Abstract
The fungally conserved subset of amino acid biosynthetic enzymes not present in humans offer exciting potential as an unexploited class of antifungal drug targets. Since threonine biosynthesis is essential in Cryptococcus neoformans, we further explored the potential of threonine biosynthetic enzymes as antifungal drug targets by determining the survival in mice of Saccharomyces cerevisiae homoserine kinase (thr1Δ) and threonine synthase (thr4Δ) mutants. In striking contrast to aspartate kinase (hom3Δ) mutants, S. cerevisiae thr1Δ and thr4Δ mutants were severely depleted after only 4 h in vivo. Similarly, Candida albicans thr1Δ mutants, but not hom3Δ mutants, were significantly attenuated in virulence. Consistent with the in vivo phenotypes, S. cerevisiae thr1Δ and thr4Δ mutants as well as C. albicans thr1Δ mutants were extremely serum sensitive. In both species, serum sensitivity was suppressed by the addition of threonine, a feedback inhibitor of Hom3p. Because mutation of the HOM3 and HOM6 genes, required for the production of the toxic pathway intermediate homoserine, also suppressed serum sensitivity, we hypothesize that serum sensitivity is a consequence of homoserine accumulation. Serum survival is critical for dissemination, an important virulence determinant: thus, together with the essential nature of C. neoformans threonine synthesis, the cross-species serum sensitivity of thr1Δ mutants makes the fungus-specific Thr1p, and likely Thr4p, ideal antifungal drug targets.
Fungal infections are an increasingly significant cause of human disease and morbidity due to an expanding immunocompromised population. However, only four main classes of broad-spectrum antifungal drugs are currently available (polyenes, azoles, echinocandins, and 5-fluorocytosine), which target only three cellular components: the cell membrane, cell wall, and nucleotide biosynthesis (55). Compared with the identification of antibacterial drug targets, an obstacle to antifungal drug target identification is the eukaryotic nature of both the fungal pathogen and the host, ensuring a considerably higher degree of conserved genes and pathways. Since a subset of amino acid biosynthetic pathways are not present in humans (46), yet are conserved in fungi, and many are required for survival in vivo and/or virulence (22, 31, 35, 36, 45, 58), various amino acid biosynthetic enzymes are an attractive, unexploited class of antifungal drug targets.
The threonine biosynthetic pathway is of particular interest for antifungal drug targets. Threonine is produced from aspartate, via the intermediate homoserine, in a series of five enzymatic steps, initiated by aspartate kinase (Hom3p). Homoserine is converted to threonine by the sequential actions of homoserine kinase (Thr1p) and threonine synthase (Thr4p). Threonine synthesis is regulated by induction of pathway genes via the general control pathway in response to amino acid starvation (26, 43) and by feedback regulation of aspartate kinase when threonine is abundant (41, 48). Homoserine and threonine are intermediates in the synthesis of methionine and isoleucine, respectively, and we have found that various fungal methionine and isoleucine auxotrophs are unable to survive in vivo and/or are avirulent (31, 36, 45, 58). Myriad phenotypes in addition to auxotrophy have been associated with Saccharomyces cerevisiae threonine biosynthetic mutants, particularly thr1Δ or thr4Δ mutants (2, 8, 14–16, 20, 33, 51) as a result of toxic homoserine accumulation (33), many phenotypes of which may also affect the ability of these mutants to survive in vivo.
Since we find that S. cerevisiae hom3Δ mutants are unable to survive in vivo (31) and that C. neoformans hom3Δ and thr1Δ mutations are lethal (34), we further investigated the potential of threonine biosynthetic enzymes as antifungal drug targets by examining the in vivo survival of thr1Δ and thr4Δ mutants constructed in a clinically derived S. cerevisiae strain. Given the severe survival defects of these mutants after only 4 h in vivo, we extended our investigations to Candida albicans, a more clinically relevant pathogen, and observed that C. albicans thr1Δ mutants had attenuated virulence. Consistent with the in vivo defects, we demonstrated that S. cerevisiae thr1Δ, thr4Δ, and C. albicans thr1Δ strains were serum sensitive. We explored the basis of the serum sensitivity and show that low serum threonine concentrations and the accumulation of the biosynthetic intermediate homoserine are key to the rapid death of thr1Δ and thr4Δ mutants in serum.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains used in this study are listed in Table 1. S. cerevisiae strains were isogenic with clinically derived YJM145 (42), and C. albicans strains were isogenic with strain SC5314 (21). Unless otherwise specified, all strains described are diploid and homozygous for the gene disruption described. Standard culture media included yeast extract-peptone-dextrose (YPD) and synthetic dextrose (SD), which were prepared as described previously (54). Dulbecco's modified Eagle's medium (DMEM) (with dextrose, l-glutamine, and sodium pyruvate; Mediatech, Inc., catalog no. 10-013-CV) was supplemented with NaHCO3 (22 mM) and HEPES (50 mM) or Na MOPS (morpholinepropanesulfonic acid; 25 mM) or, alternatively, made as specified by Mediatech, Inc., but with various components omitted. RPMI 1640 (with l-glutamine and NaHCO3) was obtained from Sigma (catalog no. R8758). Where specified, media were supplemented with l-methionine (20 μg ml−1), maltose (20 mg ml−1), nourseothricin (Nat; 100 μg ml−1 for S. cerevisiae selection and 200 μg/ml for C. albicans selection; Hans Knöll Institute für Naturstoff-Forschung, Jena, Germany), hygromycin B (300 μg ml−1; Calbiochem) and Geneticin (200 μg ml−1; Life Technologies). Unless specified otherwise, l-threonine was added to SD at a concentration of 300 μg ml−1.
