Abstract
The filamentous fungus Alternaria alternata includes seven pathogenic variants (pathotypes) which produce different host-selective toxins and cause diseases on different plants. The Japanese pear pathotype produces the host-selective AK-toxin, an epoxy-decatrienoic acid ester, and causes black spot of Japanese pear. Previously, we identified four genes, AKT1, AKT2, AKT3, and AKTR, involved in AK toxin biosynthesis. AKT1, AKT2, and AKT3 encode enzyme proteins with peroxisomal targeting signal type 1 (PTS1)-like tripeptides, SKI, SKL, and PKL, respectively, at the C-terminal ends. In this study, we verified the peroxisome localization of Akt1, Akt2, and Akt3 by using strains expressing N-terminal green fluorescent protein (GFP)-tagged versions of the proteins. To assess the role of peroxisome function in AK-toxin production, we isolated AaPEX6, which encodes a peroxin protein essential for peroxisome biogenesis, from the Japanese pear pathotype and made AaPEX6 disruption-containing transformants from a GFP-Akt1-expressing strain. The ΔAaPEX6 mutant strains did not grow on fatty acid media because of a defect in fatty acid β oxidation. The import of GFP-Akt1 into peroxisomes was impaired in the ΔAaPEX6 mutant strains. These strains completely lost AK toxin production and pathogenicity on susceptible pear leaves. These data show that peroxisomes are essential for AK-toxin biosynthesis. The ΔAaPEX6 mutant strains showed a marked reduction in the ability to cause lesions on leaves of a resistant pear cultivar with defense responses compromised by heat shock. This result suggests that peroxisome function is also required for plant invasion and tissue colonization in A. alternata. We also observed that mutation of AaPEX6 caused a marked reduction of conidiation.
Peroxisomes are single-membrane-bound organelles and have a wide range of metabolic functions, including β oxidation of fatty acids, peroxide detoxification, and glyoxylate metabolism (60, 62). Peroxisome biogenesis has been extensively studied using yeast mutants, and a number of PEX genes that encode peroxins have been identified (48, 60, 62). Peroxins are proteins required for peroxisome biogenesis and division and for import of proteins into the peroxisome matrix (48, 60). Most peroxisome matrix proteins contain peroxisomal targeting signal type 1 (PTS1), the tripeptide sequence (S/A/C)-(K/R/H)-L, at the C terminus (9). Other matrix proteins have PTS2 sequences close to the N terminus; the consensus PTS2 sequence is (R/K)-(L/V/I)-X5-(H/Q)-(L/A) (29).
Peroxisomes are required for specific functions in filamentous fungi. In Penicillium chrysogenum, peroxisomes participate in penicillin biosynthesis: acyl coenzyme A (acyl-CoA):isopenicillin N acyltransferase, which catalyzes the final step of penicillin biosynthesis, localizes to peroxisomes (38, 39). In the cucumber anthracnose pathogen Colletotrichum orbiculare (synonym, C. lagenarium) and the rice blast pathogen Magnaporthe oryzae (synonym, M. grisea), peroxisome function is necessary for plant infection (6, 8, 25, 49, 65, 66). These pathogens produce specific infection structures, called appressoria, that are used to penetrate the host plant cuticle using mechanical force (61). Their dome-shaped appressoria are darkly pigmented with dihydroxynaphthalene melanin, which is essential for the function of the appressoria. In these pathogens, peroxisome function is involved in appressorium maturation with accumulation of melanin and initial host invasion via appressoria (6, 8, 25, 49, 65, 66).
The imperfect fungus Alternaria alternata is one of the most cosmopolitan fungal species and is generally saprophytic (28, 50, 58). This species, however, does include seven pathogenic variants (pathotypes) which produce different host-selective toxins and cause necrotic diseases on different plants (28, 58). Host-selective toxins produced by fungal plant pathogens are generally low-molecular-weight secondary metabolites and are critical determinants of host-specific pathogenicity or virulence in several plant-pathogen interactions (17, 28, 58, 68).
The Japanese pear pathotype of A. alternata produces AK-toxin (Fig. 1) and causes black spot on a narrow range of susceptible Japanese pear cultivars, including the commercially important cultivar Nijisseiki (41, 45). We previously isolated the gene cluster involved in AK-toxin biosynthesis from the Japanese pear pathotype and identified four genes, AKT1, AKT2, AKT3, and AKTR (55, 56). AKTR encodes a transcription regulator of the Zn(II)2Cys6 family (56). AKT1, AKT2, and AKT3 are predicted to encode proteins with similarity to the carboxyl-activating enzymes, the estelase-lipase family enzymes, and the hydratase-isomerase family enzymes, respectively (55, 56). A PSORT II (40) analysis of the amino acid sequences of Akt1, Akt2, and Akt3 identified PTS1-like tripeptides SKI, SKL, and PKL, respectively, at the C-terminal ends of these proteins, suggesting that these enzymes are located in peroxisomes (55, 56).
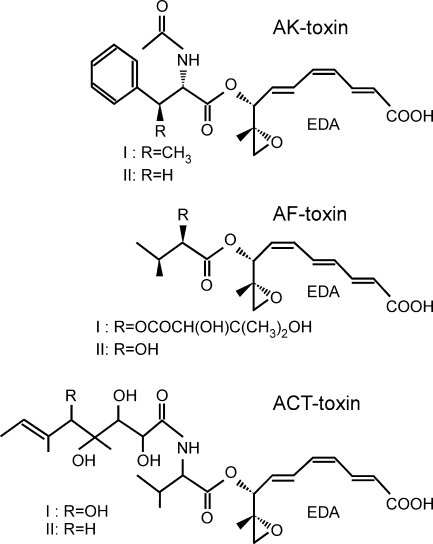
Fig. 1.
Host-selective toxins produced by three pathotypes of A. alternata. AK-toxins of the Japanese pear pathotype (41), AF-toxins of the strawberry pathotype (43), and ACT-toxins of the tangerine pathotype (27) have a moiety, EDA, in common.
Orthologs of AKT1, AKT2, AKT3, and AKTR are present in the strawberry and tangerine pathotypes of A. alternata (15, 33, 35, 55, 56). The strawberry pathotype produces AF-toxin (Fig. 1) and causes Alternaria black spot of strawberry (31, 43). The tangerine pathotype produces ACT-toxin (Fig. 1) and causes brown spot of tangerines and mandarins, a disease that has not yet occurred in Japan (27). The toxins of Japanese pear, strawberry, and tangerine pathotypes have a 9,10-epoxy-8-hydroxy-9-methyl-decatrienoic acid (EDA) structural moiety in common (Fig. 1) (27, 41, 43). The AKT orthologs were isolated from the strawberry and tangerine pathotypes and named the AFT and ACTT genes, respectively (15, 33, 35). AKT, AFT, ACTT, and their respective orthologs show more than 90% nucleotide identity, suggesting that these genes encode the enzymes for the biosynthesis of the common precursor, EDA. The PTS1-like tripeptides are also conserved in the orthologous enzymes from the strawberry and tangerine pathotypes (15, 33, 35). EDA is biosynthesized by the condensation of six molecules of acetic acid, followed by modifications including reduction, dehydration, and decarboxylation (42). It is likely that Akt1, Akt2, and Akt3 are involved in modifications of the acetyl-CoA-derived backbone of the EDA molecule. Given the predicted PTS1-like tripeptides of these orthologs, we hypothesize that peroxisomes are involved in the biosynthesis of the AK-, AF-, and ACT-toxins.
Here we report the role of peroxisomes in AK toxin biosynthesis and pathogenicity in the Japanese pear pathotype. Peroxisome localization of Akt1, Akt2, and Akt3 was verified using strains expressing each of these proteins fused to N-terminal green fluorescent protein (GFP) tags. We isolated AaPEX6 with high similarity to fungal and yeast PEX6, which encodes a peroxin protein essential for peroxisome biogenesis in eukaryotic cells (48, 60), from the Japanese pear pathotype. The strains in which AaPEX6 is disrupted completely lost the production of EDA and AK-toxin, resulting in loss of pathogenicity on host pear leaves. These results demonstrate that peroxisomes are required for the biosynthesis of EDA, a precursor of AK-toxin. In both C. orbiculare and M. oryzae, the strains in which AaPEX6 is disrupted form nonfunctional appressoria that fail to form infection hyphae (25, 49, 65). Similarly, we demonstrated that peroxisome function is also involved in plant invasion and colonization by A. alternata.
MATERIALS AND METHODS
Fungal strains and genomic library.
Strain 15A of the Japanese pear pathotype of A. alternata and its transformants were used in this study and routinely maintained on potato dextrose agar (PDA; Difco, Detroit, MI). A genomic cosmid library of strain 15A has been previously described (26).
Plasmids.
The plasmids used in this study are listed in Table 1. The integrative transformation vectors pSH75 (26), pII99 (44), and pSB116 were used for the transformation of A. alternata. The vectors pSH75 and pII99 carry hph and nptII, respectively, fused to the Aspergillus nidulans trpC promoter and terminator (26, 37, 44). The vector pSB116 was made by replacing the trpC promoter and hph region in pSH75 with a 1.0-kb fragment containing the trpC promoter and the bar open reading frame (ORF) from pBIG4MRBrev (57).
Table 1.
