Abstract
Sexual reproduction of the human pathogen Aspergillus fumigatus (teleomorph: Neosartorya fumigata) was assumed to be absent or cryptic until recently, when fertile crosses among geographically restricted environmental isolates were described. Here, we provide evidence for mating, fruiting body development, and ascosporogenesis accompanied by genetic recombination between unrelated, clinical isolates of A. fumigatus, and this evidence demonstrates the generality and reproducibility of this long-time-undisclosed phase in the life cycle of this heterothallic fungus. Successful mating requires the presence of both mating-type idiomorphs MAT1-1 and MAT1-2, as does expression of genes encoding factors presumably involved in this process. Moreover, analysis of an A. fumigatus mutant deleted for the nsdD gene suggests a role of this conserved regulator of cleistothecium development in hyphal fusion and hence heterokaryon formation.
For decades, aspergilli have served as model organisms in genetic studies, based on their multifaceted life cycle. The characteristic asexual reproductive mode of Aspergillus, during which conidiospores are abundantly formed on highly characteristic structures, the conidiophores (synonym, aspergillum) (2), defines the genus morphologically. Fundamental aspects of the genetics, such as heterokaryosis and parasexuality, were first studied in the model system Aspergillus nidulans (38). Moreover, about one-third of all known Aspergillus species are able to form fruiting bodies, the so-called cleistothecia, in which recombinant ascospores are generated by meiosis (8, 15, 48). Again, most insights into cleistothecium formation stem from studies on the homothallic ascomycete A. nidulans with its teleomorph Emericella nidulans, for which a variety of environmental as well as genetic determinants for successful fruiting were identified: surface growth in darkness and with restricted aeration generally favors sexual development, which might correspondingly be the dominant reproductive mode in the soil or within substrates. Nuclear identity is presumably achieved by a bipolar mating-type system comprising the high-mobility group (HMG) factor MatA and the α-box protein MatB (36, 46). Among regulators of cleistothecium formation that have been characterized at the molecular level, the GATA-type transcription factor NsdD plays a prominent role, as it determines the balance between asexual and sexual propagation and positively supports fruiting (18).
For Aspergillus fumigatus, the predominant human pathogen among aspergilli, sexuality had been only supposed for a long time, based on several lines of reasoning. The very first hints were deduced from the preliminary genome sequence, in which several genes likely to encode a pheromone and respective pheromone receptors could be identified (37). Moreover, the presence of a putative mating-type locus was pointed out in these primary studies (12, 53). A more comprehensive inspection of the complete and annotated genome sequence retrieved several more homologues that are involved in mating or ascosporogenesis of ascomycetous fungi (14, 32), and phylogenetic analyses also suggested the existence of a sexual reproductive mode (39). Strong evidence for the presence of an A. fumigatus sexual cycle was provided by identification of a bipolar mating-type system resembling that of A. nidulans (32) and equal distributions of either of the mating-type idiomorphs MAT1-1 and MAT1-2 among worldwide isolates (35). Gene-swapping experiments for both A. fumigatus idiomorphs accompanied by overexpression studies for a conserved regulator of fruiting body development in a fertile Aspergillus species demonstrated their functional conservation (16, 41). Ultimate proof, however, for the existence of a sexual phase within the A. fumigatus life cycle was given recently, when isolates of a limited regional sample set from Dublin City, Ireland, were successfully paired on self-made oatmeal agar plates after 6 months of incubation under specific conditions (33). Along the barrage zone of compatible pairings, cleistothecia were formed that harbored ornamented and viable ascospores, from which typical A. fumigatus colonies were grown under standard conditions. Segregation patterns of genetic markers provided evidence of meiotic recombination during the developmental process. Following the dual nomenclature, the corresponding teleomorph was coined Neosartorya fumigata.
Besides this seminal but solitary description, no further evidence for sexual reproduction among A. fumigatus isolates has been reported. In order to test the general practice of A. fumigatus mating and to characterize genetic determinants required for this process, we undertook efforts to verify cleistothecium formation among established, unrelated clinical isolates of A. fumigatus. Furthermore, by crossing mutants that carry deletions of mating-type genes or lacking the conserved regulator of cleistothecium development NsdD, the role in fruiting body formation was assessed. Noticeably, a role of NsdD in cell wall stress resistance could be established, which implies a role of this transcriptional regulator in hyphal fusion accompanying heterokaryon formation.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The bacterial strain Escherichia coli DH5α was used for general cloning procedures and cultivated in LB medium (1% Bacto tryptone, 0.5% yeast extract, 1% NaCl, pH 7.5). Fungal strains used in this study are listed in Table 1. Aspergillus fumigatus isolates were typed for their MAT idiomorph by a multiplex PCR approach as described by Paoletti et al. (35). Growth of A. fumigatus strains was carried out at 37°C on minimal medium prepared according to the methods of Barrat et al. (4) or for mating purposes at 30°C on purchased Difco oatmeal agar in the absence of light with restricted aeration (33). More specifically, strains were spot inoculated on oatmeal plates and allowed to germinate overnight before sealing the plates with parafilm to restrict aeration and wrapped in aluminum foil to prevent illumination. Parafilm seals were removed after 1 week to avoid conditions of excessive hypoxia. Phenotypes of recombinant strains, such as sterility of mating-type deletants or nsdD-related phenotypes, were validated by verification in independent mutants or by suppression analysis of reconstituted isolates, respectively.
Table 1.
A. fumigatus strains used in this study
| Strain | Description | Reference |
|---|---|---|
| D141 | A. fumigatus wild-type strain (syn. NRRL 6585), clinical isolate (MAT1-1) | 50 |
| Af293 | A. fumigatus genome sequence reference strain, clinical isolate (MAT1-2) | 32 |
| AfS41 | pabaA::loxP riboB::loxP derivative of D141 | 16 |
| AfS45 | pabaA::loxP pyroA::loxP derivative of Af293 | 16 |
| AfS14 | nsdDΔ deletant in D141 background (nsdD::loxP-Phleor/tk-loxP) | This study |
| AfS69 | Reconstituted isolate of AfS14 (pniaD::nsdD ptrA) | This study |
| AfS71 | mat1-1::ptrA deletion strain, D141 derivative | This study |
| AfS72 | mat1-2::ptrA deletion strain, Af293 derivative | This study |
| AfS81 | D141 derivative expressing GFP-H2A (pgpdA::gfp2-5::his2A ptrA) | This study |
| AfS82 | Af293 derivative expressing mCherry-H2A (pgpdA::mCherry::his2A ptrA) | This study |
| AfS83 | AfS14 expressing GFP-H2A (nsdDΔ; pgpdA::gfp2-5::his2A ptrA) | This study |
| AfS84a/b | Independent mat1-1::ptrA deletants, D141 derivatives | This study |
| AfS85a/b | Independent mat1-2::ptrA deletants, Af293 derivatives | This study |
Antibiotic concentrations were 100 μg/ml for ampicillin, 30 μg/ml for phleomycin, and 0.1 μg/ml for pyrithiamine; nutritional auxotrophies were supplemented with 1 μg/ml p-aminobenzoate, 0.5 μg/ml pyridoxine-HCl, or 2.5 μg/ml riboflavin-HCl.