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| S. cerevisiae | ||
| YAG40 | HO/hoΔ::hphMX4 | 22 |
| YAG129 | MATa/MATα gal2/gal2 HO/HO | 22 |
| YJK369b | thr4Δ::natMX4/thr4Δ::natMX4 | This study |
| YJK391b | thr4Δ::kanMX4/thr4Δ::kanMX4 | This study |
| YJK456 | THR4/thr4Δ::kanMX4 | This study |
| YJK461 | THR4/thr4Δ::natMX4 | This study |
| YJK487a | hom3Δ::kanMX4/hom3Δ::kanMX4 | 31 |
| YJK495a | hom3Δ::natMX4/hom3Δ::natMX4 | 31 |
| YJK498a | thr1Δ::kanMX4/thr1Δ::kanMX4 | This study |
| YJK506a | thr1Δ::natMX4/thr1Δ::natMX4 | This study |
| YJK547 | THR1/thr1Δ::kanMX4 | This study |
| YJK677 | hom3Δ::kanMX4/hom3Δ::kanMX4 thr4Δ::natMX4/thr4Δ::natMX4 | This study |
| YJK682 | hom3Δ::kanMX4/hom3Δ::kanMX4 thr1Δ::natMX4/thr1Δ::natMX4 | This study |
| YJK862 | hom6Δ::kanMX4/hom6Δ::kanMX4 | This study |
| YJK948 | hom6Δ::kanMX4/hom6Δ::kanMX4 thr1Δ::natMX4/thr1Δ::natMX4 | This study |
| YJK1078 | thr1Δ::natMX4/thr1Δ::natMX4 gcn4Δ::kanMX4/gcn4Δ::kanMX4 | This study |
| YJK1379 | hom6Δ::kanMX4/hom6Δ::kanMX4 thr4Δ::natMX4/thr4Δ::natMX4 | This study |
| C. albicans | ||
| SC5314 | Wild type | 21 |
| CJK18 | HOM3/hom3Δ::SAT1 | This study |
| CJK22 | THR1/thr1Δ::SAT1 | This study |
| CJK26 | HOM3/hom3Δ::FRT | This study |
| CJK30 | THR1/thr1Δ::FRT | This study |
| CJK35, CJK37 | thr1Δ::SAT1/thr1Δ::FRT | This study |
| CJK41, CJK43 | hom3Δ::SAT1/hom3Δ::FRT | This study |
| CJK59 | THR1/thr1Δ::SAT1 (THR1-complemented strain) | This study |
| CJK61 | HOM3/hom3Δ::SAT1 (HOM3-complemented strain) | This study |
| CJK77 | hom3Δ::FRT/hom3Δ::FRT thr1Δ::SAT1/thr1Δ::FRT | This study |
Gene deletions.
S. cerevisiae genes were replaced with natMX4, kanMX4, or hphMX4 cassettes by PCR-mediated gene deletion (23, 56). Since the YJM145 background is diploid and homothallic, transformants were sporulated at 30°C and tetrads were dissected to achieve homozygous gene deletions. To construct multiple deletions in a strain, separate strains containing deletions with different drug markers were sporulated and crossed, and diploids were selected by acquisition of multiple drug resistance. Strains were sporulated and dissected to obtain strains with multiple homozygous deletions. To complement mutant strains with wild-type alleles, strains were transformed with a PCR product containing the wild-type gene and flanking sequence. Gene deletions and mutant complementation were confirmed by PCR and by acquisition or loss of a phenotype.
Genes were disrupted in C. albicans using a similar PCR-mediated strategy, in which the SAT1 flipper cassette (50) was amplified using primers that contained at their 5′ ends 60 bp of sequence homologous to the region immediately flanking the gene of interest. Strains were transformed with the gene-targeting SAT1 PCR product by electroporation (50), and Nat-resistant transformants were purified and verified by PCR analysis. Transformants were grown for 2 h in YP medium containing maltose [YP(maltose)] to induce FLP-mediated excision of the SAT1 cassette, leaving an FLP recombination target (FRT) site. Nat-sensitive strains then underwent a second round of transformation to disrupt the second allele. To complement strains with a wild-type gene, homozygous deletion strains were transformed with the gene of interest, amplified using primers homologous to sequence upstream and downstream of the deleted region. Transformants were selected by reversion to prototrophy, and transformants in which the wild-type allele had replaced a disrupted allele were chosen; thus, the introduced gene was expressed from its original chromosomal location. Gene disruptions and mutant complementation were verified by PCR, phenotype where available, and Southern hybridization analysis (see Fig. S1 in the supplemental material).
All primers used in this study are listed in Table S1 in the supplemental material.
Manipulation of nucleic acids.
DNA was extracted from S. cerevisiae and C. albicans strains for PCR and Southern hybridization analyses, as described previously (27). To confirm C. albicans gene deletions by Southern hybridization analysis, 2 μg of genomic DNA was digested with various restriction enzymes, separated by agarose gel electrophoresis, denatured, and transferred to a nylon membrane (Roche) as described previously (52). Southern hybridization probes were prepared from PCR products (agarose gel purified using the QIAquick gel extraction kit; Qiagen) and labeled with [α-32P]dCTP (Perkin-Elmer) using the RediprimeII random prime labeling system (Amersham Biosciences), as described by the manufacturer. Prehybridization and hybridization were performed in ULTRAhyb buffer (Ambion), blots were washed according to the manufacturer's instructions, and hybridized bands were visualized using a Typhoon 9200 variable mode imager (Molecular Dynamics).
Serum treatments and sensitivity assays.
To test S. cerevisiae for serum sensitivity, strains were typically grown overnight in YPD, washed twice in sterile distilled water, and then added to 1 or 3 ml fetal bovine serum (FBS; Sigma catalog no. F2442) at a concentration of approximately 1 × 106 cells/ml. The volume of FBS depended on whether cell viability was estimated by spot dilution or cells were plated for absolute numbers. Strains were either added to independent serum tubes, or differently marked strains (typically the wild-type, YAG40, and two differently marked strains with the same gene disrupted) were competed in the same tube. Serum was incubated at 37°C, and at various time points, aliquots were removed, serially diluted, and plated to selective media to determine cell viability. To provide an approximate estimate of cell viability, cultures were diluted 10-fold and 5-μl spots were plated. To determine absolute numbers, 100-μl aliquots of appropriate dilutions were plated at least in duplicate. To test C. albicans serum survival, the assay was essentially the same, but strains were incubated in serum separately, and the incubation temperature was 30°C to reduce hyphal formation. Survival in other media was assayed similarly. Typically, experiments were performed with two individually constructed strains with the same gene disrupted to ensure reproducibility of results.
Serum was treated in various ways to remove individual components. Serum and YPD pH was adjusted using HCl or KOH, following the addition of 0.05 mM HEPES buffer. Serum was delipidated by using PHM-L Liposorb (Calbiochem) according to the manufacturer's instructions. To remove proteins, serum was filtered through a <3-kDa Centricon centrifugal filter and then heat inactivated by incubation at 56°C for 30 min, followed by an overnight digestion with proteinase K (InVitrogen) (50 μg ml−1) at 37°C. Calcium and iron were chelated by the addition of 1,2-bis(2-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid (BAPTA; 4 mM) or deferoxamine mesylate (25 μg ml−1), respectively. Other divalent cations were removed by incubation of serum with 5% (wt/vol) Chelex-100 (Sigma) at 4°C for 3 h. Catalase (200 μg ml−1) was added and incubated for 3 h at 25°C to remove serum peroxide. Where specified, amino acids were added to serum at the following concentrations: l-serine (250 μg ml−1), l-threonine (400 μg ml−1), l-methionine (170 μg ml−1), l-isoleucine (120 μg ml−1), l-glycine (230 μg ml−1), and l-valine (410 μg ml−1). Survival in human serum (Sigma catalog no. H4522) was also tested.