Plasmids used in this study
| Plasmid | Constructiona | Source |
|---|---|---|
| pSH75 | trpCp-hph-trpCt | 26 |
| pII99 | trpCp-nptII-trpCt | 44 |
| pSB116 | trpCp-bar-trpCt | This study |
| pYTGFPc | trpCp-GFP-trpCt | 19 |
| pYTGFP-N | trpCp-GFP-trpCt | 19 |
| pGFP-AKT1 | trpCp-GFP-AKT1-trpCt | This study |
| pGFP-AKT2 | trpCp-GFP-AKT2-trpCt | This study |
| pGFP-AKT3 | trpCp-GFP-AKT3-1-trpCt | This study |
| pGFP-AKT1Δ | trpCp-GFP-AKT1ΔPTS1-trpCt | This study |
| pGFP-AKT2Δ | trpCp-GFP-AKT2ΔPTS1-trpCt | This study |
| pGFP-AKT3Δ | trpCp-GFP-AKT3-1ΔPTS1-trpCt | This study |
| pcAaPEX6 | Cosmid containing AaPEX6 | This study |
| pDNR-PEX6 | Plasmid containing AaPEX6 (6.5 kb) | This study |
| pGDPEX6n | 5′AaPEX6::trpCp-nptII-trpCt::3′AaPEX6 | This study |
trpCp and trpCt, A. nidulans trpC promoter and terminator, respectively (37); AKT1ΔPTS1, AKT2ΔPTS1, and AKT3-1ΔPTS1, lacking nine nucleotides encoding C-terminal PTS1-like tripeptides.
The GFP expression vector pYTGFP-N was used to make a GFP N-terminal fusion vector (see Fig. S1 in the supplemental material). The vector pYTGFP-N contains the GFP (enhanced GFP) ORF fused to the A. nidulans trpC promoter and terminator (19). Because the trpC promoter is constitutively active in A. alternata, we expected it to be useful for expressing the target genes under any culture conditions and in any fungal structures. The AKT1, AKT2, and AKT3-1 cDNAs were amplified from the total RNA of strain 15A with primer pairs AKT1-f/AKT1-r, AKT2-f/AKT2-r, and AKT3-f/AKT3-r (see Table S1 in the supplemental material), respectively, using the RNA PCR kit ver. 2.1 (Takara Bio, Ohtsu, Japan), digested with restriction enzymes, and ligated into pYTGFP-N to make pGFP-AKT1, pGFP-AKT2, and pGFP-AKT3, respectively (see Fig. S1 in the supplemental material). Strain 15A was found to have at least two copies of AKT3 (AKT3-1 and AKT3-2) in its genome (56). The two genes are predicted to encode proteins with 94% amino acid sequence identity and with the same PTS1-like tripeptide, PKL, at their respective C-terminal ends (56). In this study, we used AKT3-1 to make pGFP-AKT3. The AKT1, AKT2, and AKT3-1 cDNAs lacking the nine nucleotides that encode PTS1 were amplified from pGFP-AKT1, pGFP-AKT2, and pGFP-AKT3, respectively, with primer pairs AKT1-f/AKT1-r2, AKT2-f/AKT2-r2, and AKT3-f/AKT3-r2 (see Table S1 in the supplemental material), digested with restriction enzymes, and ligated into pYTGFP-N to make pGFP-AKT1Δ, pGFP-AKT2Δ, and pGFP-AKT3Δ, respectively (see Fig. S1 in the supplemental material).
All of the PCR products cloned into the vectors were sequenced to confirm that no nucleotide substitution had occurred during amplification.
Fungal transformation.
Protoplast preparation and transformation of A. alternata were performed as previously described (20). Transformants carrying hph or nptII were selected on regeneration medium (26) containing hygromycin B (Wako Pure Chemicals, Osaka, Japan) at 100 μg/ml or Geneticin (Gibco BRL, Life Technology, Gaithersburg, MD) at 400 μg/ml (20). Protoplasts transformed with pSB116 carrying the bar cassette were regenerated on minimal agar medium (MAM) (52) supplemented with 1 M sucrose and 50 μg/ml bialaphos (Meiji Seika Kaisha, Ltd., Tokyo, Japan).
DNA and RNA manipulation.
Isolation of total DNA and RNA from A. alternata and DNA gel blot hybridization were performed as previously described (55). For analysis of nucleotide sequences, DNA was cloned into pBluescript KS+ (Stratagene, La Jolla, CA) or pGEM-T Easy (Promega, Madison, WI). DNA sequences were determined with the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom) and an automated fluorescent DNA sequencer ABI PRISM 3100 genetic analyzer (Applied Biosystems). DNA sequences were analyzed with BLAST (4). Alignment of nucleotide and amino acid sequences was done with the CLUSTAL W program (59).
Isolation of AaPEX6.
The AaPEX6 fragment was amplified from the total DNA of strain 15A by PCR using the primer pair PEX6-f/PEX6-r (see Table S1 in the supplemental material) and Taq DNA polymerase (Takara Bio). These primers were designed using the conserved regions of PEX6 genes from P. chrysogenum (24), C. orbiculare (25), and Saccharomyces cerevisiae (64). PCR products of the expected size (∼420 bp) were cloned into the pGEM-T Easy vector and found to encode a peptide with strong similarity to the corresponding regions of Pex6 proteins. This PCR product was used as a probe for the screening of a genomic cosmid library of strain 15A, and a positive clone, named pcAaPEX6, was isolated. A 6.8-kb region in pcAaPEX6 was sequenced, and a putative ORF of AaPEX6 was identified. The AaPEX6 cDNA was amplified from the total RNA of strain 15A with primer pair PEX6-6/PEX6-9 (see Table S1 in the supplemental material) and cloned into the pGEM-T Easy vector to determine the sequence.
The entire AaPEX6 gene was cloned into pDNR-CMV (Clontech, Mountain View, CA) with the In-Fusion Dry-Down PCR cloning kit (Clontech). The 6.5-kb fragment which includes all of the exons and introns of AaPEX6 and the 1.6-kb upstream and 0.5-kb downstream regions was amplified from pcAaPEX6 DNA by PCR using primer pair AaPEX6-f/AaPEX6-r (see Table S1 in the supplemental material). The In-Fusion cloning reaction and transformation in Escherichia coli Fusion-Blue competent cells (Clontech) yielded plasmid pDNR-PEX6. The AaPEX6-targeting vector pGDPEX6n was prepared by replacing a 3.2-kb BamHI-EcoRI fragment within AaPEX6 in pDNR-PEX6 with a 1.9-kb BamHI-EcoRI fragment of the nptII cassette.
Microscopy.
Transformants with GFP fusion constructs were grown for 3 days at 25°C on glass slides covered with a thin layer of potato agar medium supplemented with glucose or oleic acid. Infection-related morphogenesis of transformants was observed on onion epidermis (10). Sections of onion epidermis (about 1 cm2) floating on sterilized water were separately inoculated with drops (10 μl) of each conidial suspension (about 2 × 104 conidia/ml) and incubated at 25°C for 24 h. Confocal laser scanning fluorescence images of fungal structures were recorded on an LSM510 confocal system (Carl Zeiss, Inc., Göttingen, Germany) with a 64× numerical aperture 1.0 oil immersion lens. A krypton-argon laser was used as the source of excitation at 488 nm, and GFP fluorescence was recorded at 505 nm. The images were stored as TIF files and processed with Canvas X software (ACD Systems of America, Inc., Miami, FL).
Assay for vegetative growth and conidiation.
To test for vegetative growth, strains were grown on PDA at 25°C for 4 days. Agar blocks (3 mm in diameter) carrying mycelia were prepared from the resulting colonies and inoculated onto PDA and MAM supplemented with one carbon source, 1% glucose, 0.5% oleic acid, or 0.5% Tween 80. After incubation at 25°C for 5 days, colony growth was observed.
To test for conidiation, strains were grown on oatmeal sucrose agar at 25°C for 10 days and induced for conidiation as previously described (14, 22).
Assay for infection-related morphogenesis, AK-toxin production, and pathogenicity.
Conidial germination and appressorium formation were observed on glass. Conidial suspensions (about 1 × 105 conidia/ml) were dropped onto glass microscope slides and incubated in a moist box at 25°C for 24 h. The frequencies of germinated conidia and germinated conidia forming appressoria were measured using differential interference contrast (DIC) microscopy. In each experiment, at least 100 conidia were examined. The means and standard deviations were calculated from four independent experiments.
The ability to penetrate intact plant epidermal cells on onion epidermis was investigated (10). The frequency of appressoria forming penetration hyphae was measured using DIC microscopy. In each experiment, at least 100 appressoria were examined. The means and standard deviations were calculated from four independent experiments.
To test for AK-toxin production, strains were grown statically in 5 ml of potato dextrose broth (PDB; Difco) in test tubes at 25°C for 7 days. To assay culture filtrates for toxicity, culture filtrates were dropped onto wounded sites of leaves of susceptible Japanese pear cultivar Nijisseiki and incubated in a moist box at 25°C for 24 h. AK-toxin I and EDA in culture filtrates were quantified by reverse-phase high-performance liquid chromatography (HPLC) as previously described (12, 16). To prepare conidial germination fluids, a conidial suspension (about 5 × 105 conidia/ml) was sprinkled onto paper towels and incubated in a moist box at 25°C for 24 h (16). AK-toxin in conidial germination fluids were tested by bioassay using Nijisseiki leaves and by reverse-phase HPLC analysis.
For the pathogenicity assays, Nijisseiki leaves were spray inoculated with a conidial suspension (about 5 × 105 conidia/ml) and incubated in a moist box at 25°C for 24 h. To heat shock leaves of resistant cultivar Chojuro, detached leaves were dipped into distilled water at 50°C for 50 s and then cooled in water (46). Conidial suspensions (about 5 × 105 conidia/ml) of the test strains were spray inoculated onto the heat-shocked leaves. As a control, leaves were dipped into distilled water at 25°C for 50 s before inoculation. After incubation for 24 h, the lesions were counted. The means and standard deviations of four experiments using different leaves were calculated.
