Abstract
Genetic manipulation of mitochondrial DNA (mtDNA) is the most direct method for investigating mtDNA, but until now, this has been achieved only in the diploid yeast Saccharomyces cerevisiae. In this study, the ATP6 gene on mtDNA of the haploid yeast Candida glabrata (Torulopsis glabrata) was deleted by biolistic transformation of DNA fragments with a recoded ARG8m mitochondrial genetic marker, flanked by homologous arms to the ATP6 gene. Transformants were identified by arginine prototrophy. However, in the transformants, the original mtDNA was not lost spontaneously, even under arginine selective pressure. Moreover, the mtDNA transformants selectively lost the transformed mtDNA under aerobic conditions. The mtDNA heteroplasmy in the transformants was characterized by PCR, quantitative PCR, and Southern blotting, showing that the heteroplasmy was relatively stable in the absence of arginine. Aerobic conditions facilitated the loss of the original mtDNA, and anaerobic conditions favored loss of the transformed mtDNA. Moreover, detailed investigations showed that increases in reactive oxygen species in mitochondria lacking ATP6, along with their equal cell division, played important roles in determining the dynamics of heteroplasmy. Based on our analysis of mtDNA heteroplasmy in C. glabrata, we were able to generate homoplasmic Δatp6 mtDNA strains.
The mitochondrion is a double-membrane-bound organelle present in most eukaryotic cells and is responsible for many essential physiological functions, such as the citric acid cycle, ATP synthesis, amino acid synthesis, and signal transduction (40), apoptosis (54), and cellular proliferation (36). Mitochondria reproduce semiautonomously and are partially dependent on their own genome and transcription/translation system (19, 49). In organisms from yeasts to humans, deficiency in mitochondrial function affects the robust growth of host cells and can even lead to cell death (13, 47). Manipulation of mitochondrial function has the potential to be an efficient way to regulate the physiological processes of eukaryotic cells. The main strategy for manipulating yeast mitochondrial function is modifying the mitochondrial DNA (mtDNA). In most yeast species, this carries several key energy metabolism-related genes, such as COX1, COX2, COX3, ATP6, ATP8, and ATP9, and all mitochondrial tRNA genes (20, 30, 61), making mtDNA a critical component of the cell. Strains with mutated mtDNA (ρ−) or strains that completely lack mtDNA (ρ0) have respiratory deficiencies, leading to the formation of petite colonies (57). Cells that can grow without mtDNA are referred to as “petite positive,” and those that are inviable without mtDNA are termed “petite negative.” Most of ρ− and all ρ0 cells are respiration deficient (29). The genetic manipulation of mtDNA is the most direct route to understand the function of these genes and the physiological processes associated with the organelle (5, 48).
Mitochondria and mtDNA differ from the nucleus and the nuclear genome (nDNA) in several ways: (i) the mitochondrion is smaller than the nucleus, (ii) most cells have multiple mitochondria and mtDNA chromosomes (19), and (iii) codon usage for mitochondrial genes is different from that for nuclear genes. These special characteristics hinder mtDNA transformation techniques and restrict them to using classical antibiotic resistance markers and biosynthetic markers (1, 66). Fortunately, difficulties in mtDNA transformation can be overcome by a recoded ARG8m gene, which complements an Δarg8 deficiency in Saccharomyces cerevisiae nDNA when integrated into mtDNA with a suitable mitochondrial promoter and terminator (37). ARG8m contains two codons recognized as stop codons by the nDNA system, so it cannot be correctly translated when integrated into nDNA or when present in a yeast plasmid. The ARG8m-based mitochondrial transformation system is currently the most widely used in S. cerevisiae, allowing defined mutations and new genes to be inserted into mtDNA (5, 48).
A wide variety of defined alterations can now be generated in mtDNA, but this is confined to S. cerevisiae and the green alga Chlamydomonas reinhardtii (5). Few other successful systems for mtDNA transformation have been developed because of difficulties in mtDNA transformation. In S. cerevisiae, mtDNA transformation requires a yeast strain with multiple auxotrophies (Δarg8 Δura3/Δleu2) and that is ρ0 or ρ+ mit−. However, mit− strains are very difficult to obtain. While a ρ0 strain can be easily obtained by culturing with ethidium bromide, the newly transformed ρ0 cells must then be mated with ρ+ cells after mtDNA transformation (5). Strains with multiple auxotrophies with the ability to mate cannot always be obtained with industrial strains and clinical isolates (39).