Transformation procedures.
Calcium/manganese-treated E. coli cells were used for transformation (19); A. fumigatus recipients were transformed by polyethylene glycol-mediated fusion of protoplasts as described previously (40).
Nucleic acids manipulations and plasmid construction.
Standard protocols of recombinant DNA technology were carried out (45). Phusion high-fidelity DNA polymerase (Finnzymes) was generally used in PCRs (44), and essential cloning steps were verified by sequencing by the GATC Biotech Company (Konstanz, Germany). Sequence analyses were carried out using the Lasergene Biocomputing software package from DNAStar. Fungal genomic DNA was prepared according to the methods of Kolar et al. (23), and Southern analyses were carried out essentially as described previously (49). Recombinant plasmids used during the course of this study are listed in Table 2, with relevant oligonucleotides specified in Table S1 of the supplemental material.
Table 2.
Plasmids used in this study
| Name | Description | Reference or source |
|---|---|---|
| pBluescript II KS | General cloning plasmid (bla, MCS)a | Stratagene |
| pJET1.2 | Positive selection vector, pUC19 derivative (bla, MCS in eco47IR) | Fermentas |
| pSK275 | ptrA resistance gene flanked by SfiI sites in pBluescript II KS (syn. pME3024) | 27 |
| pSK259 | Recyclable phleomycin resistance cassette flanked by loxP attachment sites (syn. pME2891) | 25 |
| pSK393 | Integrative plasmid for expression of A. fumigatusnsdD (pniaD::nsdD::his2AtptrA)b | 16 |
| pSK379 | Integrative A. fumigatus expression plasmid carrying pgpdA-his2At cassette and ptrA | This study |
| pSK245 | nsdD genomic locus (EcoRI/BamHI) from A. fumigatus D141 in pBluescript II KS | This study |
| pSK263 | nsdD::loxP-Phleor/tk-loxP replacement cassette | This study |
| pSK491 | mat1-1::ptrA deletion cassette | This study |
| pSK492 | mat1-2::ptrA deletion cassette | This study |
| pSK494 | Synthetic, codon-optimized gfp2-5 allele for expression in A. fumigatus | This study |
| pSK496 | Synthetic, codon-optimized mCherry allele for expression in A. fumigatus | This study |
| pSK505 | Integrative plasmid for expression of GFP-H2A (pgpdA::gfp2-5::his2A ptrA) | This study |
| pSK507 | Integrative plasmid for expression of mCherry-H2A (pgpdA::mCherry::his2A ptrA) | This study |
MCS, multiple cloning site.
p, promoter; t, terminator.
In detail, replacement cassettes and expression constructs were generated as follows. Deletion cassettes for the mating-type idiomorphs were obtained by applying the fusion PCR approach (52). 5′ and 3′ regions of about 2 kb flanking the MAT1-1 locus were amplified from genomic DNA of A. fumigatus strain D141 with the oligonucleotide pairs ES001/ES017 and ES022/ES004, respectively, and fused to the pyrithiamine resistance cassette from pSK275 amplified with primers ES019/ES020. The resulting 6-kb amplicon was used directly to transform A. fumigatus D141 and also cloned into pJET1.2 to result in plasmid pSK491. The MAT1-2 deletion construct of pSK492 was isolated in a similar fashion from genomic DNA of strain Af293 using primers ES005/ES018 (5′ flank) and ES023/ES008 (3′ flank). In order to generate a replacement cassette for targeting nsdD, a 7.8-kb BamHI/EcoRI fragment comprising the nsdD gene was isolated from A. fumigatus genomic DNA and inserted into pBluescript II KS to generate plasmid pSK245, from which the 5′ and 3′ flanking regions were isolated as a 2.2-kb HindIII and 3.3-kb FspI fragment, respectively. Both homology arms were inserted sequentially into pSK259 via HindIII and HpaI to result in construct pSK263, from which an nsdD replacement cassette was released via NdeI digestion prior to A. fumigatus transformation. Expression plasmids for fluorescent labeling of A. fumigatus nuclei in live cells were constructed by fusing synthetic coding sequences for the fluorescent proteins green fluorescent protein (GFP) and mCherry (pSK494 and pSK496, respectively; custom synthesized by GeneArt, Germany) followed by a (GA)4 linker region to a sequence comprising the coding region and 579 bp of its 3′-untranslated region of the histone 2A-encoding gene. Chimeric fragments were generated by fusion PCRs with primer pairs Sv648/Sv649 (GFP-GA4), Sv653/Sv654 (mCherry-GA4), and Sv557/Sv652 (GA4-H2A), and the resulting amplicons were inserted into expression vector pSK379 after removal of a 2.0-kb PmeI/AleI fragment to yield integrative plasmids pSK505 and pSK507.
qRT-PCR.
Quantitative real-time PCRs (qRT-PCRs) were performed as described recently (6). Sequences of priming oligonucleotides are given in Table S1 of the supplemental material, and gene-specific combinations were as follows: TubFw/TubRev (AFUA_1G10910; encoding β-tubulin), ES077/78 (AFUA_6G06360; α-like pheromone precursor PpgA), ES079/80 (AFUA_5G07880; pheromone receptor PreA), ES081/82 (AFUA_3G14330; pheromone receptor PreB), ES083/84 (AFUA_3G13870; GATA-type transcriptional activator NsdD), and ES085/86 (AFUA_1G12490; sexual development regulator VeA). Melting curves were plotted to confirm specificity of each reaction, and results were quantified according to the comparative threshold cycle method (1).
HAMF assay.
Heterokaryon formation of A. fumigatus strains was monitored by histone-assisted merged fluorescence (HAMF) (42) from strains of opposite mating type that expressed fluorescent protein-labeled histone H2A (51). Cultivations were carried out in μ-Slides (ibidi, Germany) to assess the distribution of nuclear fluorescence within hyphae on a Zeiss Axiolab fluorescence microscope equipped with an AxioCam ICc1 digital camera and the AxioVision LE software.
RESULTS
Unrelated clinical isolates of Aspergillus fumigatus are able to mate.
