Abstract
A single-step protein affinity purification protocol using Aspergillus nidulans is described. Detailed protocols for cell breakage, affinity purification, and depending on the application, methods for protein release from affinity beads are provided. Examples defining the utility of the approaches, which should be widely applicable, are included.
Over 50 years ago, Richards and colleagues found that cleavage of RNase A by subtilisin resulted in two peptides, the S-peptide (residues 1 to 20) and the S-protein (residues 21 to 124), which bind tightly to each other with high affinity (dissociation constant, ∼10−9 M) (1, 8, 9). Because of this high affinity and the small size of the S-peptide (which can be reduced to 15 amino acids [aa] and still bind with high affinity), this interaction has been used as the basis for protein affinity purification (2).
We previously described methods for generating C-terminally green fluorescent protein (GFP)-tagged constructs by using fusion PCR and Aspergillus fumigatus pyrG as the nutritional transformation marker (12). To maximize the utility of the primers introducing the C-terminal GFP tag, we have generated new base cassettes which, with the use of the very same primers, allow the generation of endogenous C-terminally S-tagged versions of proteins of interest (4; http://www.fgsc.net/plasmid/atagging.htm). The focus of this report is to outline protocols for efficient protein extraction and single-step S-tag affinity purification using Aspergillus nidulans as the model system. The methods should be appropriate for other cell types.
By using previously described methods (5, 7, 12), the S-tag coding sequence is targeted via homologous recombination in frame to the 3′ or 5′ ends of target genes. Diagnostic PCR and Western blot analysis are used to confirm gene replacement and expression of the correctly sized S-tagged protein. Samples for this diagnostic analysis are generated from static petri dish liquid cultures (see Protocol S1A in the supplemental material). Cells are harvested and lyophilized overnight (3, 6). The lyophilized cells are broken (see Protocol S1A and Video S1 in the supplemental material) to generate small-scale protein samples for Western blot analysis (see Protocol S1B in the supplemental material) or DNA for diagnostic PCR (see Protocol S1C in the supplemental material).
Once correct tagging is confirmed, strains containing S- tagged proteins are grown (typically after one outcross) in a shaken liquid flask culture (see Protocol S2 in the supplemental material) to generate samples for protein extraction and affinity purification. The filamentous mode of growth of many fungi facilitates rapid harvesting by filtration followed by freezing in liquid nitrogen (1,000-ml cultures can be harvested and frozen in less than 1 min). The frozen cells are then lyophilized. A 1,000-ml culture typically yields 0.7 to 1.0 g of dried cell mass. The lyophilized cells may be stored at −80°C before protein extraction but are relyophilized for 30 min before processing.
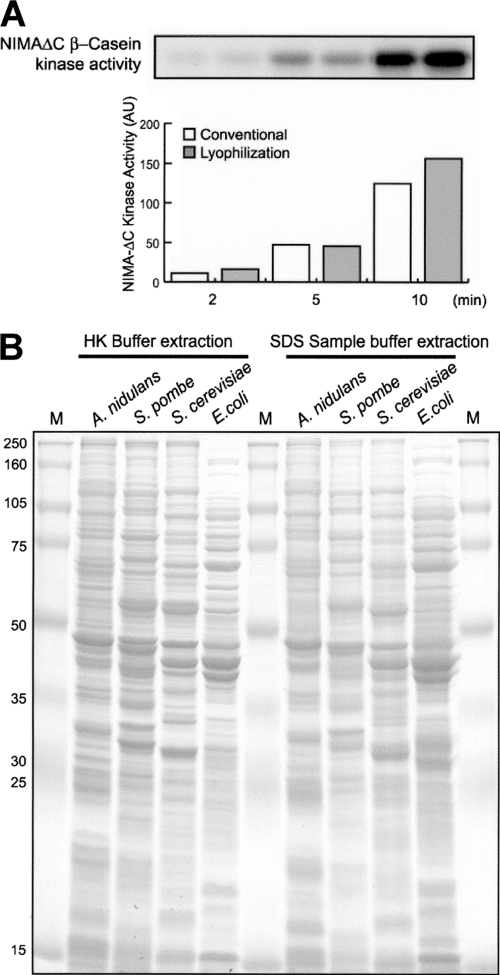
Breaking lyophilized cells is simple, quick, and efficient. It is achieved by grinding with a mortar and pestle at room temperature (see Video S2 in the supplemental material) until the lyophilized cells have become a homogenous fine powder. For protein extraction (see Protocol S2 in the supplemental material), the cell powder is mixed directly with cold HK extraction buffer and subsequent steps are carried out at 4°C. The powder is thoroughly mixed by subjection to a vortex (see Video S2 in the supplemental material). The cell extract is cleared by two rounds of centrifugation to harvest the supernatant. The protein concentration is typically 20 to 25 mg/ml as measured using a Bradford protein assay. Importantly, this method of generating protein extracts maintains sensitive biochemical activities (Fig. 1A) and has also been utilized successfully for Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Escherichia coli cells (Fig. 1B). This method has advantages in that cell breakage and efficient protein extraction are very rapid and no specialized instrumentation is required.