Rapid increases in the use of yeasts in both the industrial biotechnology and clinical research settings call for increased understanding of the physiological processes of their mitochondria and mtDNA in order to optimize engineering processes and the discovery of new drugs (47, 63). Thus, this leads to an increased demand for a more effective route of genetic manipulation of mtDNA in other non-Saccharomyces eukaryotic microorganisms. The haploid facultative aerobe yeast Candida glabrata (Torulopsis glabrata) is the yeast species most similar to S. cerevisiae according to standard phylogenetic classification (4). It has been extensively characterized for use in industrial biotechnology (32, 65) and in clinical (28, 44) and basic research (39, 51) settings. C. glabrata is a common Candida pathogen, second only to C. albicans, and causes both bloodstream and mucosal infections (56). Some noninfectious C. glabrata strains are commonly used in the industrial-scale production of pyruvic acid or α-ketoglutaric acid (67). In contrast to other Candida species, C. glabrata is a “petite-positive” yeast (12), i.e., it is viable in the absence of mtDNA, making it an attractive model for mtDNA genetic manipulation. The complete mtDNA sequence of C. glabrata is 20 kb, making it the smallest among the sequenced hemiascomycetous yeasts and similar in size to human mtDNA (30). On the other hand, the 80-kb mtDNA of S. cerevisiae is larger than any other of the sequenced yeast mtDNAs, such as those of Candida albicans (40 kb), Yarrowia lipolytica (48 kb), and Pichia canadensis (27 kb). Despite its small size, the mtDNA gene content is highly similar to that of other characterized yeast strains (30).
ATP6 is part of the mtDNA of C. glabrata and encodes subunit 6 (a) of the Fo sector of mitochondrial FoF1-ATP synthase (30). In this work, ATP6 was partially deleted in C. glabrata mtDNA heteroplasmic cells, and the dynamics of heteroplasmic mtDNA transformants under aerobic, anaerobic, and oligomycin-supplemented conditions were investigated. Also, using an anaerobic screening process, a homoplasmic Δatp6 strain was obtained from heteroplasmic transformants. This characterization of the dynamics of mtDNA heteroplasmy may lead to effective techniques for the genetic manipulation of mtDNA in other haploid or mating-negative diploid yeasts and facilitate the definition of the physiological functions associated with mtDNA.
MATERIALS AND METHODS
Strains.
C. glabrata CCTCC M202019, a pyruvate overproducer, was screened in our lab (32). The C. glabrata Δarg8 Δura3 double mutant was previously generated from C. glabrata CCTCC M202019 (66).
Culture medium.
Yeast-peptone-dextrose culture medium (YPD) contains 10 g·liter−1 yeast extract, 20 g·liter−1 peptone, and 20 g·liter−1 dextrose; yeast peptone glycerol culture medium (YPG) contains 10 g·liter−1 yeast extract, 20 g·liter−1 peptone, and 20 g·liter−1 glycerol; minimal medium (MM) contains 20 g·liter−1 glucose, 1.0 g·liter−1 KH2PO4, 0.5 g·liter−1 MgSO4·7H2O, 10 g·liter−1 (NH4)2SO4, and 100 mg·liter−1 uracil; and supplement medium with arginine (SM) consists of MM with 100 mg·liter−1 arginine. MM-S and SM-S were MM and SM with 1 mol·liter−1 sorbitol. The initial pH of all media was adjusted to 5.5. All media included 10 ml of vitamin solution (1.0 g·liter−1 niconacid, 5.0 mg·liter−1 biotin, 5.0 mg·liter−1 vitamin B1, and 50 mg·liter−1 vitamin B6 [filter sterilized]). All plates were the corresponding liquid medium with 15 g·liter−1 of agar.
Replacement of ATP6 ORF with ARG8m.