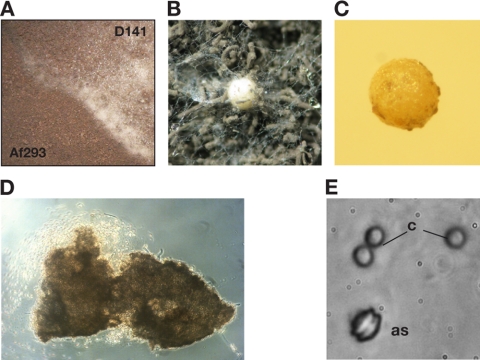
The first hard evidence for a sexual phase in the A. fumigatus life cycle was recently provided by successful crossings among environmental isolates that had been collected in Dublin City, Ireland (33). We were interested in whether this capacity for cleistothecium formation was restricted to members of this collection, which had been limited to one geographic area, or whether mating and ascosporogenesis could be observed between completely unrelated clinical isolates of A. fumigatus. In order to put this to the test, established strains carrying opposite mating types were inoculated on growth plates containing purchased oatmeal agar. After prolonged incubation under restricted aeration at 30°C in the dark, a clear barrage zone was evident after 6 weeks for the MAT1-1/MAT1-2 combination of clinical isolates D141 (syn. NRRL 6585) (MAT1-1) (50) and Af293 (MAT1-2) (32, 34) (Fig. 1A). This junction is characterized by a cotton wool-like appearance, and closer inspection revealed the presence of numerous cleistothecial balls (Fig. 1B). Upon further incubation for 3 to 4 months, the fluffy fruiting bodies matured to become more rigid, with a size of 300 to 400 μm (Fig. 1C). Squeezing these cleistothecia released ascospores of typical size and appearance (Fig. 1D and E).
Fig. 1.
Cleistothecium development and ascosporogenesis by crossing of unrelated clinical isolates of A. fumigatus. (A) Section from the crossing plate with clinical isolates D141 (MAT1-1) and Af293 (MAT1-2) on oatmeal agar. (B) These strains of opposite mating types develop a cotton-like barrage zone, in which cleistothecia develop upon further incubation. (C to E) From mature fruiting bodies (C), ascospores can be released by squeezing (D), which results in a characteristic appearance with two equatorial crests (E); for comparison, conidia (c) are shown beside one ascospore (as).
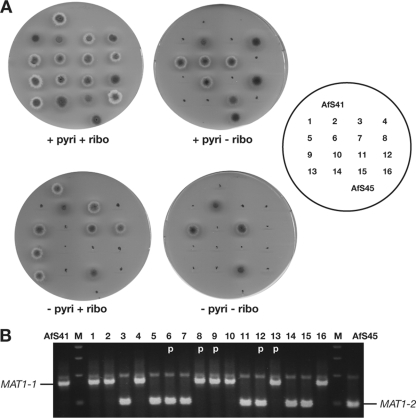
To confirm recombination between the compatible clinical isolates during mating, mutant strains carrying defined deletions in metabolic marker genes—AfS41 (MAT1-1; pabaA::loxP riboB::loxP) and AfS45 (MAT1-2; pabaA::loxP pyroA::loxP) as descendants of D141 and Af293, respectively (16)—were crossed. Ascospores were randomly isolated from several of the developing cleistothecia and inoculated for single colonies on defined growth medium. Auxotrophies of clonal isolates were determined by replica plating on appropriately supplemented medium to reveal different genotypes that were in agreement with recombination, such as riboBΔ pyroAΔ or riboB+ pyroA+ (Fig. 2A). Mating types of the recombinants were determined by multiplex PCR (35), and their segregation patterns in correlation to the auxotroph markers also supported genetic recombination during ascosporogenesis (Fig. 2B).
Fig. 2.
Distribution of auxotrophies (A) and mating types (B) among 16 descendants from a cross between A. fumigatus strains AfS41 (MAT1-1; pabaA::loxP riboB::loxP) and AfS45 (MAT1-2; pabaA::loxP pyroA::loxP). All growth media were supplemented with p-aminobenzoic acid. The presence of the MAT1-1 or MAT1-2 idiomorph was verified by diagnostic multiplex PCR (19). Marker combinations and segregation of mating types clearly indicated the genetically recombined nature of the progeny. p, parental genotype.
Mat1-1 and Mat1-2 determine A. fumigatus fruiting body formation and regulate expression of key elements of a putative mating pathway.
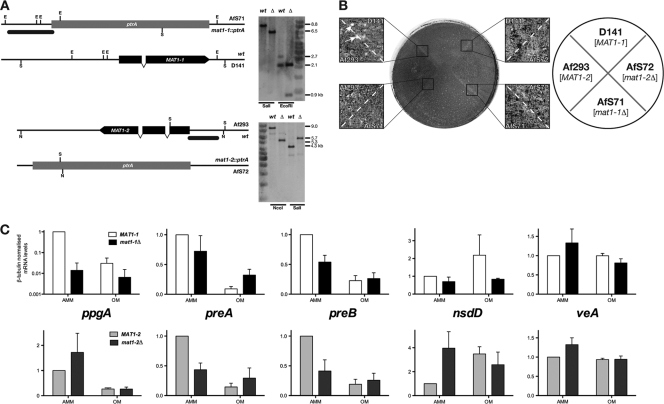
In order to assess the role of mating-type gene products in A. fumigatus cleistothecium formation, we created mutant strains deleted for either locus by targeted gene replacement. The correct genotypes of the corresponding deletants AfS71 (mat1-1Δ) and AfS72 (mat1-2Δ) were confirmed by Southern analyses (Fig. 3A), and strains were inspected for noticeable deviations from their wild-type progenitors. No obvious phenotype became evident under standard culture conditions, such as growth in or on minimal medium at 37°C (data not shown). However, in crossing experiments, neither deletant supported formation of mature fruiting bodies when confronted with a strain carrying the opposite mating-type idiomorph and incubated under the established conditions for 6 months (Fig. 3B). Although evidence of early stages of mating, such as the cotton wool-like hyphal structures, was noted in pairings of AfS71 with Af293 or of AfS72 with D141, no further development to mature, ascospore-containing cleistothecia could be observed. Testing additionally and independently generated mutants deleted for either mating-type gene confirmed the associated phenotype of sterility: when crossing strains AfS84 or AfS85 with the clinical isolates of the opposite mating type under appropriate conditions, no signs of cleistothecium formation became evident. Accordingly, both products of the mating-type locus are essential for successful fruiting of A. fumigatus.
Fig. 3.