To observe the infection-related morphogenesis of transformants on Nijisseiki leaf cells, inoculated leaves were submerged in ethanol-acetic acid (3:1, vol/vol) overnight, stained in a 55°C lactic acid-phenol-trypan blue solution (0.5 mg/ml aniline blue, 25% lactic acid, 25% phenol, 25% glycerol) for 1.5 h, and then cooled. Samples were destained in a lactic acid-phenol solution (25% lactic acid, 25% phenol, 25% glycerol) for 1 h. The frequencies of germinated conidia, germinated conidia forming appressoria, and appressoria forming penetration hyphae were measured using DIC microscopy as described above. The means and standard deviations of four experiments using different leaves were calculated.
Nucleotide sequence accession number.
The AaPEX6 sequence has been deposited in the DDBJ/EMBL/GenBank databases under accession number AB500683.
RESULTS
Intracellular localization of GFP-tagged Akt1, Akt2, and Akt3-1.
The predicted amino acid sequences of Akt1, Akt2, and Akt3-1 contain C-terminal PTS1-like tripeptides (55, 56). For each of these proteins, we made a strain expressing the protein fused to an N-terminal GFP tag to determine whether these enzymes localize to peroxisomes. We constructed GFP-AKT1, GFP-AKT2, and GFP-AKT3-1 fusions as pGFP-AKT1, pGFP-AKT2, and pGFP-AKT3, respectively (see Fig. S1 in the supplemental material). These constructs were introduced into strain 15A by cotransformation with plasmid pSH75, which confers hygromycin B resistance (26). As a control, strain 15A was transformed with plasmid pYTGFPc, which carries only GFP (see Fig. S1 in the supplemental material).
Transformants were grown on PDA, and their hyphae were observed under a fluorescence microscope. Transformation of strain 15A with pYTGFPc, pGFP-AKT1, pGFP-AKT2, and pGFP-AKT3 gave rise to 32, 40, 54, and 36 GFP-expressing transformants, respectively. In all of the pYTGFPc transformants, the localization of GFP fluorescence was cytosolic and did not appear to be associated with any specific organelle or component of hyphal cells (Fig. 2). In contrast, GFP fluorescence localized to punctate organelles in hyphal cells of all of the pGFP-AKT1, pGFP-AKT2, and pGFP-AKT3 transformants (Fig. 2), consistent with the hypothesis that these enzymes are targeted to peroxisomes. We verified the presence of the GFP fusion constructs in the transformants by DNA gel blot analysis (see Fig. S2 in the supplemental material).
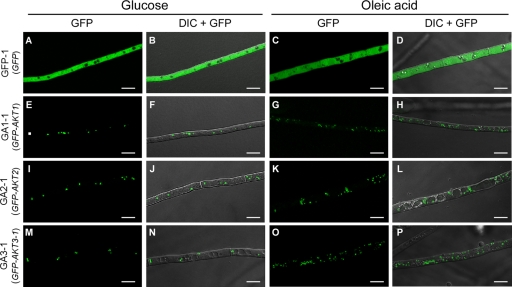
Fig. 2.
Intracellular localization of GFP-tagged Akt1, Akt2, and Akt3-1. Strains were grown for 3 days on potato agar medium supplemented with glucose or oleic acid. GFP-1, pYTGFPc transformant (A to D); GA1-1, pGFP-AKT1 transformant (E to H); GA2-1, pGFP-AKT2 transformant (I to L); GA3-1, pGFP-AKT3 transformant (M to P); GFP, GFP fluorescence images; DIC + GFP, DIC images merged with GFP fluorescence images. Bars = 10 μm.
It is known that supplementation of oleic acid as a carbon source in culture media enhances the proliferation of peroxisomes in hyphal cells (62, 63). Transformants were grown on potato agar supplemented with oleic acid instead of glucose. The number of punctate organelles with GFP fluorescence markedly increased in hyphal cells expressing GFP-Akt fusion proteins grown on oleic acid medium compared with those grown on glucose medium (Fig. 2). The cytosolic localization and intensity of GFP fluorescence in pYTGFPc transformants were similar on both media (Fig. 2). These data are consistent with the hypothesis that the punctate organelles labeled by the GFP fusion proteins are peroxisomes.
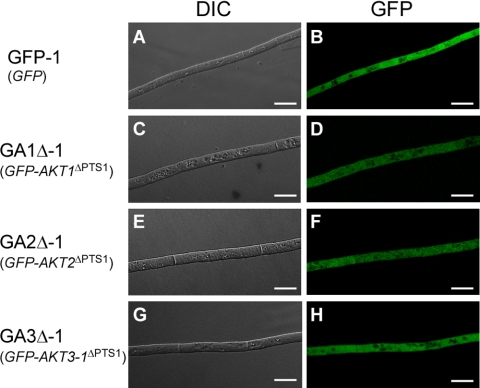
To assess whether the C-terminal tripeptides in Akt1, Akt2, and Akt3-1 act as peroxisomal targeting signals, we constructed transformation vectors pGFP-AKT1Δ, pGFP-AKT2Δ, and pGFP-AKT3Δ, in which AKT1, AKT2, and AKT3-1 lacking nine nucleotides that encode C-terminal PTS1-like tripeptides were fused to the 3′ end of GFP (see Fig. S1 in the supplemental material). These constructs were introduced into strain 15A by cotransformation with plasmid pSH75, and 20, 17, and 21 GFP-expressing transformants were obtained with pGFP-AKT1Δ, pGFP-AKT2Δ, and pGFP-AKT3Δ, respectively. We verified the presence of the GFP fusion constructs in the transformants by DNA gel blot analysis (see Fig. S3 in the supplemental material). When these transformants were grown on PDA, none of them showed GFP fluorescence in a punctate distribution in hyphal cells; all GFP fluorescence was cytosolic (Fig. 3). These results confirmed the peroxisome-targeting function of the C-terminal tripeptides in Akt1, Akt2, and Akt3-1 and suggested that peroxisomes are involved in AK-toxin production.
Fig. 3.
Intracellular localization of GFP-tagged Akt1, Akt2, and Akt3-1 lacking PTS1 sequences. Strains were grown on PDA for 3 days. GFP-1, pYTGFPc transformant (A and B); GA1Δ-1, pGFP-AKT1Δ transformant (C and D); GA2Δ-1, pGFP-AKT2Δ transformant (E and F); GA3Δ-1, pGFP-AKT3Δ transformant (G and H); DIC, DIC images; GFP, GFP fluorescence images. Bars = 10 μm.
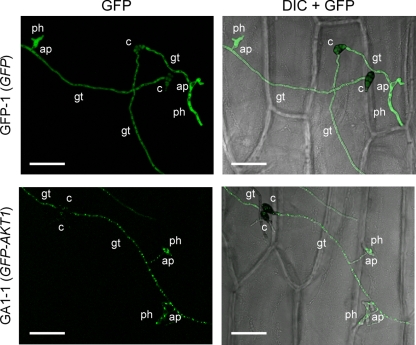
Most host-selective toxins, including AK-toxin, are produced during growth in medium and also during conidial germination (17, 28, 58, 68). Toxins released during conidial germination are required by the producing fungi to penetrate host cells and colonize tissue (17, 28, 58, 68). Conidial suspensions of transformants were dropped onto onion epidermal strips, and germinated conidia forming appressoria and penetration hyphae were observed under a fluorescence microscope at 24 h after inoculation. In pYTGFPc transformant GFP-1, the localization of GFP fluorescence was cytosolic in germ tubes, appressoria, and penetration hyphae (Fig. 4). In GA1-1 expressing the GFP-Akt1 fusion protein, abundant punctate organelles with GFP fluorescence were observed in germ tubes, appressoria, and penetration hyphae (Fig. 4). Similar patterns of GFP fluorescence distribution in germinated conidia were observed in GFP-Akt2- and GFP-Akt3-1-expressing transformants. Abundant peroxisomes in germinated conidia suggested that peroxisome function is important for the infection-related morphogenesis of A. alternata.
Fig. 4.
Intracellular localization of GFP-tagged Akt1 in infection-related structures. A sample of a conidial suspension was dropped onto onion epidermis and incubated for 24 h. GFP-1, pYTGFPc transformant; GA1-1, pGFP-AKT1 transformant; GFP, GFP fluorescence images; DIC + GFP, DIC images merged with GFP fluorescence images. c, conidium; gt, germ tube; ap, appressorium; ph, penetration hypha. Bars = 50 μm.
Isolation and identification of AaPEX6.
PEX6 encodes a peroxin protein which belongs to the AAA (ATPase associated with various cellular activities) protein family and is essential for peroxisome biogenesis in eukaryotic cells (7, 48, 60). We isolated PEX6 from A. alternata (AaPEX6) and analyzed the function of peroxisomes in AK-toxin production and pathogenicity in the Japanese pear pathotype by using strains in which AaPEX6 is disrupted.
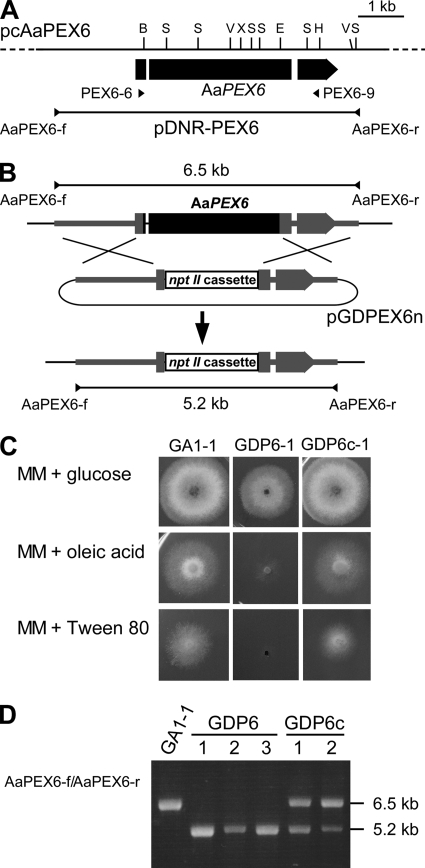
To isolate the AaPEX6 gene, part of AaPEX6 was amplified from the DNA of strain 15A by PCR. The resulting PCR product was used as a probe to screen a cosmid genomic library of strain 15A, and a positive clone, pcAaPEX6, was isolated. Sequencing of the 6.8-kb region in pcAaPEX6 detected AaPEX6, which has three exons (175, 3,469, and 688 bp) divided by two introns (57 and 89 bp) and potentially encodes a 1,444-amino acid protein (Fig. 5A). The presence of two introns was confirmed by comparison of the genomic sequence with the cDNA sequence.