The plasmid pDS24, containing an ARG8m gene (55), was used as the template for PCR amplification of Δatp6::ARG8m cassettes with primers Con-ATP6-F (GCggatccAATATTATTTATTATATAATAATATTAATTTTAATAAGTTATAATATATATTTATAA AGTATGACACATTTAGAAAGAAG) and Con-ATP6-R (GCGggatccTATTAATAATAATTAATTAAAGAATATTATAATATAATTAATTTATTTGTATTATATAAATTAAGCATATACAGCTTCG). The primers contained a BamHI site (lowercase) at their 5′ ends and regions of homology to ATP6 and ARG8m. The regions of homology (bold) to the ATP6 open reading frame (ORF) comprised 60 bp upstream of the ATP6 initiation codon and 60 bp downstream of the ATP6 stop codon (48). PCR products were digested with BamHI and inserted into pUC19. The resulting plasmid was named pUC-atp6::ARG8m. For biolistic transformation, C. glabrata Δura3 Δarg8 cells grown in 100 ml of YPD (200 rounds per minute [rpm], 30°C) to the desired growth phase were harvested and cultivated in MM-S to a cell density of 5 × 109 cells·ml−1. A 0.5-ml aliquot of cell paste was spread onto MM-S plates for precooling at 4°C before biolistic transformation (5). Plasmids were extracted, purified with an EZ Spin Column Plasmid Medi-Preps kit (Bio Basic Inc., Markham, Canada), and concentrated to 2 μg·μl−1 with DNAMate (Takara, Dalian, China). The concentration and purity of plasmids for transformation were determined with an Eppendorf BioPhotometer (Eppendorf AG, Hamburg, Germany). Gold Microcarrier or Tungsten Microcarrier (Bio-Rad, Hercules, CA) (0.6 μm or 1.0 μm) was used for mtDNA transformation. Concentrated DNA molecules were bound to microcarriers as described previously (5). Rupture disks of 900 lb/in2, 1,100 lb/in2, and 1,350 lb/in2 were used for yeast mtDNA transformation, and a stopping screen was not assembled (9). Distances between the disk with the lawn of cells and the macrocarrier assembly were 6 cm, 9 cm, and 15 cm. Biolistic transformation was performed using a PDS-1000/He system (Bio-Rad, Hercules, CA) (5), and bombarded plates were incubated at 30°C for 3 to 4 days until colonies appeared. Transformants were identified as Arg+ prototrophic colonies and subcloned on MM-S for at least three generations.
Colony PCR.
Half of a single colony or 20 μl of cultured cells was transferred to a 1.5-ml Eppendorf tube and heated in a microwave oven for 1 min (600 W) before addition of 25 μl of premixed PCR mixture. PCR was carried out as 94°C for 4 min for 1 cycle and then at 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min per kb for 30 cycles, followed by 10 min at 72°C (18).
Culture conditions for loss of transformed mtDNA.
Transformants on subcloned plates were picked, grown in 20 ml of MM (250-ml flask) for 24 h (200 rpm, 30°C), washed (4,000 × g for 1 min) with MM, and grown under the conditions described in Table 2. Anaerobic growth conditions were achieved by perfusion with purified nitrogen gas. Cells were washed with sterilized MM, diluted (10−5, 10−6, and 10−7), and spread on MM. The colonies that appeared on MM after 72 h were transferred to corresponding YPD and YPG plates. The percentage of heteroplasmic cells was calculated by the ratio of colonies on YPG to colonies on YPD.
Table 2.
Effect of culture conditions on the loss ratio of mtDNA(Δatp6::ARG8m)
| Medium | Loss ratio of mtDNA(Δatp6::ARG8m) (%) at: |
||
|---|---|---|---|
| t0 | t10 | t20 | |
| YPD | 27.0 | 64.9 | 99.6 |
| SM | 27.0 | 62.9 | 98.0 |
| MM | 27.0 | 43.1 | 61.0 |
| MM-oligomycin | 27.0 | 12.0 | 6.2 |
| MM, anaerobic | 27.0 | 14.0 | 7.1 |
| MM-oligomycin, anaerobic | 27.0 | 6.1 | 3.4 |
Effect of culture conditions on the loss ratio of mtDNA(Δatp6::ARG8m).