MAT1-1 and MAT1-2 are required for mating of A. fumigatus. (A) Schematic representation of deleted mating-type loci and Southern analyses of strains AfS71 (mat1-1::ptrA) and AfS72 (mat1-2::ptrA). Black bars correspond to probe positions, and restriction enzyme recognition sites are indicated for EcoRI (E), NcoI (N), and SalI (S), with the resulting fragment sizes displayed on the right. (B) Crossing plate for mat1 deletants with wild-type progenitors. Close-up images of barrage zones between indicated strains show the presence of mature cleistothecia only for the D141 × Af293 cross (arrows), whereas other combinations failed to produce fruiting bodies. (C) Effects of mating-type deletions on expression of genes involved in pheromone signal transduction and regulation of cleistothecium formation. Mycelia of the indicated strains were propagated at 30°C in the dark under restricted aeration on Aspergillus minimal medium (AMM) or oatmeal agar (OA) for 4 days before preparation of total RNA. Transcript levels were determined from three biological replicates by quantitative real-time PCRs with three technical replicates, using β-tubulin transcript levels as the internal standard and assigning the levels for wild-type strains on AMM as 100%.
As both gene products from the A. fumigatus mating-type idiomorphs are DNA binding factors putatively involved in regulation of gene expression, we were interested in the levels of selected transcripts that are likely involved in mating and sexual development. More precisely, genes encoding the putative sexual pheromone precursor PpgA, the pheromone receptors PreA and PreB, and the regulators of cleistothecium formation, NsdD and VeA, were analyzed (Fig. 3C). Transcript levels of the encoding genes were determined from mycelia that had been transferred from minimal medium to oatmeal agar to be further incubated under conditions that favor sexual development. In parallel, split mycelia were further propagated on minimal medium under the same conditions to assess the influence of the culture medium, and message levels in wild-type strains D141 and Af293 under these conditions served as respective benchmarks (Fig. 3C). Distinct differences became evident for the pheromone precursor-encoding gene ppgA: on minimal medium, expression of this transcript declined drastically in the absence of the MAT1-1 gene but increased if the MAT1-2 gene had been deleted. On oatmeal agar, this trend was less pronounced (D141 versus AfS71) or absent (Af293 versus AfS72), with expression levels being generally lower in either genetic background. These data suggest that expression of the α-factor-like pheromone is regulated reciprocally by the mating-type genes. Furthermore, transcript levels for the pheromone receptor genes preA and preB were determined to reveal similar patterns of regulation for both loci. On minimal culture medium, transcript levels decreased when either mating type was deleted; on oatmeal agar, this situation was counterbalanced for preB to result in transcript levels similar to the ones of either deletant on minimal medium. Accordingly, either mating-type gene product appears to be required for downregulation of preB transcription upon a shift from minimal medium to oatmeal agar. When propagated on the oatmeal culture medium, levels of the preA transcript in each mutant were slightly higher than in the wild type. In the mat1-2Δ background, this elevated level was equal to that on minimal medium, whereas in the mat1-1Δ strain a downregulation of preA transcript levels upon the shift from minimal to oatmeal medium was still evident, although to a lesser degree than in the parental wild-type strain.
To analyze any influence of Mat1-1 or Mat1-2 on expression of regulators of Aspergillus sexual development, transcript levels for the GATA-type transcription factor NsdD and the velvet complex subunit VeA (5, 16) were determined. No significant changes were evident for either transcript in the mat1-1 deletion strain on either culture medium, except for a reduction of elevated nsdD transcript levels in the presence of oatmeal. In the mat1-2Δ strain, when grown on minimal medium, an increase in nsdD transcription could be detected, whereas elevated transcript levels did not change significantly on the complex medium. Levels of the veA transcript did not show any significant regulation with respect to genotype or culture medium.
The GATA-type factor NsdD is required at an early stage of mating and contributes to resistance toward cell wall stress.
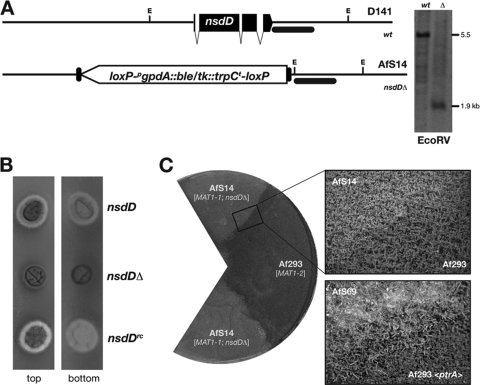
To assess the role of an established determinant of Aspergillus fruiting body formation for A. fumigatus mating, we investigated the GATA-type transcriptional activator NsdD in more detail. Crossing strains that overexpress the nsdD gene (16) on oatmeal agar containing the inducing N source nitrate did not speed up cleistothecium development or skew the balance between asexual and sexual sporulation (data not shown), as it is the case in homothallic A. nidulans (18). In further mating experiments, the Af293 isolate (MAT1-2) was crossed to the mutant strain AfS14 (MAT1-1; nsdDΔ), which is derived from D141 and carries a deletion of the conserved nsdD gene (18), which was introduced by gene targeting and confirmed by Southern hybridization analyses (Fig. 4A). One obvious phenotype of this deletant became evident when the strain was grown on minimal medium, on which reduced hyphal extension and darkening of the colony's reverse side was observed (Fig. 4B). Both phenotypes were revoked by reintroducing the nsdD gene at basal levels from the niaD promoter (16). To test for its ability to mate and to form cleistothecia, AfS14 (nsdDΔ; MAT1-1) was crossed with the MAT1-2 strain Af293 on oatmeal agar. No fruiting bodies could be retrieved from this strain combination when propagated under the validated conditions, even after prolonged incubation for 6 months. When taking a closer look at the barrage zone that is formed between these crossed strains, a difference from pairings with the sterile mating-type deletants became evident: while the latter formed an intimate hyphal mesh from which aerial hyphae emerged, no such structures between AfS14 and Af293 were observed (Fig. 4C). Again, this particular phenotype was reversed when crossing the reconstituted strain AfS69 to a strain of the opposite mating type.
Fig. 4.
The conserved GATA factor NsdD is required for mating. (A) Deletion of nsdD and Southern analysis after EcoRV (E) digestion; recognition sites are schematically indicated together with calculated fragment sizes, and the black bar corresponds to the hybridization probe position. (B) Growth phenotypes of an nsdDΔ strain on minimal medium, characterized by reduced hyphal extension (top view) and darkening of mycelium (bottom view), which is reversed after reconstitution (nsdDrc). (C) Mating capacities of the nsdDΔ deletant. Neither aerial hyphae nor fruiting bodies were formed in the barrage zone (top close-up image) between the deletant AfS14 (nsdDΔ; MAT1-1) and the MAT1-2 strain Af293. For comparison, the barrage zone of the reconstituted strain AfS69 with a MAT1-2 mating-type partner is shown (bottom close-up image), in which aerial hyphae as early indicators of the mating process are formed.