Experimental mouse infections.
The in vivo survival of S. cerevisiae strains was compared following infection by lateral tail vein injection of 4-week-old male CD-1 mice (outbred, immune competent; Charles River Laboratories), as described previously (10, 22). For each gene disruption tested, 15 mice were injected with 2 × 107 CFU of an equal ratio of one reference strain (YAG40) and two experimental strains, each containing different markers disrupting the same gene, that had been grown to mid-log phase in YPD medium, washed in sterile phosphate-buffered saline (PBS), and resuspended in PBS. To determine the exact proportion of each strain present, the inocula were diluted and plated to selective media. At times 4 h, 1 week, and 2 weeks postinfection, five mice per time point were euthanized by CO2 inhalation. Since the brain is the predominant organ inhabited by S. cerevisiae in CD-1 mice (10), the brains were recovered, homogenized in 5 ml PBS supplemented with 100 μg ml−1 ampicillin and streptomycin, pelleted by centrifugation (700 × g for 10 min), resuspended in 1 to 2 ml PBS, and plated to selective media to determine the relative numbers of each strain present. Results at each time point (t = x) were expressed as competitive index (CI) values (7, 9), a measure of (experimental strain/reference strain)t = x/(experimental strain/reference strain)t = 0.
To test virulence of C. albicans strains, 7-week-old male CD-1 mice were given a lateral tail vein injection of 1 × 106 cells suspended in PBS buffer. Mice were observed twice daily, and animals that appeared moribund (>15% loss of body weight, lethargic, or not accessing food) were sacrificed. Mice that remained healthy throughout the course of the experiment were euthanized after 28 days, and their organs were recovered, homogenized in 5 ml PBS plus ampicillin plus streptomycin, and then plated to plates containing YPD plus nourseothricin (YPD + Nat) to determine if the infection had been cleared. Mouse survival data were analyzed using the Kaplan-Meier test.
Mice were fed ad libitum for the course of the experiments. All animal experiments met with institutional guidelines and were approved by the Institutional Animal Care and Use Committee.
MIC assays.
To determine the MICs of fluconazole (Pfizer), 5-fluorocytosine (Sigma), amphotericin B (Gibco), and caspofungin (Merck), 10-μl volumes of a 2-fold dilution series of each drug were dispensed into the wells of a flat-bottomed microdilution plate (Corning). Subsequently, 90-μl volumes of cells resuspended in 1.1 × YPD or SD plus methionine plus threonine (YPD + Met + Thr) to a concentration of approximately 2 × 103 CFU ml−1 were added to the microdilution plate wells containing the drug dilutions, and the microdilution plates were incubated for 2 days at 30°C. Experiments were typically performed in triplicate on two independent mutants. MICs were defined as the minimum concentration of drug that inhibited growth to ≤20% of the OD600 measurement of the no-drug control, following background correction (MIC80). To determine minimum fungicidal concentrations (MFCs), the entire contents of wells containing no visible growth were plated on YPD. The MFC was defined as the lowest concentration of drug that resulted in a 95% reduction in CFU from the initial inoculums (MFC95).
RESULTS
S. cerevisiae thr1Δ and thr4Δ mutants are very rapidly eliminated in vivo.
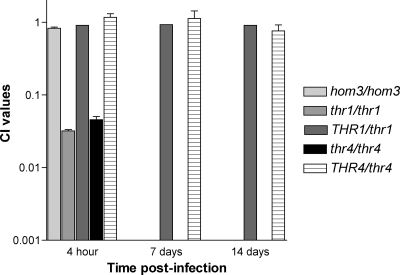
Having determined that S. cerevisiae hom3Δ mutants are eliminated by 1 week in vivo (31), we decided to assess the in vivo survival requirement of other threonine biosynthetic genes. The survival in the brains of mice of two differently marked thr1Δ (YJK498a and YJK506a) and thr4Δ (YJK369b and YJK391b) strains, constructed in a clinically derived S. cerevisiae strain, was compared with that of the wild-type strain (YAG40). Like the hom3Δ mutants, the thr1Δ and thr4Δ mutants were unrecoverable after 1 week in vivo, with average CI values of <0.0009 (thr1Δ mutants) and <0.0015 (thr4Δ mutants) (Fig. 1). However, in contrast to the hom3Δ mutants, the thr1Δ and thr4Δ mutants were severely attenuated after only 4 h in vivo, with average CI values of 0.032 and 0.045, respectively. Complementation of the mutants with the wild-type THR1 (YJK547) and THR4 (YJK456 and YJK461) genes restored survival to wild-type levels (average CI values of 0.91 and 0.76 after 2 weeks, respectively); thus, in vivo survival was indeed dependent upon THR1 and THR4.
Fig. 1.
Survival of S. cerevisiae thr1Δ and thr4Δ strains in vivo. Results were the average of experiments with two independent mutants with the same gene deleted. hom3Δ mutant results were previously published (31) and were included as a comparison. CI values = (experimental strain/reference strain)t = x/(experimental strain/reference strain)t = 0.
C. albicans thr1Δ mutants have attenuated virulence.
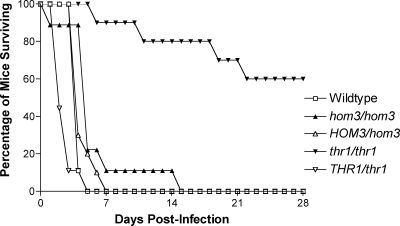
Given the severe in vivo survival defects of S. cerevisiae threonine pathway mutants and the essential nature of these genes in C. neoformans (34), we investigated the virulence phenotypes of hom3Δ and thr1Δ mutants constructed in the highly clinically relevant pathogen C. albicans. We chose to focus on C. albicans thr1Δ rather than thr4Δ mutants since S. cerevisiae thr1Δ mutants have phenotypes similar to, to more severe than, those of thr4Δ mutants (33). The mice infected with the hom3Δ mutant (CJK41) survived a similar length of time to those infected with the wild type (SC5314) and the HOM3-complemented mutant (CJK61), with mean survival times of 4.9 ± 3.8, 3.1 ± 0.3, and 3.6 ± 1.1 days, respectively (Fig. 2). In contrast, the virulence of the thr1Δ (CJK35) mutant was considerably attenuated compared with the wild type (P < 0.0001), with 60% of the infected mice still surviving after 28 days. thr1Δ mutants were still recovered from the kidneys of all of the surviving mice; therefore, mice had not cleared the infection. The attenuated virulence of the thr1Δ mutant was confirmed to be due to the absence of THR1, as the THR1-complemented strain (CJK59) had similar virulence to the wild type (mean survival time of 1.7 ± 1.0 days).
