Microbiol. 8:1134–1146 [DOI] [PubMed] [Google Scholar]
- 25.Spielmann T., Beck H. P. 2000. Analysis of stage-specific transcription in Plasmodium falciparum reveals a set of genes exclusively transcribed in ring stage parasites. Mol. Biochem. Parasitol. 111:453–458 [DOI] [PubMed] [Google Scholar]
- 26.Spielmann T., Fergusen D. J., Beck H. P. 2003. etramps, a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite-host cell interface. Mol. Biol. Cell 14:1529–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spielmann T., Gardiner D. L., Beck H. P., Trenholme K. R., Kemp D. J. 2006. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol. Microbiol. 59:779–794 [DOI] [PubMed] [Google Scholar]
- 28.Tarun A. S., Dumpit R. F., Camargo N., Labaied M., Liu P., Takagi A., Wang R., Kappe S. H. 2007. Protracted sterile protection with Plasmodium yoelii pre-erythrocytic genetically attenuated parasite malaria vaccines is independent of significant liver-stage persistence and is mediated by CD8+ T cells. J. Infect. Dis. 196:608–616 [DOI] [PubMed] [Google Scholar]
- 29.Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. 2004. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 137:13–21 [DOI] [PubMed] [Google Scholar]
- 30.Trager W., Jensen W. 1978. Cultivation of malaria parasites. Nature 273:621–622 [DOI] [PubMed] [Google Scholar]
- 31.Vaughan A. M., Aly A. S., Kappe S. H. 2008. Malaria parasite pre-erythrocytic stage infection: gliding and hiding. Cell Host Microbe 4:209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaughan A. M., O'Neill M. T., Tarun A. S., Camargo N., Phuong T. M., Aly A. S., Cowman A. F., Kappe S. H. 2009. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell. Microbiol. 11:506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vignali M., McKinlay A., LaCount D. J., Chettier R., Bell R., Sahasrabudhe S., Hughes R. E., Fields S. 2008. Interaction of an atypical Plasmodium falciparum ETRAMP with human apolipoproteins. Malar. J. 7:211. [DOI] [PMC free article] [PubMed] [Google Scholar]