Fig. 5.
Transformation-mediated disruption of AaPEX6. (A) Map of the AaPEX6 locus. The arrowed bar indicates the protein coding region, with introns (white segments), of AaPEX6. Plasmid pDNR-PEX6 contains the entire AaPEX6 region amplified by PCR from cosmid clone pcAaPEX6 DNA using the primer pair AaPEX6-f/AaPEX6-r. Arrowheads (PEX6-6 and PEX6-9) denote the orientations and locations of the oligonucleotide primers used in reverse transcription-PCR experiments. B, BamHI; E, EcoRI; V, EcoRV; H, HindIII; S, SalI; X, XhoI. (B) Structure of the AaPEX6 locus before and after homologous integration of the targeting vector pGDPEX6n. To make pGDPEX6n, a 3.2-kb BamHI-EcoRI fragment within AaPEX6 was replaced with a 1.9-kb BamHI-EcoRI fragment of the nptII cassette. (C) Growth of AaPEX6 disruption-containing and complemented transformants on fatty acid medium. Strains were grown for 4 days on MAM supplemented with glucose, oleic acid, or Tween 80 as the sole carbon source. GA1-1, GFP-Akt1-expressing strain; GDP6-1, AaPEX6 disruption-containing transformant made from GA1-1; GDP6c-1, AaPEX6-complemented transformant made from GDP6-1. (D) PCR analysis of AaPEX6 disruption-containing and complemented transformants. The total DNA of each strain was used as the template for a PCR with the primer pair AaPEX6-f/AaPEX6-r. GDP6-1 to GDP6-3, AaPEX6 disruption-containing transformants; GDP6c-1 and GDP6c-2, AaPEX6-complemented transformants.
A database search using the BLAST algorithm revealed that the deduced amino acid sequence shows 53, 52, 50, and 30% identity with those of the Pex6 proteins from M. oryzae (49), C. orbiculare (25), P. chrysogenum (24), and S. cerevisiae (64), respectively (see Fig. S4 in the supplemental material). AaPex6 contains Walker A and B motifs and a signature AAA protein family motif (7) (see Fig. S4 in the supplemental material).
To examine the function of AaPEX6 in peroxisome biogenesis and AK-toxin production, homologous recombination was employed to replace AaPEX6 with plasmid pGDPEX6n containing an allele in which a 3.2-kb region within AaPEX6 had been replaced with the nptII cassette (Fig. 5B). Strain GA1-1 expressing the GFP-Akt1 fusion protein was transformed with pGDPEX6n, and 10 Geneticin-resistant transformants were isolated. It has been reported in yeast and other fungal species that most peroxisome-defective mutants are unable to grow on media containing fatty acids as sole carbon sources because of their defect in fatty acid β oxidation (62, 63). We observed the growth ability of GA1-1 and the GA1-1 pGDPEX6n transformants on MAM supplemented with oleic acid or Tween 80. GA1-1 and seven transformants could grow on fatty acid medium. However, the remaining three transformants (GDP6-1 to GDP6-3) exhibited very poor growth on the medium (Fig. 5C), suggesting that these transformants were deficient in fatty acid utilization due to the disruption of AaPEX6.
A replacement event at the AaPEX6 locus in three transformants showing poor growth on fatty acid medium was confirmed by PCR analysis. The AaPEX6 locus was amplified from the total DNA of GA1-1 and transformants by PCR with the primer pair AaPEX6-f/AaPEX6-r (Fig. 5A). A PCR product of the expected size, ∼6.5 kb DNA, was amplified from GA1-1, and an ∼5.2-kb product, corresponding to the mutated AaPEX6 locus, was amplified from GDP6 transformants (Fig. 5B and D).
To further confirm the disruption of AaPEX6 in these transformants, genetic complementation of ΔAaPEX6 mutant strain GDP6-1 with wild-type AaPEX6 was performed. Plasmid pDNR-PEX6, containing the entire AaPEX6 gene (including the exons, introns and upstream and downstream regulatory sequences) (Fig. 5A), was introduced into GDP6-1 by cotransformation with plasmid pSB116, conferring resistance to bialaphos. When bialaphos-resistant transformants were tested for growth on oleic acid or Tween 80 medium, two transformants (GDP6c-1 and GDP6c-2) could grow on fatty acid medium (Fig. 5C). PCR analysis of these transformants with the primer pair AaPEX6-f/AaPEX6-r verified the presence of the introduced 6.5-kb fragment containing the wild-type AaPEX6 gene in addition to the 5.2-kb fragment corresponding to the mutated AaPEX6 locus (Fig. 5D).
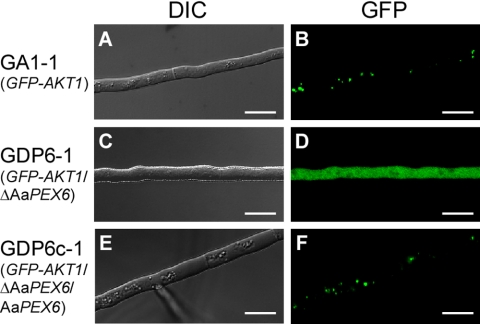
We investigated the intracellular localization of GFP-Akt1 in ΔAaPEX6 mutant strains (GDP6-1 to GDP6-3) and AaPEX6-complemented strains (GDP6c-1 and GDP6c-2). Mycelia grown on PDA were observed under a fluorescence microscope. All of the ΔAaPEX6 mutant strains showed diffuse GFP fluorescence with no punctate pattern in hyphal cells (Fig. 6). This localization was similar to that seen for GFP or GFP-AKT1ΔPTS1 expression in the wild-type strain (Fig. 2 and 3). In contrast, abundant punctate fluorescence was observed in AaPEX6-complemented strains, similar to that seen in the parent strain, GA1-1 (Fig. 6). These results support the hypothesis that AaPEX6 is essential for peroxisome biogenesis in A. alternata.
Fig. 6.
Intracellular localization of the GFP-Akt1 fusion protein in hyphal cells of a ΔAaPEX6 mutant strain. Strains were grown on PDA for 3 days. GA1-1, GFP-Akt1-expressing strain (A and B); GDP6-1, ΔAaPEX6 mutant strain made from GA1-1 (C and D); GDP6c-1, AaPEX6-complemented strain made from GDP6-1 (E and F); DIC, DIC images; GFP, GFP fluorescence images. Bars = 10 μm.
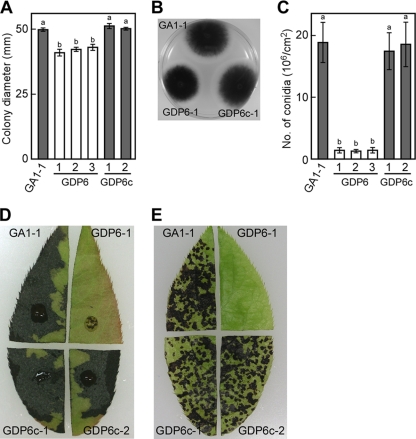
We also investigated colony growth and conidiation of ΔAaPEX6 mutant strains on nutrient-rich media. The growth rates of ΔAaPEX6 mutant strains on PDA were lower than those of the GA1-1 and AaPEX6-complemented strains (Fig. 7A). The melanization of colonies of the mutants was similar to that of GA1-1 (Fig. 7B). Conidiation on oatmeal sucrose medium was dramatically reduced in ΔAaPEX6 mutant strains relative to that of strain GA1-1; the ΔAaPEX6 mutant strains produced 13.2 to 14.4 times fewer conidia than did GA1-1 (Fig. 7C).
Fig. 7.
Phenotypic analysis of a ΔAaPEX6 mutant strain. (A to C) Colony growth, colony morphology, and conidiation of a ΔAaPEX6 mutant strain. Strains were grown on PDA for 5 days, and colony diameter was measured (A and B). Strains were grown on oatmeal agar for 10 days, and conidiation was induced (C). Data represent the means and standard deviations of four replications. Columns with the same letters are not significantly different; columns with different letters are different at a significance level of P ≤ 0.01 according to the Tukey-Kramer multiple-comparison test. GA1-1, GFP-Akt1-expressing strain; GDP6-1 to GDP6-3, ΔAaPEX6 mutant strains made from GA1-1; GDP6c-1 and GDP6c-2, AaPEX6-complemented strains made from GDP6-1. (D and E) AK-toxin production and pathogenicity of ΔAaPEX6 mutant strain. Leaves of Japanese pear cultivar Nijisseiki were wounded slightly, treated with culture filtrate of each strain, and incubated for 24 h (D). Leaves were spray inoculated with a conidial suspension (about 5 × 105 conidia/ml) of each strain and incubated for 24 h (E).
AK-toxin production and pathogenicity in ΔAaPEX6 mutant strains.
Strains were grown in PDB, and AK-toxin production was evaluated on the basis of the toxicity of culture filtrates to leaves of susceptible pear cultivar Nijisseiki. Although culture filtrates of parent strain GA1-1 showed marked toxicity to pear leaves, those of ΔAaPEX6 mutant strains showed no toxicity (Fig. 7D). Culture filtrates of AaPEX6-complemented strains had toxicity similar to that of GA1-1 (Fig. 7D).