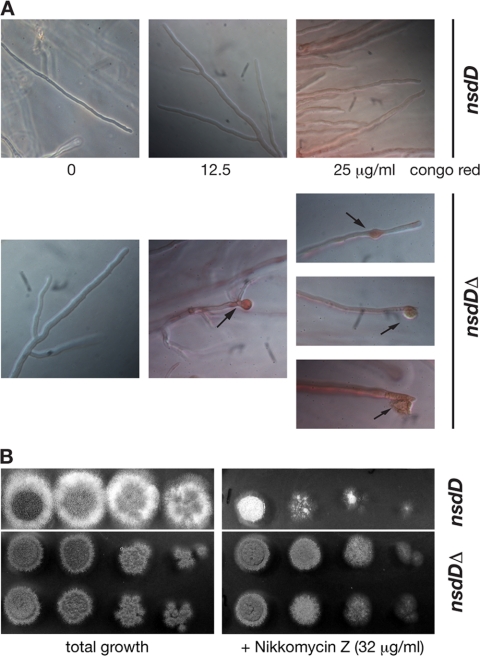
The fact that internucleic complementation of NsdD function is apparently not feasible when crossing the nsdDΔ deletion strain to a wild-type mating partner points to impaired heterokaryon formation, a process tightly linked to hyphal fusion and cell wall remodeling. To test this aspect, the nsdD deletant AfS14 was subjected to the cell wall-interfering agents calcoflour white and Congo red to reveal a pronounced morphological phenotype in dependence on the nsdD gene; whereas the wild-type progenitor displayed straight hyphal extension in the presence of the cell wall stressors, hyphae of the nsdDΔ deletion mutant appeared stunted, sometimes with swollen hyphal tips forming bulky structures that occasionally burst (Fig. 5A). This phenotype was restored to wild type in reconstituted strain AfS69, which carries a plasmid for expression of nsdD and thus displays straight hyphae in the presence of cell wall stress-inducing agents (data not shown). To test any associated phenotype with respect to cell wall synthesis-inhibiting agents, susceptibilities for the antimycotics caspofungin, a β-1,3-glucan synthase inhibitor, and nikkomycin Z, a chitin synthase-inhibiting agent, were analyzed. Etests quantifying the sensitivities of strains D141 and AfS14 toward caspofungin did not uncover any significant differences (data not shown), whereas growth tests on minimal medium plates in the presence of nikkomycin Z revealed an increase in resistance of the nsdDΔ deletant toward this antibiotic (Fig. 5B). This was also reflected in liquid cultures when increasing concentrations of nikkomycin Z were applied: mutants deleted for the nsdD gene were able to germinate and grow profoundly, accompanied by conidiation at concentrations of up to 2,048 μg/ml, in contrast to the wild-type isolate D141 or the reconstituted strain AfS69, which were able to germinate at 512 μg/ml and conidiated at a concentration of 8 μg/ml or less.
Fig. 5.
NsdD is involved cell wall stress resistance. (A) An nsdDΔ strain displays hyphal tip swelling (arrows) in the presence of Congo red, which occasionally results in bursting and the apparent release of hyphal contents. (B) The nsdD deletion mutant is less sensitive toward nikkomycin Z. Aliquots of 5 μl from serial dilutions of conidial suspensions (4 × 106 to 4 × 103 ml−1) were spotted on minimal medium containing 32 μg/ml nikkomycin Z and allowed to grow at 37°C for 2 days. Two independent nsdDΔ isolates are shown.
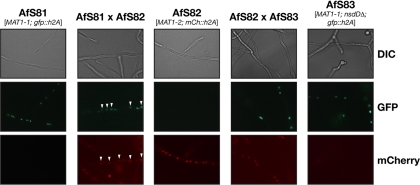
In summary, these data indicate that the GATA-type transcription factor NsdD plays a role in cell wall synthesis and that it is required during an early stage of mating. Accordingly, it might influence initial cellular processes, such as formation of anastomoses resulting in heterokaryosis. In order to support this hypothesis we assessed the ability of the nsdDΔ strain AfS14 to form heterokarya when cocultured with a mating partner by HAMF (42). Nuclei of AfS14 and its parental strain D141 were labeled with the green fluorescent protein by constitutively expressing a GFP-H2A fusion protein, whereas strain Af293 was accordingly labeled with the red fluorescent variant mCherry. Strains of opposite mating types were inoculated in minimal medium, and the formation of heterokaryotic hyphae that contained both types of fluorescent nuclei was monitored (Fig. 6). Either strain alone displayed patterns of uniformly fluorescing nuclei. When scanning the pairing of the D141 descendant AfS81 with the Af293 derivative AfS82, few hyphae could be spotted in which both kinds of nuclei were present. In comparison, the mixture of AfS82 with the nsdDΔ deletion strain AfS83, which expresses the GFP-H2A allele, exclusively yielded homokaryotic hyphae, indicating that deletion of the nsdD gene impairs heterokaryon formation.
Fig. 6.
NsdD is required for heterokaryon formation as assessed by HAMF. Strains of opposite mating types that express fluorescent protein-tagged histone 2A variants were cocultivated to spot mycelial compartments that contained labeled nuclei of either type and, in the case of heterokaryon formation, both kinds. When derivatives of the wild-type strains D141 and Af293 were coincubated, hyphae with both fluorescently labeled nuclei could be identified (AfS81 × AfS82; white arrowheads), whereas exclusively uniform hyphae were detected when a labeled nsdDΔ strain was crossed to the Af293 descendant.
DISCUSSION
We were able to demonstrate mating capacities of unrelated clinical A. fumigatus strains. Possible differences between environmental and clinical isolates of this saprophytic pathogen have been under debate for a long time (3, 9, 10, 17, 29, 43); as with most studies, our finding argues against such a difference, as we could validate that clinical specimens were able to mate and produce fruiting bodies containing recombinant ascospores in a similar manner as the environmental isolates described in the seminal study of O'Gorman et al. (33). Accordingly, the mating behavior and the existence of a sexual phase in the life cycle of A. fumigatus are not restricted to a locally constrained subpopulation but appear to be a general trait. However, Kwon-Chung and Sugui recently described their failed efforts to mate the reference strain Af293 to other established isolates, such as CE10 or B-5233 (30), and further and more comprehensive studies will have to answer the question of how universal mating is among the A. fumigatus population. Nutritional requirements of mating partners might have an influence, as they have been described to hamper cleistothecium formation of the homothallic relative A. nidulans (13, 26). The fact that we were able to produce recombinant offspring from two genetically marked strains indicates that at least the employed auxotrophies for riboflavin and pyridoxin do not influence fertility of A. fumigatus or its ability to produce fruiting bodies. One additional aspect of our mating studies lies in the fact that cleistothecia were already formed after 4 months, which is 2 months sooner than described for the Irish isolates. Given that even in this collection variations in the abilities to mate and produce fruiting bodies were evident, the quicker formation of cleistothecia in our experimental setup seems reasonable. Mating of N. fumigata appears to be determined by a variety of genetic determinants and results in differing characteristics of fruiting body formation between different isolates.