Fig. 1.
Extraction of proteins from A. nidulans, yeast, and bacterial cells after lyophilization. (A) Protein extracts were generated from frozen mycelia before (conventional) or after lyophilization. The activity of the NIMA protein kinase was determined by the ability of S-tag affinity-purified NIMA (eluted with S-peptide) to phosphorylate β-casein in standard protein kinase assays. The radioactivity depicted in the autoradiograph was quantified by ImageQuant (Molecular Dynamics). NIMAΔC, NIMA lacking the C terminus; AU, arbitrary units. (B) Cells of the indicated organisms were grown and frozen in liquid nitrogen and then lyophilized. Samples of equal weights were extracted using HK extraction buffer or SDS sample buffer, the extracted proteins were run through SDS-PAGE, and the gel was stained with Coomassie blue. M, molecular weight markers.
To minimize proteolysis, protein extraction and affinity purification are completed sequentially without delay. For affinity purification (see Protocol S3 in the supplemental material), a batch-wise protocol utilizing S-protein agarose beads is used. After 1 h of incubation, beads are collected by centrifugation and washed extensively (see Protocol S3 in the supplemental material).
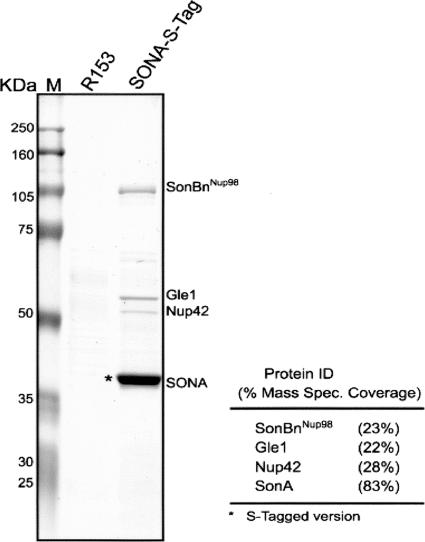
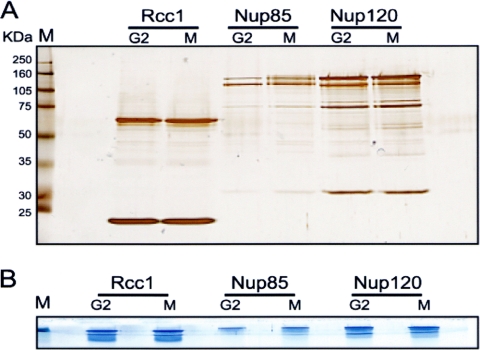
It is possible to release purified proteins from the S-protein beads by using different methods depending on the experimental demands. For mass spectrometry (MS) analysis, purified proteins are released by boiling the beads in sodium dodecyl sulfate (SDS) sample buffer (see Protocol S3A in the supplemental material). The released proteins are run through SDS-PAGE. To identify the main proteins purified, the gel is stained with Coomassie blue and visible bands can be excised and processed for MS analysis by standard methodologies (Fig. 2). Alternatively, to identify all proteins purified, including those in low abundance, 1/10 of the purified sample is run on an analytical SDS-PAGE gel and the gel is silver stained to record the pattern of proteins purified (Fig. 3A). For MS analysis, the remaining 9/10 of the sample is subjected to preparative SDS-PAGE but the proteins are run through the stacking gel and just into the separating gel. This results in all purified proteins' being compressed into a tight series of bands (Fig. 3B). The total protein bands are excised as one and processed for MS in the normal manner. By using this approach, all proteins purified can be identified in a single MS analysis.
Fig. 2.
Identification of proteins copurified with SONA (suppressor of nimA1 A) by S-tag affinity purification. Endogenously S-tagged SONA was subjected to affinity purification along with a control extract from a strain not expressing an S-tagged protein, and the purified proteins were separated by SDS-PAGE. After Coomassie blue staining, the indicated bands were excised and identified using liquid chromatography-tandem MS. M, molecular mass markers; R153, untagged wild-type strain; SonBnNup98, orthologue of human Nup98; mass spec., mass spectrometry.