The Japanese pear pathotype produces two related molecular species, AK-toxins I and II (Fig. 1), with toxin I being the most abundant and biologically activity species (41, 45). Toxin I in culture filtrates was quantified by reverse-phase HPLC. Culture filtrates of the GA1-1 and AaPEX6-complemented strains contained toxin I (Table 2). However, culture filtrates of ΔAaPEX6 mutant strains contained no detectable toxin I (Table 2). EDA, a precursor of AK-toxin, in culture filtrates was also quantified by HPLC. EDA was detected in culture filtrates of the GA1-1 and AaPEX6-complemented strains but not in those of ΔAaPEX6 mutant strains (Table 2). We also tested for AK-toxin production in ΔAaPEX6 mutant strains during conidial germination. Although germinated conidia of the GA1-1 and AaPEX6-complemented strains produced toxin I, those of ΔAaPEX6 mutant strains did not produce detectable toxin I (data not shown). Thus, it appeared that the presence of functional AaPEX6 is essential for AK-toxin production. These results also indicated that the peroxisome localization of peroxisomal enzymes involved in AK-toxin production is required for the biosynthesis of EDA and AK-toxin.
Table 2.
AK-toxin I and EDA production and pathogenicity of ΔAaPEX6 mutant strains
| Strain | Production (μg/ml)a of: |
Pathogenicityb | |
|---|---|---|---|
| AK- toxin I | EDA | ||
| GA1-1 (GFP-AKT1) | 3.3 | 24.8 | + |
| GFP-AKT1/ΔAaPEX6 mutants | |||
| GDP6-1 | NDc | ND | − |
| GDP6-2 | ND | ND | − |
| GDP6-3 | ND | ND | − |
| GFP-AKT1/ΔAaPEX6/AaPEX6 mutants | |||
| GDP6c-1 | 2.9 | 25.6 | + |
| GDP6c-2 | 2.5 | 22.0 | + |
AK-toxin I and EDA in culture filtrates were analyzed by reverse-phase HPLC. Each value represents the average of two determinations.
+, pathogenic; −, nonpathogenic.
ND, not detected.
AK-toxin-minus mutants completely lose pathogenicity on host pear leaves (47, 55, 56). The ΔAaPEX6 mutant strains were tested for pathogenicity to Nijisseiki leaves by spray inoculation of conidial suspensions. Although the GA1-1 and AaPEX6-complemented strains caused a number of lesions on pear leaves within 24 h after inoculation, all of the ΔAaPEX6 mutant strains caused no lesions on pear leaves (Fig. 7E). This result suggested that these mutants lost pathogenicity due to loss of AK-toxin.
We attempted to restore pathogenicity to the ΔAaPEX6 mutant strain by the addition of AK-toxin I to conidia. Conidia of the Japanese pear pathotype germinate within 6 h after inoculation on host leaves, subsequently form appressoria, and begin to form penetration hyphae as early as 8 h after inoculation (16, 47). Lesions begin to appear as early as 18 h after the inoculation of conidial suspensions and become obvious after 24 h (47). We previously observed that this pathogen produced about 0.1 μg AK-toxin I per ml in the first 12 h when a conidial suspension (about 5 × 105 conidia/ml) was sprinkled onto paper towels and incubated at 25°C (16). AK-toxin I constantly causes necrosis on Nijisseiki leaves at a concentration of more than 0.005 μg/ml within 24 h when a toxin solution is dropped onto wounded sites of the leaves (45). To assess whether the addition of AK-toxin can restore pathogenicity to the ΔAaPEX6 mutant strain, conidia of GA1-1 and GDP6-1 suspended in water or AK-toxin I solution (0.1 μg/ml) were tested for the ability to infect Nijisseiki leaves. We could not evaluate whether the addition of AK-toxin I can restore the ability of GDP6-1 to cause lesions because spray inoculation of AK-toxin I solution with and without conidia developed approximately the same numbers of similar-size lesions on Nijisseiki leaves within 24 h, due to the toxicity of AK-toxin I. Thus, we observed the elaboration of penetration hyphae at 12 h after inoculation.
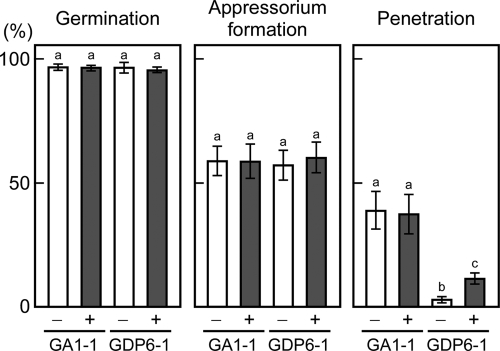
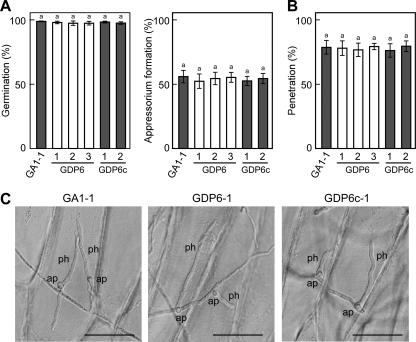
Conidia of GDP6-1 germinated and formed appressoria on pear leaves at a frequency similar to that of conidia of GA1-1, and the addition of AK-toxin I to the conidia had no significant effect on conidial germination and appressorium formation (Fig. 8). In GA1-1, about 38% of the appressoria formed penetration hyphae after 12 h when conidia were suspended in water or AK-toxin I solution (Fig. 8). The penetration frequency was dramatically reduced in GDP6-1 relative to GA1-1: only 3.2% of the appressoria formed penetration hyphae in water (Fig. 8). The penetration ability of GDP6-1 was partially, but not completely, restored by the addition of AK-toxin I to conidia: 11.2% of the appressoria formed penetration hyphae in the presence of AK-toxin I (Fig. 8). Restoration of the penetration ability of GDP6-1 was not observed in the presence of 0.01 μg/ml AK-toxin I. These results suggested that peroxisome function is involved in the penetration ability, in addition to AK-toxin production, of the Japanese pear pathotype.
Fig. 8.
Penetration-related morphogenesis of ΔAaPEX6 mutant strain. Leaves of Japanese pear cultivar Nijisseiki were spray inoculated with conidia suspended in water (−) or an AK-toxin I solution (0.01 μg/ml) (+) and incubated for 12 h. Inoculated leaves were stained with aniline blue, and the percentages of germinated conidia, germinated conidia forming appressoria, and appressoria forming penetration hyphae were determined using DIC optics. Data represent the means and standard deviations of four replications using different leaves. Columns with the same letters are not significantly different; columns with different letters are different at a significance level of P ≤ 0.01 according to the Tukey-Kramer multiple-comparison test. GA1-1, GFP-Akt1-expressing strain; GDP6-1, ΔAaPEX6 mutant strain made from GA1-1.
Infection-related morphogenesis of ΔAaPEX6 mutant strains.
We further observed the infection-related morphogenesis in ΔAaPEX6 mutant strains on glass and onion epidermis. To observe conidial germination and appressorium formation, conidial suspensions in water were dropped onto glass and incubated for 24 h. There were no significant differences in conidial germination, germ tube elongation, or appressorium formation among the parent, AaPEX6 disruption-containing, and AaPEX6-complemented strains (Fig. 9A; see Fig. S5 in the supplemental material). To determine the ability of ΔAaPEX6 mutant strains to penetrate plant epidermal cells, conidial suspensions were dropped onto onion epidermal strips and monitored to detect the elaboration of penetration hyphae at 24 h. The ΔAaPEX6 mutant strains successfully formed penetration hyphae from appressoria at a frequency similar to those of the parent and AaPEX6-complemented strains (Fig. 9B and C), suggesting that peroxisome function is not essential for the penetration of onion epidermal cells by A. alternata.
Fig. 9.
Penetration-related morphogenesis of ΔAaPEX6 mutant strain. (A and B) Conidial germination, appressorium formation, and penetration hypha formation. A sample of a conidial suspension was dropped onto glass and incubated for 24 h; the percentages of germinated conidia and germinated conidia forming appressoria were determined using DIC optics (A). A sample of a conidial suspension was dropped onto onion epidermis and incubated for 24 h; the percentages of appressoria forming penetration hyphae were determined using DIC optics (B). Data represent the means and standard deviations of four replications. Columns with the same letters are not significantly different according to the Tukey-Kramer multiple-comparison test. GA1-1, GFP-Akt1-expressing strain; GDP6-1 to GDP6-3, ΔAaPEX6 mutant strains made from GA1-1; GDP6c-1 and GDP6c-2, AaPEX6-complemented strains made from GDP6-1. (C) Appressoria and penetration hyphae formed on onion epidermis. ap, appressorium; ph, penetration hypha. Bars = 50 μm.
A. alternata conidia are known to invade leaf tissue and produce visible lesions on leaves of resistant pear cultivars with defense responses partially compromised by heat shock (46). Leaves of the resistant pear cultivar Chojuro, which are insensitive to AK-toxin and completely resistant to the Japanese pear pathotype, were dipped in water at 50°C for 50 s, spray inoculated with a conidial suspension, and then incubated for 24 h. The parent, AaPEX6 disruption-containing, and AaPEX6-complemented strains caused no lesions on untreated leaves (Fig. 10A). The parent and complemented strains caused many visible lesions on heat-shocked leaves (Fig. 10B). In contrast, the ΔAaPEX6 mutant strains produced fewer and smaller lesions than the parent strain (Fig. 10B). Together with the finding that the addition of AK-toxin I to conidia partially, but not completely, restored the penetration ability of the ΔAaPEX6 mutant strain, this result suggested that loss of peroxisome function results in a reduction in the penetration into pear leaf epidermal cells and the proliferation in leaf tissue of this A. alternata pathotype.