A. fumigatus is not the only anamorphic fungus for which a long-time-undisclosed sexual cycle has recently been demonstrated. Aspergillus parasiticus as well as A. flavus produces recombinant progeny in fruiting bodies within several months when crossed under appropriate conditions (20–22), and for several other fungal species a bipolar mating-type system or even the existence of mating and/or sexual reproduction was confirmed in recent times (28). From a technical point of view, mating competence of the A. fumigatus sequence reference strain Af293 assists molecular characterization of its heterothallic breeding system, as mutant generation by means of gene targeting is facilitated in such a defined and established isolate. The existence of isogenic auxotrophs, for instance, in this genetic background (54) will support the generation of recombinant strains and will surely contribute to molecular biology of this fungal pathogen (7, 24). However, given the long period of time that is required to obtain mature cleistothecia harboring recombinant ascospores from N. fumigata, further studies are mandatory to enhance feasibility of genetic crosses for routine research purposes. Molecular determinants that support mating and sexual differentiation of A. fumigatus need to be defined, and moreover, any relevance of its restricted sexuality for virulence has to be addressed (31).
Mating-type loci and their products are considered master regulators of sexual propagation, especially in heterothallic species. MAT genes determine sexual (and probably nuclear) identity, and as DNA binding factors they do so by orchestrating transcriptional programs in the respective cellular compartment. The generated deletion strains confirm a presumed role of both mating-type loci in A. fumigatus sexuality, as cleistothecium formation was blocked in compatible crosses of mutant strains. This situation clearly resembles findings for other heterothallic and also homothallic ascomycetes (11, 36) and emphasizes the role of MAT gene products as master regulators for sexual development. We have gained initial information on the regulatory targets by quantitative analyses of gene transcript levels putatively involved in pheromone signaling and regulation of fruiting body formation. Most interestingly, we were able to demonstrate a role of both Mat proteins in regulation of pheromone expression: whereas Mat1-1 is required for proper ppgA gene transcription, Mat1-2 appears to repress its expression. The latter finding is in line with data from the homothallic species A. nidulans, for which a negative influence of the HMG protein MatA on ppgA transcript levels has been suggested (36). Our finding that ppgA transcription is regulated in a reciprocal manner in the different mating-type backgrounds illustrates the identity-establishing function of the Mat proteins and might assist in identifying the so-far-ambiguous ppgB pheromone-encoding gene. Both putative pheromone receptor genes, however, appear to be regulated by the MAT loci in a similar fashion, being less transcribed in deletants grown on minimal medium but stably transcribed when grown on oatmeal agar. This pattern reveals a pronounced effect of the mating-inducing growth substrate but also implies balanced fine-tuning of receptor expression. The preA or preB transcript levels are similar in either mating-type deletion strain irrespective of the culture medium, which indicates that environmental regulation of pheromone receptor expression is in part mediated by the mating-type factors. We were unable to detect any significant regulation of the MAT gene products on veA transcription, an established regulator of Aspergillus cleistothecium formation, which suggests that this factor acts upstream of or parallel to any Mat1-directed signal transduction. For the mat1-2Δ deletion strain, an increase in nsdD transcript levels was evident on minimal medium; any significance of this negative regulation of NsdD expression by Mat1-2 for sexual development, however, remains to be demonstrated. Our quantitative data demonstrate that expression profiles per se do not mirror the capacity to propagate sexually: transcript levels of genes involved in the mating process were elevated on minimal medium, which does not support cleistothecium formation, but appeared to be downregulated on the complex and conducive oatmeal culture medium. The differing patterns of transcription reflect the completely dissimilar culture conditions (synthetic minimal versus complex rich medium), a situation that apparently is overcome by an unknown oatmeal component to support cleistothecium formation.
We and others had shown previously that functional determinants of sexual development in the homothallic relative A. nidulans are expressed in A. fumigatus, among them the GATA-type factor NsdD (18). Phenotypes of the nsdDΔ deletant AfS14 reveal conserved functions of this transcription factor in both species, illustrated by accumulation of dark mycelial pigments and the absence of sexual development. Our finding that this conserved activator of cleistothecium formation appears to be required for mating in A. fumigatus and that crosses of the deletant with a compatible wild-type isolate do not result in fruiting bodies implies that it plays a role in early steps of heterothallic reproduction. This reflects the situation in homothallic A. nidulans, in which an early role for NsdD to act on initiation of fruiting body formation has been proposed (47). Compatible fertile strains usually fuse hyphae at the zone of confrontation. The presence of mixed nuclei within the resulting heterokaryon would allow intergenic complementation and therefore fruiting body formation in an nsdDΔ × nsdD+ cross. The observation that this is not the case implies a defect in anastomosis for this strain combination. Alternatively, other cellular processes might require NsdD in either mating partner, or levels of NsdD expression might not be sufficient to support fruiting body formation. Our data from histone-assisted merged fluorescence reveal a defect in hyphal fusion of the nsdDΔ deletion mutant. In line with this is the observed sensitivity against cell wall stressors and apparently weakened hyphal tips of the nsdDΔ deletant. This particular phenotype is in accordance with the one observed when nsdD is overexpressed, which results in hyphal curling (16). Hence, NsdD appears to be necessary for balanced cell wall synthesis and may act on proper expression of proteins involved in this fundamental process, such as chitin synthases, α-1,3-glucan synthases, or cell wall mannoproteins. The fact that sensitivity of A. fumigatus toward the antimycotic nikkomycin Z, an inhibitor of chitin synthesis, is altered when NsdD is lacking supports this supposed role of the regulator in cell wall maintenance. Accordingly, NsdD appears to affect the cell wall integrity pathway and thus might be required for hyphal fusion events, during which cell wall remodeling has to occur. The scope of the NsdD-dependent regulon, however, awaits determination by comprehensive analyses of its transcriptome to provide further information on the cellular function of this regulator of Aspergillus sexual development.
In summary, we were able to demonstrate that determinants of Aspergillus mating and fruiting body formation, mainly characterized in the homothallic species A. nidulans, are required for A. fumigatus mating and are therefore functionally conserved in this heterothallic species.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of YIG2 and of the Institute for Molecular Infection Biology for support, suggestions, and discussions. We are indebted to Thomas Hartmann for indispensable help with expression analyses, and highly valuable technical assistance was carried out by Michaela Dümig. Ulrich Kück, Birgit Hoff, and David Löper from the University of Bochum are thanked for stimulating and helpful discussions.