Transformants from a single colony in liquid MM were inoculated into YPD, SM, MM, or MM with oligomycin under aerobic conditions or into MM or MM with oligomycin under anaerobic conditions. All six cultures had the same transformed mtDNA loss ratio at the starting point (t0). Cultures were further incubated and diluted with fresh medium to prevent cell densities in excess of 5 × 107 cells·ml−1 and to maintain exponential growth. Samples from three cultures were plated for single colonies onto YPD after 10 (t10) and 20 (t20) generations and then replica plated onto MM and SM plates. Numbers are the percentages of Arg− clones that could grow on YPD or SM, but not MM, to Arg+ clones. At least 500 colonies were counted for each condition.
Fluorescence microscopy analysis.
Cells for staining were washed with phosphate-buffered saline (PBS) (5.84 g NaCl, 11.5 g Na2HPO4·7H2O, and 5.84 g NaH2PO4 per liter) and suspended in PBS containing 0.5 μM MitoTracker Green FM (Molecular Probes, Eugene, OR), a fluorescent green dye that localizes to mitochondria regardless of mitochondrial membrane potential (46, 60). After incubation at 30°C for 10 min, cells were washed and resuspended in PBS. Stained cells were observed with a Nikon Eclipse E600 fluorescence microscope connected to a DXM1200C camera and ACT-1 Nikon software (Nikon, Tokyo, Japan).
qPCR.
The relative mtDNA copy number (RCN) was determined by quantitative PCR (qPCR) with SYBR green (62). Nuclear (qPCR-ACT1-F/R) (39) and mtDNA (qPCR-COX1-F/R) primers (Table 1) were used to amplify ACT1 and COX1 with SYBR Premix Ex Taq II (Takara, Dalian, China). PCRs were performed on a Bio-Rad iCycler and analyzed with iCycler IQ software version 3.0a (Bio-Rad, Hercules, CA). At least three experiments were run for each condition analyzed. The relative amounts of ATP6 and ARG8m were determined with a qPCR assay similar to that used for RCN. ATP6 and ARG8m were amplified with primers qPCR-ATP6-F/R and qPCR-ARG8m-F/R (Table 1).
Table 1.
Oligonucleotide primers
| Function and name | Sequence (5′ → 3′) | Primer pair target or use |
|---|---|---|
| qPCR | ||
| qPCR-ACT1-F | AGTTGCTGCTTTAGTTATTG | ACT1 |
| qPCR-ACT1-R | CTTGGTGTCTTGGTCTAC | |
| qPCR-COX1-F | TGAGAACTAATGGTATGACAATGC | COX1 |
| qPCR-COX1-R | GTAACACCTGCTGATAATACTGG | |
| qPCR-ATP6-F | CTTATGTTGCTAGAGCTTTCT | ATP6 |
| qPCR-ATP6-R | AATACCAAATTCTAAGCACAT | |
| qPCR-ARG8 m-F | CACCAGTTGTACTACGAAGTTCTC | ARG8m |
| qPCR-ARG8 m-R | TGATAAAGCACCCATTGTTCTACC | |
| Strain confirmation | ||
| P1a | ATGAATCAATTAACATATGG | ATP6/ARG8m and flanking sequence |
| P2 | CTTCTGATAAAGCCATTC | |
| P3b | ATTCTTATTAACTTCACC | ATP6/ARG8m and left flanking sequence |
| P4 | ATGTATTCTGCATACTTT | |
| P4′ | TAGGATTAGCATGACCTA | |
| ARG8 m-F | ATGACACATTTAGAAAGAAG | ORF of ARG8m |
| ARG8 m-R | TTAAGCATATACAGCTTCG |
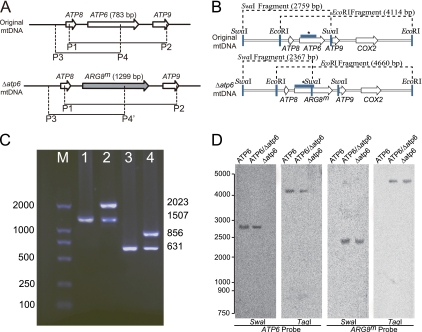
The products of primer pair P1/P2 were 1,507 bp and 2,023 bp with mtDNA(ATP6) and mtDNA(Δatp6::ARG8m), respectively.