Fig. 3.
SDS-PAGE conditions for identification of all purified proteins. (A) Fractions of 1/10 of the samples of the indicated S-tagged proteins purified from cells arrested in G2 or mitosis (M) were run through SDS-PAGE, and the gel was stained with silver to record the pattern of proteins purified. (B) To identify the proteins purified in a single MS analysis, the remaining purified samples were run just into the separating gel during SDS-PAGE and the gel was stained with Coomassie blue. A single gel fragment containing all purified proteins can then be excised and processed for MS analysis.
To release native biochemically active proteins, elution with free S-peptide is employed (see Protocol S3B in the supplemental material), enabling biochemical analysis of purified proteins in solution.
Enzymatic assays can also be carried out utilizing the purified protein still bound to the beads. For such on-bead assays, the washed beads are equilibrated in the appropriate assay buffer. We have confirmed that cell breakage at room temperature after lyophilization is compatible with biochemical activity. For example, comparable NIMA kinase activities were purified from extracts generated from lyophilized cells and from extracts generated by the more normal method of grinding frozen nonlyophilized cells in liquid nitrogen (Fig. 1A). Notably, however, significantly more total protein was extracted using lyophilization-based breakage (48% more in this experiment).
We have also used in vitro phosphatase treatment of purified S-tagged proteins on beads to confirm that mobility shifts observed upon SDS-PAGE are caused by phosphorylation (10, 11).
We have successfully utilized the methods described herein for purification of a wide range of S-tagged proteins, demonstrating the general applicability of these methods. If necessary, conditions of cell growth and buffer stringencies during purification can readily be optimized on a protein-by-protein basis.
Supplementary Material
Acknowledgments
We thank Bo Liu (UC—Davis) for discussions regarding SDS-PAGE purification for MS analysis.
This work was supported by the National Institutes of Health grant GM 42564.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Kalman S. M., Linderstrom-Lang K., Ottesen M., Richards F. M. 1955. Degradation of ribonuclease by subtilisin. Biochim. Biophys. Acta 16:297–299 [DOI] [PubMed] [Google Scholar]
- 2.Kim J. S., Raines R. T. 1993. Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 2:348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S. B., Taylor J. W. 1990. Isolation of DNA from fungal mycelia and single spores, p. 282–287 InMichael D. H. G., Innis A., Sninsky J. J., White T. J. (ed.), PCR protocols: a guide to methods and applications Academic Press, Inc., Harcourt Brace Jovanovich, New York, NY [Google Scholar]
- 4.Liu H.-L., De Souza C. P. C., Osmani A. H., Osmani S. A. 2009. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol. Biol. Cell 20:616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayak T., Szewczyk E., Oakley C. E., Osmani A., Ukil L., Murray S. L., Hynes M. J., Osmani S. A., Oakley B. R. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172:1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osherov N., May G. S. 1998. Optimization of protein extraction from Aspergillus nidulans for gel electrophoresis. Fungal Genet. Newsl. 45:38–40http://www.fgsc.net/fgn45/45osherov.html [Google Scholar]
- 7.Osmani A. H., Davies J., Liu H. L., Nile A., Osmani S. A. 2006. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17:4946–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards F. M. 1955. On an active intermediate produced during the digestion of ribonuclease by subtilisin. C. R. Trav. Lab. Carlsberg Chim. 29:329–346 [PubMed] [Google Scholar]
- 9.Richards F. M. 1955. Titration of amino groups released during the digestion of ribonuclease by subtilisin. C. R. Trav. Lab. Carlsberg Chim. 29:322–328 [PubMed] [Google Scholar]
- 10.Son S., Osmani S. A. 2009. Analysis of all protein phosphatase genes in Aspergillus nidulans identifies a new mitotic regulator, Fcp1. Eukaryot. Cell 8:573–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ukil L., Varadaraj A., Govindaraghavan M., Liu H. L., Osmani S. A. 2008. Copy number suppressors of the Aspergillus nidulans nimA1 mitotic kinase display distinctive and highly dynamic cell cycle-regulated locations. Eukaryot. Cell 7:2087–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L., Ukil L., Osmani A., Nahm F., Davies J., De Souza C. P., Dou X., Perez-Balaguer A., Osmani S. A. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3:1359–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.