Financial support was granted by the German Research Foundation, the Free State of Bavaria, and the University of Würzburg.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Aarskog N. K., Vedeler C. A. 2000. Real-time quantitative polymerase chain reaction. A new method that detects both the peripheral myelin protein 22 duplication in Charcot-Marie-Tooth type 1A disease and the peripheral myelin protein 22 deletion in hereditary neuropathy with liability to pressure palsies. Hum. Genet. 107:494–498 [DOI] [PubMed] [Google Scholar]
- 2.Adams T. H., Wieser J. K., Yu J. H. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aufauvre-Brown A., Brown J. S., Holden D. W. 1998. Comparison of virulence between clinical and environmental isolates of Aspergillus fumigatus. Eur. J. Clin. Microbiol. Infect. Dis. 17:778–780 [DOI] [PubMed] [Google Scholar]
- 4.Barratt R. W., Johnson G. B., Ogata W. N. 1965. Wild-type and mutant stocks of Aspergillus nidulans. Genetics 52:233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayram Ö., Krappmann S., Ni M., Bok J. W., Helmstaedt K., Valerius O., Braus-Stromeyer S., Kwon N. J., Keller N. P., Yu J. H., Braus G. H. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506 [DOI] [PubMed] [Google Scholar]
- 6.Bergmann A., Hartmann T., Cairns T., Bignell E. M., Krappmann S. 2009. A regulator of Aspergillus fumigatus extracellular proteolytic activity is dispensable for virulence. Infect. Immun. 77:4041–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brakhage A. A., Langfelder K. 2002. Menacing mold: the molecular biology of Aspergillus fumigatus. Annu. Rev. Microbiol. 56:433–455 [DOI] [PubMed] [Google Scholar]
- 8.Braus G. H., Krappmann S., Eckert S. E. 2002. Sexual development in ascomycetes: fruit body formation of Aspergillus nidulans, p. 215–244 InOsiewacz H. D. (ed.), Molecular biology of fungal development Marcel Dekker, Inc., New York, NY [Google Scholar]
- 9.Chazalet V., Debeaupuis J. P., Sarfati J., Lortholary J., Ribaud P., Shah P., Cornet M., Vu Thien H., Gluckman E., Brucker G., Latgé J.-P. 1998. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J. Clin. Microbiol. 36:1494–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debeaupuis J. P., Sarfati J., Chazalet V., Latgé J.-P. 1997. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect. Immun. 65:3080–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debuchy R., Turgeon B. G. 2006. Mating-type structure, evolution, and function in Euascomycetes, p. 293–323 InKües U., Fischer R.(ed.), The Mycota I: growth, differentiation and sexuality Springer-Verlag, Berlin, Germany [Google Scholar]
- 12.Dyer P. S., Paoletti M., Archer D. B. 2003. Genomics reveals sexual secrets of Aspergillus. Microbiology 149:2301–2303 [DOI] [PubMed] [Google Scholar]
- 13.Eckert S. E., Hoffmann B., Wanke C., Braus G. H. 1999. Sexual development of Aspergillus nidulans in tryptophan auxotrophic strains. Arch. Microbiol. 172:157–166 [DOI] [PubMed] [Google Scholar]
- 14.Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R., Batzoglou S., Lee S. I., Basturkmen M., Spevak C. C., Clutterbuck J., Kapitonov V., Jurka J., Scazzocchio C., Farman M., Butler J., Purcell S., Harris S., Braus G. H., Draht O., Busch S., D'Enfert C., Bouchier C., Goldman G. H., Bell-Pedersen D., Griffiths-Jones S., Doonan J. H., Yu J., Vienken K., Pain A., Freitag M., Selker E. U., Archer D. B., Penalva M. A., Oakley B. R., Momany M., Tanaka T., Kumagai T., Asai K., Machida M., Nierman W. C., Denning D. W., Caddick M., Hynes M., Paoletti M., Fischer R., Miller B., Dyer P., Sachs M. S., Osmani S. A., Birren B. W. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 15.Geiser D. M. 2009. Sexual structures in Aspergillus: morphology, importance and genomics. Med. Mycol. 47Suppl. 1:S21–S26 [DOI] [PubMed] [Google Scholar]
- 16.Große V., Krappmann S. 2008. The asexual pathogen Aspergillus fumigatus expresses functional determinants of Aspergillus nidulans sexual development. Eukaryot. Cell 7:1724–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guinea J., Pelaez T., Alcala L., Ruiz-Serrano M. J., Bouza E. 2005. Antifungal susceptibility of 596 Aspergillus fumigatus strains isolated from outdoor air, hospital air, and clinical samples: analysis by site of isolation. Antimicrob. Agents Chemother. 49:3495–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han K. H., Han K. Y., Yu J. H., Chae K. S., Jahng K. Y., Han D. M. 2001. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 41:299–309 [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D., Jessee J., Bloom F. R. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63–113 [DOI] [PubMed] [Google Scholar]
- 20.Horn B. W., Moore G. G., Carbone I. 2009. Sexual reproduction in Aspergillus flavus. Mycologia 101:423–429 [DOI] [PubMed] [Google Scholar]
- 21.Horn B. W., Ramirez-Prado J. H., Carbone I. 2009. Sexual reproduction and recombination in the aflatoxin-producing fungus Aspergillus parasiticus. Fungal Genet. Biol. 46:169–175 [DOI] [PubMed] [Google Scholar]
- 22.Horn B. W., Ramirez-Prado J. H., Carbone I. 2009. The sexual state of Aspergillus parasiticus. Mycologia 101:275–280 [DOI] [PubMed] [Google Scholar]
- 23.Kolar M., Punt P. J., van den Hondel C. A., Schwab H. 1988. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62:127–134 [DOI] [PubMed] [Google Scholar]
- 24.Krappmann S. 2006. Tools to study molecular mechanisms of Aspergillus pathogenicity. Trends Microbiol. 14:356–364 [DOI] [PubMed] [Google Scholar]
- 25.Krappmann S., Bayram Ö., Braus G. H. 2005. Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot. Cell 4:1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krappmann S., Braus G. H. 2003. Deletion of Aspergillus nidulans aroC using a novel blaster module that combines ET cloning and marker rescue. Mol. Genet. Genomics 268:675–683 [DOI] [PubMed] [Google Scholar]
- 27.Krappmann S., Jung N., Medic B., Busch S., Prade R. A., Braus G. H. 2006. The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis. Mol. Microbiol. 61:76–88 [DOI] [PubMed] [Google Scholar]