The product of P3/P4 was 631 bp with mtDNA(ATP6), and there was no product with mtDNA(Δatp6::ARG8m). The product of P3/P4′ was 856 bp with mtDNA(Δatp6::ARG8m), and there was no product with mtDNA(ATP6).
PCR for strain confirmation.
To avoid the interference of nDNA, mtDNA was purified as described previously (15). Potential residual linear nDNA was eliminated by digestion with λ exonuclease and RecJf (New England Biolabs, Ipswich, MA) at 37°C for 16 h and inactivated at 65°C for 10 min (2). The λ exonuclease is an exo-DNase that digests double-stranded DNA from the 5′ end and forms single-stranded DNA (59), while RecJf is a single-strand-specific exonuclease that digests the remaining complementary single strand to mononucleotides (34). Combining the two removes linear DNA from a mixture of linear and supercoiled DNAs, leaving the supercoiled mtDNA intact (2). The replacement of ATP6 and existence of ARG8m were validated with primer pairs ATP869-F/R and ARG8m-F/R, using Takara LA Taq (Takara, Dalian, China) (Table 1).
Southern blot analysis.
The mtDNA for Southern blot analysis was prepared as described for a previous study (15) from C. glabrata Δura3 Δarg8 grown to exponential phase in SM and from mtDNA heteroplasmic transformants and final homoplasmic transformants grown in MM. mtDNA (5 μg) was digested with SwaI or TaqI, electrophoresed on a horizontal 1% agarose gel in 0.5× TAE buffer (20 mM Tris-acetate, 0.5 mM EDTA), and transferred to a Nytran membrane (Pall, East Hills, NY). Hybridizations with the ATP6 and ARG8m probes were carried out in 6× saline sodium citrate buffer (SSC) (175.3 g·liter−1 NaCl, 88.2 g·liter−1 sodium citrate), 1× Denhardt's solution (0.2 g·liter−1 Ficoll 400, 0.2 g·liter−1 PVP-360, 0.2 g·liter−1 bovine serum albumin [BSA] fraction V), 0.1% SDS, and 100 μg·ml−1 carrier DNA at 60°C. DNA probes were labeled with digoxigenin (DIG) using the DIG DNA labeling and detection kit (Roche, Indianapolis, IN) according to the manual.
ROS measurement.
Reactive oxygen species (ROS) production was measured by intracellular deacylation and oxidation of 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) (Sigma-Aldrich, St. Louis, MO) to the fluorescent compound 2′,7′-dichlorofluorescein (DCF). Cells grown for 24 h were collected, diluted to 107 cells·ml−1 in YPD medium with 40 μM DCFH-DA at 30°C for 60 min, washed, and resuspended in 100 μl of PBS (100 mM, pH 7.2). The fluorescence intensity of 100 μl of cell suspension containing 107 cells was measured directly, in arbitrary units, using a Hitachi 650-60 fluorescence spectrometer (Shimadzu, Kyoto, Japan) with excitation at 480 nm and emission at 530 nm (35). The ROS production was expressed as the ratio of arbitrary units relative to C. glabrata CCTCC M202019 cells grown for the same time under the same culture conditions.
RESULTS
Transformation of mtDNA.