Fig. 2.
Virulence phenotypes of C. albicans threonine pathway mutants.
thr1Δ and thr4Δ mutants are extremely serum sensitive.
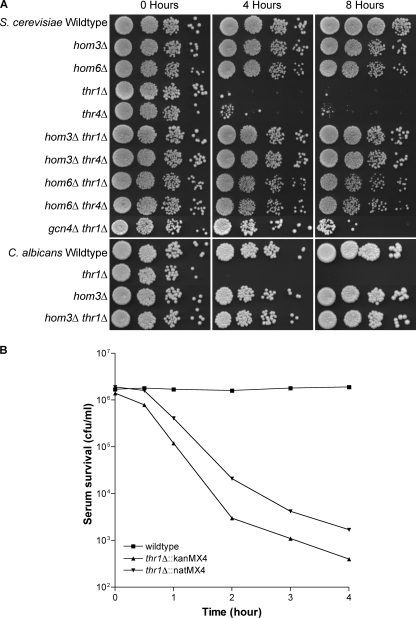
The considerable difference in survival between the S. cerevisiae thr1Δ and thr4Δ mutants and the wild type and hom3Δ mutants after only 4 h in vivo was far too high to be attributable to differences in growth rate and was not due to auxotrophy per se. The results instead indicate that the in vivo environment is more toxic to thr1Δ and thr4Δ mutants. Since an initial stage in this experimental infection process, and likely most natural infections, involves transit through the bloodstream, we investigated whether S. cerevisiae thr1Δ and thr4Δ mutants were serum sensitive. Two each of the hom3Δ, thr1Δ, and thr4Δ mutants were incubated with the wild type in fetal bovine serum (FBS) at 37°C and then serially diluted and plated to selective media to determine survival. As judged by semiquantitative spot dilution assays (Fig. 3A), the wild-type viability increased over time and hom3Δ mutant viability remained at the inoculated level, while thr1Δ and thr4Δ mutant numbers declined dramatically, with an approximately 100- to 1,000-fold reduction after 4 h. Since thr1Δ (and thr4Δ) mutants were already highly depleted after 4 h, a separate competition experiment to determine how rapidly mutants are killed in serum compared overall numbers of the wild type (YAG40) and two differently marked thr1Δ mutants (YJK498a and YJK506a) surviving serum incubation at earlier time points. As can be seen in Fig. 3B, thr1Δ mutant viability declined rapidly, with average reductions from input values of 8-fold, 279-fold, 863-fold, and 2,309-fold, following 1, 2, 3, or 4 h of incubation, respectively.
Fig. 3.
(A) Serum sensitivity of S. cerevisiae and C. albicans threonine pathway mutants and the S. cerevisiae thr1Δ gcn4Δ mutant. Strains were incubated in serum for the time designated, and 5-μl volumes of 10-fold dilutions were plated on YPD medium. (B) Time course for serum killing of S. cerevisiae thr1Δ mutants.
To investigate whether serum sensitivity is a conserved phenomenon of thr1Δ mutants, the serum survival rates of C. albicans, thr1Δ (CJK35 and CJK37), hom3Δ (CJK41 and CJK43), and wild-type (SC5314) strains were also compared. All strains were incubated at 30°C to minimize hyphal formation, but microscopic examination at 4 h revealed that the wild-type and the hom3Δ mutants still produced some hyphae. Since even low levels of hyphal formation in serum by the wild-type and hom3Δ mutants resulted in some clumping in serum, their overall cell numbers were greater than those represented by spot dilution. However, no hyphal formation was observed for the thr1Δ mutant. While the C. albicans wild-type and hom3Δ mutant numbers increased over time, thr1Δ mutants were found to also be extremely serum sensitive, with an approximately 100- to 1,000-fold reduction in viability after 4 h (Fig. 3A).
Serum toxicity is due to low threonine levels.
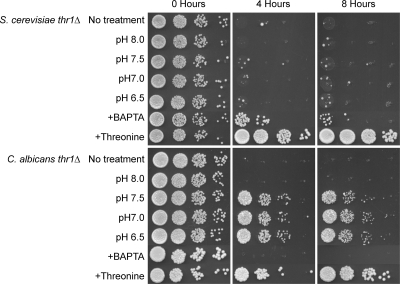
Microscopic examination of serum-incubated cells showed that S. cerevisiae thr1Δ mutants were predominantly unbudded cells, thus ruling out cell clumping as an explanation for low numbers of recoverable colonies. Therefore, to identify the toxic feature of serum, S. cerevisiae thr1Δ mutants were incubated in serum that had been treated in various ways to remove serum components, individually and in combination, and viability at different time points was estimated by spot dilution on YPD. Reduction of incubation temperature to 30°C had no influence on S. cerevisiae thr1Δ mutant survival, nor did serum pH, serum source (with FBS and human serum tested), proteins, peroxide, lipids, iron, or other trivalent and divalent ions other than calcium (see Table S2 in the supplemental material). Chelation of calcium resulted in an approximately 10-fold increase in S. cerevisiae thr1Δ mutant survival after 4 h but had no benefit at later time points (Fig. 4). The calcium effect was serum specific, as addition of CaCl2 at levels equivalent to calcium in serum (3.1 mM) had no effect on the viability of S. cerevisiae thr1Δ mutants in SD + Met + Thr (data not shown).
Fig. 4.
Effect of pH, calcium chelation, and threonine addition on serum sensitivity of S. cerevisiae and C. albicans thr1Δ mutants. Strains were incubated in serum for the time designated, and 5-μl volumes of 10-fold dilutions were plated onto YPD.
Interestingly, incubation in two defined serum-substitute media, DMEM (plus Na MOPS plus NaHCO3) and RPMI 1640, was also toxic to S. cerevisiae thr1Δ mutants, with an approximately 10-fold reduction in RPMI 1640 and a 100- to 1,000-fold reduction in DMEM after 4 h (see Fig. S2 in the supplemental material). Therefore, the thr1Δ-inhibitory feature of serum is not serum specific. Various versions of DMEM were prepared that were deficient in each inorganic salt individually or in combination, amino acids, or vitamins. Aside from a slight protective effect after 4 h in the absence of calcium chloride, consistent with calcium chelation in serum, the S. cerevisiae thr1Δ mutant died at a similar level in all of the other medium formulations.