- 28.Kück U., Pöggeler S. 2009. Cryptic sex in fungi. Fungal Biol. Rev. 23:86–90 [Google Scholar]
- 29.Kupfahl C., Michalka A., Lass-Flörl C., Fischer G., Haase G., Ruppert T., Geginat G., Hof H. 2008. Gliotoxin production by clinical and environmental Aspergillus fumigatus strains. Int. J. Med. Microbiol. 298:319–327 [DOI] [PubMed] [Google Scholar]
- 30.Kwon-Chung K. J., Sugui J. A. 2009. Sexual reproduction in Aspergillus species of medical or economical importance: why so fastidious? Trends Microbiol. 17:481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen K., Heitman J. 2007. Sex and virulence of human pathogenic fungi. Adv. Genet. 57:143–173 [DOI] [PubMed] [Google Scholar]
- 32.Nierman W. C., Pain A., Anderson M. J., Wortman J. R., Kim H. S., Arroyo J., Berriman M., Abe K., Archer D. B., Bermejo C., Bennett J., Bowyer P., Chen D., Collins M., Coulsen R., Davies R., Dyer P. S., Farman M., Fedorova N., Feldblyum T. V., Fischer R., Fosker N., Fraser A., Garcia J. L., Garcia M. J., Goble A., Goldman G. H., Gomi K., Griffith-Jones S., Gwilliam R., Haas B., Haas H., Harris D., Horiuchi H., Huang J., Humphray S., Jimenez J., Keller N., Khouri H., Kitamoto K., Kobayashi T., Konzack S., Kulkarni R., Kumagai T., Lafon A., Latgé J.-P., Li W., Lord A., Lu C., Majoros W. H., May G. S., Miller B. L., Mohamoud Y., Molina M., Monod M., Mouyna I., Mulligan S., Murphy L., O'Neil S., Paulsen I., Penalva M. A., Pertea M., Price C., Pritchard B. L., Quail M. A., Rabbinowitsch E., Rawlins N., Rajandream M. A., Reichard U., Renauld H., Robson G. D., Rodriguez de Cordoba S., Rodriguez-Pena J. M., Ronning C. M., Rutter S., Salzberg S. L., Sanchez M., Sanchez-Ferrero J. C., Saunders D., Seeger K., Squares R., Squares S., Takeuchi M., Tekaia F., Turner G., Vazquez de Aldana C. R., Weidman J., White O., Woodward J., Yu J. H., Fraser C., Galagan J. E., Asai K., Machida M., Hall N., Barrell B., Denning D. W. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 33.O'Gorman C. M., Fuller H. T., Dyer P. S. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474 [DOI] [PubMed] [Google Scholar]
- 34.Pain A., Woodward J., Quail M. A., Anderson M. J., Clark R., Collins M., Fosker N., Fraser A., Harris D., Larke N., Murphy L., Humphray S., O'Neil S., Pertea M., Price C., Rabbinowitsch E., Rajandream M. A., Salzberg S., Saunders D., Seeger K., Sharp S., Warren T., Denning D. W., Barrell B., Hall N. 2004. Insight into the genome of Aspergillus fumigatus: analysis of a 922 kb region encompassing the nitrate assimilation gene cluster. Fungal Genet. Biol. 41:443–453 [DOI] [PubMed] [Google Scholar]
- 35.Paoletti M., Rydholm C., Schwier E. U., Anderson M. J., Szakacs G., Lutzoni F., Debeaupuis J. P., Latge J.-P., Denning D. W., Dyer P. S. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15:1242–1248 [DOI] [PubMed] [Google Scholar]
- 36.Paoletti M., Seymour F. A., Alcocer M. J., Kaur N., Calvo A. M., Archer D. B., Dyer P. S. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17:1384–1389 [DOI] [PubMed] [Google Scholar]
- 37.Pöggeler S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 42:153–160 [DOI] [PubMed] [Google Scholar]
- 38.Pontecorvo G., Roper J. A., Hemmons L. M., Macdonald K. D., Bufton A. W. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141–238 [DOI] [PubMed] [Google Scholar]
- 39.Pringle A., Baker D. M., Platt J. L., Wares J. P., Latgé J.-P., Taylor J. W. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886–1899 [PubMed] [Google Scholar]
- 40.Punt P. J., van den Hondel C. A. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216:447–457 [DOI] [PubMed] [Google Scholar]
- 41.Pyrzak W., Miller K. Y., Miller B. L. 2008. Mating type protein Mat1-2 from asexual Aspergillus fumigatus drives sexual reproduction in fertile Aspergillus nidulans. Eukaryot. Cell 7:1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rech C., Engh I., Kück U. 2007. Detection of hyphal fusion in filamentous fungi using differently fluorescence-labeled histones. Curr. Genet. 52:259–266 [DOI] [PubMed] [Google Scholar]
- 43.Rosehart K., Richards M. H., Bidochka M. J. 2002. Microsatellite analysis of environmental and clinical isolates of the opportunist fungal pathogen Aspergillus fumigatus. J. Med. Microbiol. 51:1128–1134 [DOI] [PubMed] [Google Scholar]
- 44.Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. 1985. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350–1354 [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46.Scazzocchio C. 2006. Aspergillus genomes: secret sex and the secrets of sex. Trends Genet. 22:521–525 [DOI] [PubMed] [Google Scholar]
- 47.Seo J. A., Han K. H., Yu J. H. 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53:1611–1623 [DOI] [PubMed] [Google Scholar]
- 48.Sohn K. T., Yoon K. S. 2002. Ultrastructural study on the cleistothecium development in Aspergillus nidulans. Mycobiology 30:117–127 [Google Scholar]
- 49.Southern E. 2006. Southern blotting. Nat. Protoc. 1:518–525 [DOI] [PubMed] [Google Scholar]
- 50.Staib F., Mishra S. K., Rajendran C., Voigt R., Steffen J., Neumann K. H., Hartmann C. A., Heins G. 1980. A notable Aspergillus from a mortal aspergilloma of the lung. New aspects of the epidemiology, serodiagnosis and taxonomy of Aspergillus fumigatus. Zentralbl. Bakteriol. A 247:530–536 [PubMed] [Google Scholar]
- 51.Su W., Li S., Oakley B. R., Xiang X. 2004. Dual-color imaging of nuclear division and mitotic spindle elongation in live cells of Aspergillus nidulans. Eukaryot. Cell 3:553–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szewczyk E., Nayak T., Oakley C. E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S. A., Oakley B. R. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 53.Varga J. 2003. Mating type gene homologues in Aspergillus fumigatus. Microbiology 149:816–819 [DOI] [PubMed] [Google Scholar]
- 54.Xue T., Nguyen C. K., Romans A., Kontoyiannis D. P., May G. S. 2004. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch. Microbiol. 182:346–353 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.