Fig. 10.
Lesion formation by the ΔAaPEX6 mutant strain on resistant Japanese pear leaves with defense responses compromised by heat shock. Detached leaves of resistant cultivar Chojuro were dipped into distilled water at 25°C (A) or 50°C (B) for 50 s and cooled in water. They were spray inoculated with a conidial suspension (about 5 × 105 conidia/ml) of each strain and incubated for 24 h, and then the lesions were counted. The values at the bottom represent the means and standard deviations of four replications using different leaves. Values with different letters are different at a significance level of P ≤ 0.01 according to the Tukey-Kramer multiple-comparison test. GA1-1, GFP-Akt1-expressing strain; GDP6-1, ΔAaPEX6 mutant strain made from GA1-1; GDP6c-1, AaPEX6-complemented strain made from GDP6-1.
DISCUSSION
Contribution of peroxisomes to AK-toxin biosynthesis and pathogenicity.
In this study of the Japanese pear pathotype of A. alternata, we verified that the AK-toxin biosynthetic enzymes Akt1, Akt2, and Akt3-1, which have PTS1 tripeptides at their C-terminal ends, are localized in peroxisomes. To assess the role of peroxisome function in AK-toxin biosynthesis, we isolated AaPEX6, which encodes a peroxin protein essential for peroxisome biogenesis, from the Japanese pear pathotype of A. alternata. ΔAaPEX6 mutant strains failed to form functional peroxisomes and completely lost AK-toxin production and pathogenicity. Because the strawberry and tangerine pathotypes have closely related orthologs of AKT1, AKT2, and AKT3 which are involved in the biosynthesis of AF-toxin and ACT-toxin (15, 33, 35), Akt1, Akt2, and Akt3 of the Japanese pear pathotype and their orthologs in the strawberry and tangerine pathotypes participate in the biosynthesis of EDA, a common moiety of AK-, AF-, and ACT-toxins. Indeed, the ΔAaPEX6 mutant strains of the Japanese pear pathotype failed to produce not only AK-toxin but also EDA.
EDA is detected in culture filtrates of the Japanese pear, strawberry, and tangerine pathotypes (12, 15, 34). When 3H-labeled EDA was added to a growing liquid culture of the Japanese pear pathotype strain, it was efficiently converted to AK-toxin (12). This result clearly shows that EDA is an intermediate in the AK-toxin pathway. A 13C nuclear magnetic resonance analysis of EDA purified from culture filtrates supplemented with [2-13C]sodium acetate suggested that EDA is biosynthesized by the condensation of six molecules of acetic acid, followed by modifications, including reduction, dehydration, and decarboxylation (42). We identified AFT9, which encodes a polyketide synthase (PKS), in the AF-toxin biosynthetic gene cluster of the strawberry pathotype and found the AFT9 homologues from the Japanese pear and tangerine pathotypes by DNA gel blot analysis (51). We verified that the AFT9 ortholog of the Japanese pear pathotype, AKT9, resides in the AK-toxin biosynthetic gene cluster (S. Takaoka and T. Tsuge, unpublished data). It is likely that the acetyl-CoA-derived backbone of the EDA molecule is produced by the activity of PKS and then modified by other enzymes, including Akt1, Akt2, and Akt3.
In addition to AFT9 and AKT9, we have identified several other genes involved in toxin production by the three pathotypes (1, 15, 20, 34, 35, 36, 51). They include genes common to all three pathotypes, which are probably involved in EDA biosynthesis, and genes specific to each pathotype. Analysis of their predicted amino acid sequences by various sorting algorithms did not identify potential PTS sequences in any of these proteins other than Akt1, Akt2, and Akt3 of the Japanese pear pathotype and their orthologs in the strawberry and tangerine pathotypes. While these data suggest that only part of the EDA biosynthetic pathway takes place in peroxisomes, peroxisomal proteins lacking either of the two known PTS sequences have been found (48, 60), and a definitive subcellular localization of the other proteins involved in toxin production in hyphal cells remains to be determined.
The importance of peroxisomes in secondary metabolism has been reported in penicillin biosynthesis in P. chrysogenum (38, 39). Penicillin biosynthesis consists of at least three enzymatic steps (23, 24, 38, 39). The first step is a nonribosomal condensation of activated l-α-aminoadipic acid, l-cysteine, and l-valine to δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine tripeptide (ACV), catalyzed by a single enzyme, ACV synthetase. In the second step, isopenicillin N synthetase converts ACV to isopenicillin N. The final step is the exchange of the α-aminoadipyl side chain of isopenicillin N for a hydrophobic side chain catalyzed by acyl-CoA:isopenicillin N acyltransferase. It was suggested that ACV synthetase and isopenicillin N synthetase are membrane-associated and cytosolic enzymes, respectively (39), while acyl-CoA:isopenicillin N acyltransferase has a PTS1 sequence and is located in peroxisomes (38, 39). Thus, the three steps of penicillin biosynthesis take place in different cellular domains.
The possible involvement of peroxisomes has also been suggested for the biosynthesis of two polyketide mycotoxins, aflatoxin produced by Aspergillus flavus and A. parasiticus and sterigmatocystin produced by A. nidulans (32). Sterigmatocystin is the penultimate intermediate in aflatoxin biosynthesis (23). Although peroxisomal localization of any of aflatoxin biosynthetic enzymes has not been observed, a precursor, norsolorinic acid (NOR), was found to accumulate in peroxisomes of all three Aspergillus species (32). NOR, a fluorescent compound, is produced by the activity of PKS (StcA) and fatty acid synthase (α subunit StcJ and β subunit StcK), which form a complex (67). These enzymes lack either of the two known PTS sequences, and their peroxisomal localization has not been verified. However, NOR ketoreductase, which lacks either of the two PTS sequences and catalyzes the next step of NOR synthesis in the aflatoxin pathway, has been observed in the cytoplasm of A. parasiticus (30). Thus, it is likely that biosynthetic pathways of the penicillins of P. chrysogenum, aflatoxin of Aspergillus spp., and EDA ether toxins of A. alternata require intracellular transport steps for intermediates and/or products.
Host-selective toxin biosynthetic genes of A. alternata pathotypes appeared to be clustered on small chromosomes of <2.0 Mb in most of the strains tested (2, 3, 13, 15, 20, 21, 33, 35). We demonstrated that small chromosomes that contain toxin biosynthetic genes from the strawberry, apple, and tomato pathotypes are conditionally dispensable (CD) chromosomes (2, 15, 21). We are now focusing on the structural and functional analysis of the 1.05-Mb CD chromosome that contains the AFT genes of the strawberry pathotype to identify the entire gene cluster and to understand the evolution of toxin biosynthesis and the origin of CD chromosomes. Identification of the entire gene cluster and subsequent analysis of the predicted amino acid sequences of AF-toxin pathway enzymes by sorting algorithms should provide more information concerning the subcellular localization of EDA and EDA ether toxin biosynthesis in A. alternata.
Contribution of peroxisomes to plant infection.
ΔPEX6 mutant strains of C. orbiculare and M. oryzae completely lose pathogenicity (25, 49, 65). These pathogens produce dome-shaped appressoria which are darkly pigmented with dihydroxynaphthalene melanin (61). ΔPEX6 mutant strains of these pathogens form smaller appressoria with severely reduced melanization that fail to form infection hyphae (25, 49, 65). The reduced melanization of ΔPEX6 mutant strains suggests that melanin biosynthesis in appressoria depends on acetyl-CoA produced by fatty acid β oxidation in peroxisomes. A. alternata also produces dihydroxynaphthalene melanin, which accumulates in the cell walls of conidia and hyphae (22, 26, 54). However, its appressoria are smaller than those of C. orbiculare and M. oryzae and are colorless. We observed that melanin-deficient mutants and the wild-type strain of the Japanese pear pathotype developed approximately the same numbers of similar-size lesions on susceptible pear leaves in laboratory tests (22, 54). Thus, the ability to produce melanin is probably not relevant to plant infection by the Japanese pear pathotype of A. alternata.
We examined the infection-related morphogenesis of the ΔAaPEX6 mutant strains of the Japanese pear pathotype. Conidia of the ΔAaPEX6 mutant strains germinated and formed appressoria as did those of the wild type on glass and host pear leaves. When the ability to penetrate onion epidermal cells was tested, the frequency of successful production of penetration hyphae from appressoria of the ΔAaPEX6 mutant strains was not significantly different from that of the wild type, suggesting that the mutants retain the basic appressorium-mediated penetration ability. However, the ability of the ΔAaPEX6 mutant strains to penetrate host epidermal cells could be partially, but not completely, restored by the addition of AK-toxin I to a conidial suspension prior to inoculation. When conidia of the ΔAaPEX6 mutant strains were used to inoculate leaves of a resistant pear cultivar with defense responses partially compromised by heat shock, they caused fewer and smaller lesions on the leaves than did those of the wild-type strain. We also observed that germ tubes, appressoria, and penetration hyphae of this pathogen contained abundant peroxisomes. These results suggest that peroxisome function is also necessary for initial plant infection and for tissue colonization by this A. alternata pathotype.