The results implied that rather than the inhibitory feature being something that was present in serum and defined serum substitute media, it was in fact the absence or smaller amount of a compound. Since amino acids are present in serum and tissue culture media at levels lower than those added to yeast minimal media (54), threonine, and the threonine-related amino acids serine, glycine, methionine, isoleucine, and valine were added individually to serum at 10-fold-higher concentrations than those present in mouse serum (11). Interestingly, addition of threonine (Fig. 4), but no other amino acid tested (see Table S2 in the supplemental material), completely suppressed S. cerevisiae thr1Δ mutant serum sensitivity. Similarly, when S. cerevisiae thr1Δ mutants were incubated in SD + Met + low Thr (39 μg ml−1; equivalent to threonine levels in mouse serum [11]) medium, viability was reduced at least 100-fold after 8 h of incubation. In addition, when threonine was completely omitted from the medium, S. cerevisiae thr1Δ mutant numbers were reduced at least 100-fold after 4 h of incubation (Fig. 5). Consistent with serum results, incubation for 8 h in threonine-deficient medium had no effect on hom3Δ mutant viability. Therefore, the principal feature of serum toxic to S. cerevisiae thr1Δ mutants is the low threonine concentration.
Fig. 5.
Threonine starvation is cytocidal for S. cerevisiae and C. albicans thr1Δ mutants. Strains were incubated in SD + Met or SD + Met + low Thr (39 μg ml−1; equivalent to serum levels), and at the designated times, aliquots were removed and dilutions plated to YPD.
Similar experiments were also performed to ascertain why C. albicans thr1Δ mutants were serum sensitive. In contrast to S. cerevisiae thr1Δ mutants, calcium chelation had no effect on C. albicans thr1Δ mutant survival (see Table S2 in the supplemental material). Also contrasting with the S. cerevisiae thr1Δ results, buffering the serum pH to 7.5 or below substantially ameliorated C. albicans thr1Δ serum sensitivity, with an at least 100-fold increase in survival after 4 h (Fig. 4). The C. albicans thr1Δ mutants were observed to form hyphae in serum only when pH levels were conducive to survival. The pH sensitivity of C. albicans thr1Δ mutants was medium dependent, since there was no cytocidal effect when mutants were incubated in YPD medium at pH 8 (see Fig. S3 in the supplemental material). Importantly, as with the S. cerevisiae thr1Δ mutant, C. albicans thr1Δ mutant serum sensitivity could be completely overcome by threonine addition (Fig. 4), and mutants were similarly sensitive to incubation in minimal medium lacking threonine (Fig. 5).
Serum sensitivity is a consequence of intermediate accumulation.
The cidal nature of serum and threonine starvation on thr1Δ mutants in both S. cerevisiae and C. albicans is not merely a consequence of threonine auxotrophy since these phenotypes are not observed in hom3Δ mutants. Furthermore, we have shown that S. cerevisiae thr1Δ and thr4Δ as well as C. albicans thr1Δ mutants are sensitive to exogenous homoserine, and the starvation-cidal phenotype of S. cerevisiae thr1Δ and thr4Δ mutants is due to the inability to detoxify homoserine (33). To investigate whether the S. cerevisiae thr1Δ and thr4Δ and C. albicans thr1Δ serum and threonine starvation sensitivity are also due to intermediate accumulation, we compared the serum and threonine starvation survival phenotypes of S. cerevisiae hom6Δ (YJK862), hom3Δ thr1Δ (YJK682), hom3Δ thr4Δ (YJK677), hom6Δ thr1Δ (YJK948), hom6Δ thr4Δ (YJK1379), and C. albicans hom3Δ thr1Δ (CJK77) mutants. As observed for the S. cerevisiae thr1Δ and thr4Δ threonine starvation sensitivity (33), the C. albicans threonine starvation sensitivity and the serum sensitivity in both species were suppressed by disruption of HOM3 and/or HOM6, indicating that these phenotypes are also due to the accumulation of a toxic intermediate (Fig. 3A and 5A). Because mutation of genes required for the production of the intermediate homoserine blocks serum and threonine starvation sensitivity and mutation of the genes required for homoserine conversion to threonine confers serum and threonine starvation sensitivity, we predict that homoserine is the toxic intermediate accumulating.
Conditions that increase the flux through the threonine biosynthetic pathway, such as the induction of general control upon amino acid starvation, are toxic to thr1Δ mutants due to increased homoserine production (33). We investigated whether disruption of GCN4, encoding the master regulator of general control (26), influenced S. cerevisiae thr1Δ mutant serum survival. Although gcn4Δ thr1Δ (YJK1078) mutants were reduced in viability over time, survival was substantially higher than that of thr1Δ mutants at early time points, with an approximately 100-fold increased survival after 4 h of serum incubation and 10-fold increased survival after 8 h, as estimated by spot dilution (Fig. 3A). Therefore, results are consistent with low threonine levels in serum contributing to thr1Δ mutant serum sensitivity by increasing homoserine production through the elicitation of general control.
Drug sensitivity of threonine auxotrophs.
To test if Thr1p or Thr4p inhibitors would have synergistic action with other classes of antifungal drugs, the MICs of 5-fluorocytosine and the MICs and MFCs of fluconazole, amphotericin B, and caspofungin were determined for S. cerevisiae and C. albicans threonine pathway mutants in SD + Met + Thr and YPD media. Mutation of threonine pathway genes had little to no effect on fluconazole or caspofungin sensitivity (data not shown). While C. albicans strains were equally sensitive to amphotericin B, S. cerevisiae thr1Δ and thr4Δ strains were modestly (2-fold) more sensitive to amphotericin B in YPD, but not in SD + Met + Thr medium. Interestingly, while S. cerevisiae thr1Δ and thr4Δ strains were equally sensitive to 5-fluorocytosine in SD + Met + Thr medium, relative to the wild type, hom3Δ, and hom3Δ thr1Δ and hom3 thr4Δ double mutants, thr1Δ and thr4Δ mutants were at least four times more sensitive to 5-fluorocytosine in YPD (Table 2). Similarly, relative to the wild type, hom3Δ, and hom3Δ thr1Δ mutants, C. albicans thr1Δ mutants were four times more sensitive to 5-fluorocytosine in SD + Met + Thr medium and at least 30-fold more sensitive in YPD. In both species, since hom3Δ blocked the 5-fluorocytosine sensitivity of thr1Δ and thr4Δ mutants, sensitivity is a consequence of toxic intermediate accumulation. Therefore, even in the presence of abundant exogenous threonine, intermediate accumulation has deleterious, clinically relevant effects.
Table 2.
MIC80s of 5-fluorocytosine
| Strain genotype | MIC80 of 5-fluorocytosine on medium: |
|
|---|---|---|
| SD + Met + Thr (ng ml−1) | YPD (μg ml−1) | |
| S. cerevisiae | ||
| Wild type | 10–20 | 250 |
| hom3Δ | 10 | 250 |
| thr1Δ | 10 | 62.5 |
| thr4Δ | 10 | 62.5 |
| hom3Δ thr1Δ | 10 | 250 |
| hom3Δ thr4Δ | 10 | 250 |
| C. albicans | ||
| Wild type | 15.6 | >250 |
| hom3Δ | 15.6 | >250 |
| thr1Δ | 3.9 | 7.81 |
| hom3Δ thr1Δ | 15.6 | >250 |
DISCUSSION
The ability to survive in serum represents a crucial aspect for fungal virulence since travel through the bloodstream is necessary for fungal dissemination and systemic infection, and many components and conditions present in the bloodstream likely mimic those found in other body niches occupied during fungal infection. However, serum is a hostile environment for fungal growth due to various components of innate immunity, high pH, ionic composition, and the low concentrations of many nutrients. Consequently, we observed only modest serum proliferation by wild-type S. cerevisiae, a rarely observed pathogen. Conversely, the robustly pathogenic C. albicans proliferates profusely in serum and undergoes a yeast-to-hypha morphological transition upon serum exposure, enabling tissue invasion and escape from the bloodstream, which is mediated by a number of environmental cues, such as pH and temperature (reviewed in references 6, 24, and 39). Not surprisingly, the loss of the ability to survive in serum, mediated by disruption of the calcium-activated phosphatase calcineurin, correlates with attenuated virulence in both C. albicans and Aspergillus fumigatus (5, 13). Despite the importance of serum survival as a virulence factor, much of what is known about the genetic requirements for fungal proliferation and survival in serum is implied through fungal transcriptional responses observed following serum exposure (18, 19). Consistent with a commonly observed lack of correlation between transcription and phenotype (for example, see reference 2), threonine biosynthetic pathway genes were not identified in these screens.
Since some threonine biosynthetic mutants have deleterious phenotypes in addition to auxotrophy (2, 8, 14–16, 20, 33, 51), S. cerevisiae hom3Δ mutants do not survive in vivo (31), and C. neoformans threonine biosynthetic genes are essential (34), we were interested in further exploring the potential of threonine biosynthetic enzymes as antifungal drug targets. To this end, we demonstrated that S. cerevisiae thr1Δ and thr4Δ mutants were not only unable to survive in vivo, they were also highly attenuated after only 4 h postinfection. Consistent with this attenuation, serum incubation for the same period of time resulted in a severe decline in viability of thr1Δ and thr4Δ, but not hom3Δ, mutants. Furthermore, while disruption of HOM3 in the clinically relevant pathogen C. albicans did not influence virulence, C. albicans thr1Δ mutants were considerably attenuated in virulence and were also serum sensitive.
Calcium was shown to cause the serum sensitivity of C. albicans calcineurin mutants (4), and we also identified calcium as a minor contributor to the serum- and DMEM-mediated killing of S. cerevisiae thr1Δ mutants. While S. cerevisiae thr4Δ mutants have been reported to be sensitive to calcium chloride at higher concentrations (0.7 M in YPD) (8), we observed no effect on thr1Δ mutant growth when calcium was added to SD + Met + Thr at levels equivalent to those found in serum (9.1 to 9.5 mg 100 ml−1) (11). Similar results were observed by Blankenship et al. (4), who attributed differences to the presence of a possible calcium chelator in YPD, reducing the effective calcium concentration, or different metabolic rates in the different types of media affecting the calcium response.
High pH was determined to be a major contributor to C. albicans thr1Δ serum lethality. At pHs greater than 7.5, thr1Δ mutants were also deficient in germ tube formation, a process that occurs soon after serum exposure; thus, thr1Δ mutants that managed to survive serum exposure would also be deficient in tissue invasion, a potential clinically relevant benefit of Thr1p inhibitors. The pH of mouse serum can range from 7.3 to 7.55 (11), thus straddling the pH limit to which C. albicans thr1Δ mutants survive in serum and form hyphae, which might explain why these mutants are not completely eradicated in vivo, compared with S. cerevisiae thr1Δ mutants. Any localized pH drop caused by fungal metabolism within a microcolony that managed to establish in vivo, could also provide a protective effect. While lowering serum pH did not support S. cerevisiae thr1Δ mutant survival in serum, thr1 mutants have been reported to be pH sensitive (20), and indeed, we observed that incubation of thr1Δ mutants in YPD at pHs above 7 was cidal. Thus, pH could be cidal in combination with an additional component of serum for S. cerevisiae thr1Δ mutants.
The major feature of serum demonstrated to be toxic to both S. cerevisiae and C. albicans thr1Δ mutants was its low threonine concentration (25 to 40 μg ml−1) (11, 12), which equates to approximately one-tenth of the concentration typically added to yeast minimal media (54). Since hom3Δ and hom6Δ mutants are not serum sensitive, and disruption of HOM3 or HOM6 blocks thr1Δ and thr4Δ mutant serum sensitivity, toxicity is not a consequence of altered expression of neighboring genes, threonine auxotrophy, or the loss of a second function of Thr1p or Thr4p; rather, we hypothesize that serum sensitivity is due to the accumulation of the biosynthetic intermediate homoserine, which we find to be toxic (33). Our model predicts that the low levels of threonine present in serum reduce threonine feedback repression of Hom3p (41, 48), increase transcription of threonine pathway genes by the induction of general control (26, 43), and hence increase pathway flux and homoserine formation. While the threonine biosynthetic intermediate β-aspartate semialdehyde has also been shown to be toxic (1), we find that hom6Δ mutants are not serum sensitive and hom6Δ suppresses thr1Δ mutant serum sensitivity; therefore, we ruled out a role for this intermediate in the serum sensitivity of thr1Δ and thr4Δ strains. Consistent with our toxic homoserine accumulation hypothesis, there is precedence for homoserine toxicity in mammalian (49) and bacterial (37, 44) cells. Increased homoserine levels, even in the presence of exogenous threonine, may enhance the sensitivity of thr1Δ and thr4Δ strains to various stresses, such as calcium and 5-fluorocytosine, and explain why C. neoformans THR1 repression is more deleterious than HOM3 repression (34). Furthermore, we determined that both endogenously produced homoserine and exogenously added homoserine are toxic in both S. cerevisiae thr1Δ and thr4Δ mutants and C. albicans thr1Δ mutants, and this toxicity contributes to various other phenotypes described for S. cerevisiae thr1Δ and thr4Δ mutants, including pH sensitivity, likely by acting as a threonine analog (33). One hypothesis is that homoserine may act as a toxic threonine analog, inhibiting or misincorporating in processes that utilize threonine as a substrate.