The ΔPEX6 mutant strains of M. oryzae retained the capacity to form polarized infection hyphae on onion epidermal layers, albeit at a reduced frequency (65). The loss of pathogenicity of ΔPEX6 mutant strains of M. oryzae was not solely attributable to the lack of appressorium function because wounded seedlings inoculated with the mutants did not develop rice blast symptoms or support fungal growth (65). Virulence could be partially, but not completely, restored to the mutants by the addition of glucose to a conidial suspension (65). We also observed that the AaPEX6 deletion mutation caused a marked reduction of conidiation in A. alternata. The ΔAaPEX6 mutant strains produced about 13 times fewer conidia than the wild type. Because the conidiation medium contains sucrose as a carbon source, peroxisome functions other than fatty acid β oxidation and glyoxylate metabolism are likely to contribute to conidiation. Conidiation in the M. oryzae ΔPEX6 mutant strains was also dramatically reduced; ΔPEX6 mutant strains produced about 40 times fewer conidia than wild-type strains (65). These data suggest that the diverse functions of the peroxisomes, including β oxidation, glyoxylate metabolism, and peroxide detoxification, are required for plant pathogenesis in fungi.
In M. oryzae and C. orbiculare, the glyoxylate cycle, the enzymes of which are localized in peroxisomes, has been shown to be necessary for full virulence by using ΔICL1 mutants, which lack isocitrate lyase (6, 66). The importance of the glyoxylate cycle in plant pathogenesis has also been shown in the Brassica pathogen Leptosphaeria maculans (18) and the wheat pathogen Stagonospora nodorum (53). Peroxisome-associated carnitine acetyltransferase is required for the elaboration of penetration hyphae during infection by M. oryzae (8, 49). Carnitine acetyltransferase catalyzes the conversion of acetyl-CoA into acetylcarnitine prior to the transport of acetyl units from peroxisomes to the correct intracellular compartment for subsequent utilization (11). The ΔCRAT1 (ΔPTH2) mutants, which lack carnitine acetyltransferase, had weakened cell walls, probably due to defects in cell wall biosynthesis (49). Recently, it has been reported that host invasion by C. orbiculare requires Atg26, a sterol glucosyltransferase that activates pexophagy (5). It is likely that plant pathogenesis in fungi depends on several peroxisome functions, including fatty acid metabolism, acetyl-CoA generation, secondary metabolism, cell wall biogenesis, and peroxisome homeostasis.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Yasuyuki Kubo and Gento Tsuji for providing pBIG4MRBrev and Yoshitaka Takano for providing the GFP vector. We also thank Motoichiro Kodama and Hitoshi Mori for valuable suggestions and the Radioisotope Research Center, Nagoya University, for technical assistance.
This work was supported by Grants-in-Aid for Scientific Research (S) (19108001 for T.T. and 21228001 for K.A.) from the Japanese Society for Promotion of Sciences and Special Coordination Funds for Promoting Sciences from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.T.).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Ajiro N., Miyamoto Y., Masunaka A., Tsuge T., Yamamoto M., Ohtani K., Fukumoto T., Gomi K., Peever T. L., Izumi Y., Tada Y., Akimitsu K. 2010. Role of the host-selective ACT-toxin synthesis gene ACTTS2 encoding an enoyl-reductase in pathogenicity of the tangerine pathotype of Alternaria alternata. Phytopathology 100:120–126 [DOI] [PubMed] [Google Scholar]
- 2.Akagi Y., Akamatsu H., Otani H., Kodama M. 2009. Horizontal chromosome transfer: a mechanism for the evolution and differentiation of a plant pathogenic fungus. Eukaryot. Cell 8:1732–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akagi Y., Taga M., Yamamoto M., Tsuge T., Fukumasa-Nakai Y., Otani H., Kodama M. 2009. Chromosome constitution of hybrid strains constructed by protoplast fusion between the tomato and strawberry pathotypes of Alternaria alternata. J. Gen. Plant Pathol. 75:101–109 [Google Scholar]
- 4.Altschul S. F., Madden T. L., Schäfer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asakura M., Ninomiya S., Sugimoto M., Oku M., Yamashita S., Okuno T., Sakai Y., Takano Y. 2009. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 21:1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asakura M., Okuno T., Takano Y. 2006. Multiple contribution of peroxisomal metabolic function to fungal pathogenicity in Colletotrichum lagenarium. Appl. Environ. Microbiol. 72:6345–6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyer A. 1997. Sequence analysis of the AAA protein family. Protein Sci. 6:2043–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhambra G. K., Wang Z.-Y., Soanes D. M., Wakley G. E., Talbot N. J. 2006. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 61:46–60 [DOI] [PubMed] [Google Scholar]
- 9.Brocard C., Harting A. 2006. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763:1565–1573 [DOI] [PubMed] [Google Scholar]
- 10.Chida T., Sisler H. D. 1987. Restoration of appressorial penetration ability by melanin precursors in Pyricularia oryzae treated with anti-penetrants and in melanin-deficient mutants. J. Pestic. Sci. 12:49–55 [Google Scholar]
- 11.Elgersma Y., van Roermund C. W., Wanders R. J., Tabak H. F. 1995. Peroxisomal and mitochondrial carnitine acetyl-transferases of Saccharomyces cerevisiae are encoded by a single gene. EMBO J. 14:3472–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng B.-N., Nakatsuka S., Goto T., Tsuge T., Nishimura S. 1990. Biosynthesis of host-selective toxins produced by Alternaria alternata pathogens. I. (8R,9S)-9,10-epoxy-8-hydroxy-9-methyl-deca-(2E,4Z,6E)-trienoic acid as a biological precursor of AK-toxins. Agric. Biol. Chem. 54:845–848 [Google Scholar]
- 13.Harimoto Y., Hatta R., Kodama M., Yamamoto M., Otani H., Tsuge T. 2007. Expression profiles of genes encoded by the supernumerary chromosome controlling AM-toxin biosynthesis and pathogenicity in the apple pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 20:1463–1476 [DOI] [PubMed] [Google Scholar]
- 14.Harimoto Y., Tanaka T., Kodama M., Yamamoto M., Otani H., Tsuge T. 2008. Multiple copies of AMT2 are prerequisite for the apple pathotype of Alternaria alternata to produce enough AM-toxin for expressing pathogenicity. J. Gen. Plant Pathol. 74:222–229 [Google Scholar]
- 15.Hatta R., Ito K., Hosaki Y., Tanaka T., Tanaka A., Yamamoto M., Akimitsu K., Tsuge T. 2002. A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata. Genetics 161:59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi N., Tanabe K., Tsuge T., Nishimura S., Kohmoto K., Otani H. 1990. Determination of host-selective toxin production during spore germination of Alternaria alternata by high-performance liquid chromatography. Phytopathology 80:1088–1091 [Google Scholar]
- 17.Howlett B. J. 2006. Secondary metabolite toxins and nutrition of plant pathogenic fungi. Curr. Opin. Plant Biol. 9:371–375 [DOI] [PubMed] [Google Scholar]
- 18.Idnurm A., Howlett B. J. 2002. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot. Cell 1:719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue I., Namiki F., Tsuge T. 2002. Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell 14:1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito K., Tanaka T., Hatta R., Yamamoto M., Akimitsu K., Tsuge T. 2004. Dissection of the host range of the fungal plant pathogen Alternaria alternata by modification of secondary metabolism. Mol. Microbiol. 52:399–411 [DOI] [PubMed] [Google Scholar]
- 21.Johnson L., Johnson R. D., Akamatsu H., Salamiah A., Otani H., Kohmoto K., Kodama M. 2001. Spontaneous loss of a conditionally dispensable chromosome from Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr. Genet. 40:65–72 [DOI] [PubMed] [Google Scholar]
- 22.Kawamura C., Tsujimoto T., Tsuge T. 1999. Targeted disruption of a melanin biosynthesis gene affects conidial development and UV tolerance in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 12:59–63 [DOI] [PubMed] [Google Scholar]
- 23.Keller N. P., Turner G., Bennett J. W. 2005. Fungal secondary metabolism—biochemistry to genomics. Nat. Rev. Microbiol. 3:937–947 [DOI] [PubMed] [Google Scholar]
- 24.Kiel J. A., Hilbrands R. E., Bovenberg R. A., Veenhuis M. 2000. Isolation of Penicillium chrysogenum PEX1 and PEX6 encoding AAA proteins involved in peroxisome biogenesis. Appl. Microbiol. Biotechnol. 54:238–242 [DOI] [PubMed] [Google Scholar]
- 25.Kimura A., Takano Y., Furusawa I., Okuno T. 2001. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13:1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura N., Tsuge T. 1993. Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J. Bacteriol. 175:4427–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohmoto K., Itoh Y., Shimomura N., Kondoh Y., Otani H., Kodama H. M., Nishimura S., Nakatsuka S. 1993. Isolation and biological activities of two host-specific toxins from the tangerine pathotype of Alternaria alternata. Phytopathology 83:495–502 [Google Scholar]
- 28.Kohmoto K., Otani H., Tsuge T. 1995. Alternaria alternata pathogens, p. 51–63 InKohmoto K., Singh U. S., Singh R. P. (ed.), Pathogenesis and host specificity in plant diseases: histopathological biochemical, genetic and molecular bases, vol. II Eukaryotes; Pergamon, Oxford, United Kingdom [Google Scholar]
- 29.Lazarow P. B. 2006. The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 1763:1599–1604 [DOI] [PubMed] [Google Scholar]
- 30.Lee L. W., Chiou C. H., Klomparens K. L., Cray J. W., Linz J. E. 2004. Subcellular localization of aflatoxin biosynthetic enzyme Nor-1, Ver-1, and OmtA in time-dependent fractionated colonies of Aspergillus parasiticus. Arch. Microbiol. 181:204–214 [DOI] [PubMed] [Google Scholar]
- 31.Maekawa N., Yamamoto M., Nishimura S., Kohmoto K., Kuwata M., Watanabe Y. 1984. Studies on host-specific AF-toxins produced by Alternaria alternata strawberry pathotype causing Alternaria black spot of strawberry. (1) Production of host-specific toxins and their biological activity. Ann. Phytopathol. Soc. Jpn. 50:600–609 [Google Scholar]
- 32.Maggio-Hall L. A., Wilson R. A., Keller N. P. 2005. Fundamental contribution of β-oxidation to polyketide mycotoxin production in planta. Mol. Plant Microbe Interact. 18:783–793 [DOI] [PubMed] [Google Scholar]
- 33.Masunaka A., Ohtani K., Peever T. L., Timmer L. W., Tsuge T., Yamamoto M., Yamamoto H., Akimitsu K. 2005. An isolate of Alternaria alternata that is pathogenic to both tangerines and rough lemon and produces two host-selective toxins, ACT- and ACR-toxins. Phytopathology 95:241–247 [DOI] [PubMed] [Google Scholar]
- 34.Miyamoto Y., Isshiki Y., Honda Y. A., Masunaka A., Tsuge T., Yamamoto M., Ohtani K., Fukumoto T., Gomi K., Peever T. L., Akimitsu K. 2009. Function of genes encoding acyl-CoA synthetase and enoyl-CoA hydratase for host-selective ACT-toxin biosynthesis in the tangerine pathotype of Alternaria alternata. Phytopathology 99:369–377 [DOI] [PubMed] [Google Scholar]
- 35.Miyamoto Y., Masunaka A., Tsuge T., Yamamoto M., Ohtani K., Fukumoto T., Gomi K., Peever T. L., Akimitsu K. 2008. Functional analysis of a multi-copy host-selective ACT-toxin biosynthesis gene in the tangerine pathotype of Alternaria alternata using RNA silencing. Mol. Plant Microbe Interact. 21:1591–1599 [DOI] [PubMed] [Google Scholar]
- 36.Miyamoto Y., Masunaka A., Tsuge T., Yamamoto M., Ohtani K., Fukumoto T., Gomi K., Peever T. L., Tada Y., Ichimura K., Akimitsu K. 2010. ACTTS3 encoding a polyketide synthase is essential for the biosynthesis of ACT-toxin and pathogenicity in the tangerine pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 23:406–414 [DOI] [PubMed] [Google Scholar]
- 37.Mullaney E. J., Hamer J. E., Roberti K. A., Yelton M. M., Timberlake W. E. 1985. Primary structure of the trpC gene from Aspergillus nidulans. Mol. Gen. Genet. 199:37–45 [DOI] [PubMed] [Google Scholar]
- 38.Müller W. H., Bovenberg R. A., Groothuis M. H., Kattevilder F., Smaal E. B., van der Voort L. H., Verkleij A. J. 1992. Involvement of microbodies in penicillin biosynthesis. Biochim. Biophys. Acta 1116:210–213 [DOI] [PubMed] [Google Scholar]
- 39.Müller W. H., van der Krift T. P., Krouwer A. J. J., Wösten H. A. B., van der Voort L. H. M., Smaal E. B., Verkleij A. J. 1991. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 10:489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakai K., Horton P. 1999. PSORT: a program for detecting sorting signals in proteins and determining their subcellular localization. Trends Biochem. Sci. 24:34–35 [DOI] [PubMed] [Google Scholar]
- 41.Nakashima T., Ueno T., Fukami H., Taga T., Masuda H., Osaki K., Otani H., Kohmoto K., Nishimura S. 1985. Isolation and structures of AK-toxin I and II, host-specific phytotoxic metabolites produced by Alternaria alternata Japanese pear pathotype. Agric. Biol. Chem. 49:807–815 [Google Scholar]
- 42.Nakatsuka S., Feng B.-N., Goto T., Tsuge T., Nishimura S. 1990. Biosynthetic origin of (8R,9S)-9,10-epoxy-8-hydroxy-9-methyl-deca-(2E,4Z,6E)-trienoic acid, a precursor of AK-toxins produced by Alternaria alternata Japanese pear pathotype. Phytochemistry 29:1529–1531 [Google Scholar]
- 43.Nakatsuka S., Ueda K., Goto T., Yamamoto M., Nishimura S., Kohmoto K. 1986. Structure of AF-toxin II, one of the host-specific toxins produced by Alternaria alternata strawberry pathotype. Tetrahedron Lett. 27:2753–2756 [Google Scholar]
- 44.Namiki F., Matsunaga M., Okuda M., Inoue I., Nishi K., Fujita Y., Tsuge T. 2001. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant Microbe Interact. 14:580–584 [DOI] [PubMed] [Google Scholar]
- 45.Otani H., Kohmoto K., Nishimura S., Nakashima T., Ueno T., Fukami H. 1985. Biological activities of AK-toxins I and II, host-specific toxins from Alternaria alternata Japanese pear pathotype. Ann. Phytopathol. Soc. Jpn. 51:285–293 [Google Scholar]
- 46.Otani H., Nishimura S., Kohmoto K. 1983. Different responses of heat-shocked pear leaves to Alternaria alternata Japanese pear pathotype and AK-toxin. J. Fac. Agric. Tottori Univ. 18:1–8 [Google Scholar]
- 47.Otani H., Nishimura S., Kohmoto K., Yano K., Seno T. 1975. Nature of specific susceptibility to Alternaria kikuchiana in Nijisseiki cultivar among Japanese pears (V). Role of host-specific toxin in early step of infection. Ann. Phytopathol. Soc. Jpn. 41:467–476 [Google Scholar]
- 48.Platta H. W., Erdmann R. 2007. Peroxisomal dynamics. Trends Cell Biol. 17:474–484 [DOI] [PubMed] [Google Scholar]
- 49.Ramos-Pamplona M., Naqvi N. I. 2006. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acyl-CoA. Mol. Microbiol. 61:61–75 [DOI] [PubMed] [Google Scholar]
- 50.Rotem J. 1994. The genus Alternaria: biology, epidemiology, and pathogenicity The American Phytopathological Society Press, St. Paul, MN [Google Scholar]
- 51.Ruswandi S. R., Kitani K., Akimitsu K., Tsuge T., Shiraishi T., Yamamoto M. 2005. Structural analysis of cosmid clone pcAFT-2 carrying AFT10-1 encoding an acyl-CoA dehydrogenase involved in AF-toxin production in the strawberry pathotype of Alternaria alternata. J. Gen. Plant Pathol. 71:107–116 [Google Scholar]
- 52.Sanderson K. E., Srb A. M. 1965. Heterokaryosis and parasexuality in the fungus Ascochyta imperfecta. Am. J. Bot. 42:72–81 [PubMed] [Google Scholar]
- 53.Solomon P. S., Lee R. C., Wilson T. C. J., Oliver R. P. 2004. Pathogenicity of Stagonospora nodorum requires malate synthase. Mol. Microbiol. 53:1065–1073 [DOI] [PubMed] [Google Scholar]
- 54.Tanabe K., Park P., Tsuge T., Kohmoto K., Nishimura S. 1995. Characterization of the mutants of Alternaria alternata Japanese pear pathotype deficient in melanin production and their pathogenicity. Ann. Phytopathol. Soc. Jpn. 61:27–33 [Google Scholar]
- 55.Tanaka A., Shiotani H., Yamamoto M., Tsuge T. 1999. Insertional mutagenesis and cloning of the genes required for biosynthesis of the host-specific AK-toxin in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 12:691–702 [DOI] [PubMed] [Google Scholar]
- 56.Tanaka A., Tsuge T. 2000. Structural and functional complexity of the genomic region controlling AK-toxin biosynthesis and pathogenicity in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 13:975–986 [DOI] [PubMed] [Google Scholar]
- 57.Tanaka S., Yamada K., Yabumoto K., Fujii S., Huser A., Tsuji G., Koga H., Dohi K., Mori M., Shiraishi T., O'Connel R., Kubo Y. 2007. Saccharomyces cerevisiae SSD1 orthologues are essential for host infection by the ascomycete plant pathogens Colletotrichum lagenarium and Magnaporthe grisea. Mol. Microbiol. 64:1332–1349 [DOI] [PubMed] [Google Scholar]
- 58.Thomma B. P. H. J. 2003. Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4:225–236 [DOI] [PubMed] [Google Scholar]
- 59.Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Titorenko V. I., Rachubinski R. A. 2001. The life cycle of the peroxisome. Nat. Rev. Mol. Cell Biol. 2:357–368 [DOI] [PubMed] [Google Scholar]
- 61.Tucker S. L., Talbot N. J. 2001. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39:385–417 [DOI] [PubMed] [Google Scholar]
- 62.van den Bosch H., Schutgens R. B. H., Wanders R. J. A., Tager J. M. 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61:157–197 [DOI] [PubMed] [Google Scholar]
- 63.van Roermund C. W. T., Waterham H. R., Ijlst L., Wanders R. J. A. 2003. Fatty acid metabolism in Saccharomyces cerevisiae. Cell. Mol. Life Sci. 60:1838–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voorn-Brouwer T., van der Leij I., Hemrika W., Distel B., Tabak H. F. 1993. Sequence of the PAS8 gene, the product of which is essential for biogenesis of peroxisomes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1216:325–328 [DOI] [PubMed] [Google Scholar]
- 65.Wang Z.-Y., Soanes D. M., Kershaw M. J., Talbot N. J. 2007. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid β-oxidation during appressorium-mediated plant infection. Mol. Plant Microbe Interact. 20:475–491 [DOI] [PubMed] [Google Scholar]
- 66.Wang Z.-Y., Thornton C. R., Kershaw M. J., Debao L., Talbot N. J. 2003. The glyoxylate cycle is required for correct temporal regulation of virulence by the rice blast fungus Magnaporthe grisea. Mol. Microbiol. 47:1601–1612 [DOI] [PubMed] [Google Scholar]
- 67.Watanabe C. M. H., Townsend C. A. 2002. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B1. Chem. Biol. 9:981–988 [DOI] [PubMed] [Google Scholar]
- 68.Wolpert T. J., Dunkle L. D., Ciuffetti L. M. 2002. Host-selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40:251–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.