The extreme survival defects of S. cerevisiae thr1Δ and thr4Δ mutants in vivo, the inhibition of hyphal formation, the attenuated survival of C. albicans thr1Δ mutants, and the essentiality of these genes in C. neoformans (34) all validate the potential of Thr1p and Thr4p as antifungal drug targets. Several inhibitors, including phosphohomoserine analogs (17) and the phosphohomoserine analog-containing peptides rhizocticin and plumbemycin (38, 40), have been identified that target bacterial threonine synthase; however, the therapeutic efficacy of each is limited since many peptides are subject to serum protease digestion (30, 47), various phosphohomoserine analogs are also N-methyl-d-aspartate (NMDA) receptor antagonists (57), and we found no inhibition of yeast by d,l-threo-β-hydroxyaspartic acid or d,l-2-amino-5-phosphonovaleric acid (data not shown). Nonetheless, the rapidity and profound lethality observed upon exposure of thr1Δ and thr4Δ mutants to serum and threonine starvation conditions indicate that Thr1p and Thr4p inhibitors would be fungicidal in vivo. Such a fungicidal effect would be advantageous over fungistatic agents since fungistatic agents require immune function to clear existing fungi from the body so that the infection does not recrudesce upon drug removal, and many fungal infections occur in immunocompromised individuals. Since both S. cerevisiae and C. albicans thr1Δ are sensitive to other inhibitors of amino acid biosynthesis, likely due to general control-mediated increased flux through the threonine biosynthetic pathway that results in increased homoserine production (33), we predict that inhibitors of Thr1p (and Thr4p) would have a therapeutically beneficial synergistic action with inhibitors of other amino acid biosynthetic pathways. For example, thr1Δ mutants are hypersensitive to the herbicide sulfometuron methyl (33), which targets the isoleucine and valine biosynthetic enzyme acetolactate synthase, also an interesting drug target, since it is also required for fungal virulence and/or survival in vivo (31, 32, 36). An additional virtue for targeting Thr1p and Thr4p is the demonstration that thr1Δ and thr4Δ mutants are hypersensitive to the clinically relevant drug 5-fluorocytosine, even under conditions of high threonine. Since 5-fluorocytosine is converted into 5-fluorouracil, which inhibits RNA and DNA metabolism (25, 28, 29, 53), and thr1Δ mutants are hypersensitive to DNA-damaging agents (2, 3), 5-fluorocytosine-induced DNA damage may explain the observed hypersensitivity. Thus, novel Thr1p or Thr4p inhibitors would have great potential as sole (C. neoformans) and combination (C. albicans) therapies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the laboratory of Joseph Heitman for the plasmid pSFS2A with permission from Joachim Morschhäuser, Anders Esberg for statistical advice, and Joseph Heitman for critical evaluation of the manuscript.
This study was funded by the National Institutes of Health R01 grant GM070541 and R21 grant AI070247.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 19 March 2010.
REFERENCES
- 1.Arevalo-Rodriguez M., Pan X., Boeke J. D., Heitman J. 2004. FKBP12 controls aspartate pathway flux in Saccharomyces cerevisiae to prevent toxic intermediate accumulation. Eukaryot. Cell 3:1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birrell G. W., Brown J. A., Wu H. I., Giaever G., Chu A. M., Davis R. W., Brown J. M. 2002. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. U. S. A. 99:8778–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birrell G. W., Giaever G., Chu A. M., Davis R. W., Brown J. M. 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. U. S. A. 98:12608–12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blankenship J. R., Heitman J. 2005. Calcineurin is required for Candida albicans to survive calcium stress in serum. Infect. Immun. 73:5767–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blankenship J. R., Wormley F. L., Boyce M. K., Schell W. A., Filler S. G., Perfect J. R., Heitman J. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell 2:422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown A. J., Gow N. A. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333–338 [DOI] [PubMed] [Google Scholar]
- 7.Brown J. S., Aufauvre-Brown A., Brown J., Jennings J. M., Arst H., Jr., Holden D. W. 2000. Signature-tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol. Microbiol. 36:1371–1380 [DOI] [PubMed] [Google Scholar]
- 8.Care A., Vousden K. A., Binley K. M., Radcliffe P., Trevethick J., Mannazzu I., Sudbery P. E. 2004. A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics 166:707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang S. L., Mekalanos J. J. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 27:797–805 [DOI] [PubMed] [Google Scholar]
- 10.Clemons K. V., McCusker J. H., Davis R. W., Stevens D. A. 1994. Comparative pathogenesis of clinical and nonclinical isolates of Saccharomyces cerevisiae. J. Infect. Dis. 169:859–867 [DOI] [PubMed] [Google Scholar]
- 11.Crispens C. G. 1975. Handbook on the laboratory mouse, section IV, p. 93–123 Charles C. Thomas, Springfield, IL [Google Scholar]
- 12.Cynober L. A. 2002. Plasma amino acid levels with a note on membrane transport: characteristics, regulation, and metabolic significance. Nutrition 18:761–766 [DOI] [PubMed] [Google Scholar]
- 13.da Silva Ferreira M. E., Heinekamp T., Hartl A., Brakhage A. A., Semighini C. P., Harris S. D., Savoldi M., de Gouvea P. F., de Souza Goldman M. H., Goldman G. H. 2007. Functional characterization of the Aspergillus fumigatus calcineurin. Fungal Genet. Biol. 44:219–230 [DOI] [PubMed] [Google Scholar]
- 14.Deutschbauer A. M., Williams R. M., Chu A. M., Davis R. W. 2002. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 99:15530–15535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn C. D., Lee M. S., Spencer F. A., Jensen R. E. 2006. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol. Biol. Cell 17:213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enyenihi A. H., Saunders W. S. 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrington G. K., Kumar A., Shames S. L., Ewaskiewicz J. I., Ash D. E., Wedler F. C. 1993. Threonine synthase of Escherichia coli: inhibition by classical and slow-binding analogues of homoserine phosphate. Arch. Biochem. Biophys. 307:165–174 [DOI] [PubMed] [Google Scholar]
- 18.Fradin C., De Groot P., MacCallum D., Schaller M., Klis F., Odds F. C., Hube B. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56:397–415 [DOI] [PubMed] [Google Scholar]
- 19.Fradin C., Kretschmar M., Nichterlein T., Gaillardin C., d'Enfert C., Hube B. 2003. Stage-specific gene expression of Candida albicans in human blood. Mol. Microbiol. 47:1523–1543 [DOI] [PubMed] [Google Scholar]
- 20.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Guldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kotter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 21.Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 22.Goldstein A. L., McCusker J. H. 2001. Development of Saccharomyces cerevisiae as a model pathogen. A system for the genetic identification of gene products required for survival in the mammalian host environment. Genetics 159:499–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein A. L., McCusker J. H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- 24.Gow N. A., Brown A. J., Odds F. C. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366–371 [DOI] [PubMed] [Google Scholar]
- 25.Gustavsson M., Ronne H. 2008. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA 14:666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnebusch A. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae, p. 319–414 InJones E. W., Pringle J. R., Broach J. R. (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Hoffman C. S., Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:265–272 [DOI] [PubMed] [Google Scholar]
- 28.Hoskins J., Butler J. S. 2008. RNA-based 5-fluorouracil toxicity requires the pseudouridylation activity of Cbf5p. Genetics 179:323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoskins J., Scott Butler J. 2007. Evidence for distinct DNA- and RNA-based mechanisms of 5-fluorouracil cytotoxicity in Saccharomyces cerevisiae. Yeast 24:861–870 [DOI] [PubMed] [Google Scholar]
- 30.Jenssen H., Aspmo S. I. 2008. Serum stability of peptides. Methods Mol. Biol. 494:177–186 [DOI] [PubMed] [Google Scholar]
- 31.Kingsbury J. M., Goldstein A. L., McCusker J. H. 2006. Role of nitrogen and carbon transport, regulation, and metabolism genes for Saccharomyces cerevisiae survival in vivo. Eukaryot. Cell 5:816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kingsbury J. M., McCusker J. H. 2010. Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2Δ) mutants is influenced by the carbon source and rapamycin. Microbiology 156:929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingsbury J. M., McCusker J. H. 2010. Homoserine toxicity in Saccharomyces cerevisiae and Candida albicans homoserine kinase (thr1Δ) mutants. Eukaryot. Cell 9:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kingsbury J. M., McCusker J. H. 2008. Threonine biosynthetic genes are essential in Cryptococcus neoformans. Microbiology 154:2767–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kingsbury J. M., Yang Z., Ganous T. M., Cox G. M., McCusker J. H. 2004. A novel chimeric spermidine synthase-saccharopine dehydrogenase (SPE3-LYS9) gene in the human pathogen Cryptococcus neoformans. Eukaryot. Cell 3:752–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kingsbury J. M., Yang Z., Ganous T. M., Cox G. M., McCusker J. H. 2004. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37°C and in vivo. Microbiology 150:1547–1558 [DOI] [PubMed] [Google Scholar]
- 37.Kotre A. M., Sullivan S. J., Savageau M. A. 1973. Metabolic regulation by homoserine in Escherichia coli B-r. J. Bacteriol. 116:663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kugler M., Loeffler W., Rapp C., Kern A., Jung G. 1990. Rhizocticin A, an antifungal phosphono-oligopeptide of Bacillus subtilis ATCC 6633: biological properties. Arch. Microbiol. 153:276–281 [DOI] [PubMed] [Google Scholar]
- 39.Kumamoto C. A., Vinces M. D. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7:1546–1554 [DOI] [PubMed] [Google Scholar]
- 40.Laber B., Gerbling K. P., Harde C., Neff K. H., Nordhoff E., Pohlenz H. D. 1994. Mechanisms of interaction of Escherichia coli threonine synthase with substrates and inhibitors. Biochemistry 33:3413–3423 [DOI] [PubMed] [Google Scholar]
- 41.Martin-Rendon E., Farfan M. J., Ramos C., Calderon I. L. 1993. Isolation of a mutant allele that deregulates the threonine biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 24:465–471 [DOI] [PubMed] [Google Scholar]
- 42.McCusker J. H., Clemons K. V., Stevens D. A., Davis R. W. 1994. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics 136:1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mountain H. A., Bystrom A. S., Larsen J. T., Korch C. 1991. Four major transcriptional responses in the methionine/threonine biosynthetic pathway of Saccharomyces cerevisiae. Yeast 7:781–803 [DOI] [PubMed] [Google Scholar]
- 44.O'Barr T. P., Everett K. A. 1971. Effect of l-homoserine on the growth of Mycobacterium tuberculosis. Infect. Immun. 3:328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pascon R. C., Ganous T. M., Kingsbury J. M., Cox G. M., McCusker J. H. 2004. Cryptococcus neoformans methionine synthase: expression analysis and requirement for virulence. Microbiology 150:3013–3023 [DOI] [PubMed] [Google Scholar]
- 46.Payne S. H., Loomis W. F. 2006. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot. Cell 5:272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powell M. F., Grey H., Gaeta F., Sette A., Colon S. 1992. Peptide stability in drug development: a comparison of peptide reactivity in different biological media. J. Pharm. Sci. 81:731–735 [DOI] [PubMed] [Google Scholar]
- 48.Ramos C., Calderon I. L. 1992. Overproduction of threonine by Saccharomyces cerevisiae mutants resistant to hydroxynorvaline. Appl. Environ. Microbiol. 58:1677–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rees W. D., Grant S. D., Hay S. M., Saqib K. M. 1994. Threonine synthesis from homoserine as a selectable marker in mammalian cells. Biochem. J. 299:637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuss O., Vik A., Kolter R., Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 51.Roberg K. J., Bickel S., Rowley N., Kaiser C. A. 1997. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by Sec13, Lst4, Lst7 and Lst8. Genetics 147:1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 53.Seiple L., Jaruga P., Dizdaroglu M., Stivers J. T. 2006. Linking uracil base excision repair and 5-fluorouracil toxicity in yeast. Nucleic Acids Res. 34:140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman F., Fink G. R., Lawrence C. W. 1974. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 55.Tkacz J. S., DiDomenico B. 2001. Antifungals: what's in the pipeline. Curr. Opin. Microbiol. 4:540–545 [DOI] [PubMed] [Google Scholar]
- 56.Wach A., Brachat A., Pohlmann R., Philippsen P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808 [DOI] [PubMed] [Google Scholar]
- 57.Watkins J. C., Evans R. H. 1981. Excitatory amino acid transmitters. Annu. Rev. Pharmacol. Toxicol. 21:165–204 [DOI] [PubMed] [Google Scholar]
- 58.Yang Z., Pascon R. C., Alspaugh A., Cox G. M., McCusker J. H. 2002. Molecular and genetic analysis of the Cryptococcus neoformans MET3 gene and a met3 mutant. Microbiology 148:2617–2625 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.