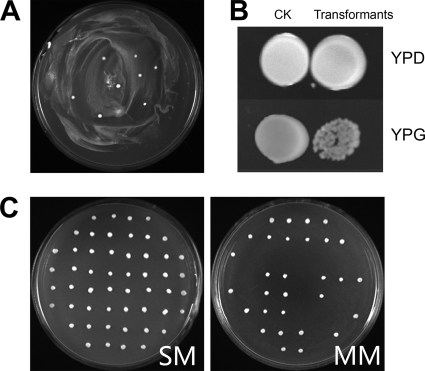
A plasmid for constructing the Δatp6::ARG8m strain was made and purified as described in Materials and Methods. After transformation of the double auxotroph Δura3 Δarg8 strain with pUC19-atp6::ARG8m, mtDNA transformants were identified as Arg+ prototrophic colonies. A series of experiments were performed to optimize the mtDNA transformation parameters, growth phase for transformation, type of rupture disk, type of microcarrier, and bombardment distance. The optimum transformation conditions were determined to be late log phase, 1,100-lb/in2 rupture disks, 29.5-in. Hg vacuum tightness, and 6-cm distance. The average microcarrier diameter of 1.0 μm or 0.6 μm for either gold or tungsten microcarriers had little effect on the transformation efficiency. However, even under optimum conditions, the transformation generated only a few transformants. A typical screening result is illustrated in Fig. 1A.
Fig. 1.
Biolistic transformation and characteristics of mtDNA transformants. (A) Transformants on bombarded plates. A YPD culture of 20 ml (30°C, 200 rpm, 24 h) of C. glabrata Δura3 Δarg8 cells was concentrated, washed with 1 mol·liter−1 sorbitol, spread on MM plates, and transformed with 5 μl of 2-μg·μl−1 pUC19-atp6::ARG8m using a particle gun. Plates were incubated at 30°C for 72 h until transformants appeared. (B) mtDNA transformants on MM and YPG plates. Overnight cultures (10 μl) of C. glabrata Δura3 Δarg8 (CK) and mtDNA transformants were spread on YPD and YPG and cultured at 30°C for 72 h. mtDNA transformants yielded colonies similar to those of CK. mtDNA transformants also grew on YPG, although more weakly. (C) mtDNA transformants lose the ability to grow on MM. After aerobic growth in YPD, some colonies lost the ability to grow on MM while retaining the ability to grow on SM. The ratio of MM to SM colonies was 32.5% based on the analysis of a total of 500 colonies.
Growth of mtDNA transformants on YPG plates.
Replicas of Arg+ transformants were picked and grown in liquid MM medium for 24 h (30°C, 200 rpm), and 10 μl of culture broth was dropped on MM and YPG and grown for 72 h. Based on previous reports, Δatp6::ARG8m transformants were expected to be respiration deficient and unable to use glycerol as a sole energy source, resulting in no growth on YPG (48). Double-auxotrophic cells not transformed with ARG8m should not be viable when grown on MM plates. However, all mtDNA transformants with the expected Δatp6::ARG8m genotype could grow on both MM and YPG plates (Fig. 1B).
Selective loss of mtDNA(Δatp6::ARG8m).
A single colony of mtDNA transformant was picked from an MM plate, grown in liquid YPD medium, diluted, and spread on YPD plates. Single colonies appearing after 72 h were replicated to SM and MM plates. All replicates grew on SM plates. Fewer than 70% of the MM replicas were viable (Fig. 1C). These results indicate that some mtDNA transformants lost their ability to grow on medium lacking arginine after aerobic growth on YPD.
Existence of heteroplasmy.
Possible explanations for above results were (i) contamination by other yeast strains and (ii) instability of transformed mtDNA. The results were reproducible under strict aseptic conditions, and mtDNA transformed by empty pUC19 did not yield similar results. Thus, we concluded that the data were not the result of contaminants. To extend our understanding of these transformants, colony PCR was performed on single colonies from the bombarded plates. The results showed that ATP6 was replaced with ARG8m, as shown by use of primer pairs P1/P2 and P3/P4 (P4′) (Table 1; Fig. 2A). However, both ARG8m and ATP6 could be amplified from the colonies, each of which represented a single transformant (Fig. 2C). Heteroplasmy was also confirmed by Southern blot analysis (Fig. 2D). These results strongly suggested that both the original and transformed mtDNAs coexisted in individual yeast cells.
Fig. 2.
Heteroplasmy. (A) Primers for confirming ATP6 or ARG8m mtDNA. (B) Probes and restriction enzyme sites for Southern blot analysis. (C) Electrophoresis of a typical colony using the primers shown in panel A. ATP6 or ARG8m was amplified using P1/P2, giving PCR products of 1,507 bp for the original mtDNA or 2,023 bp for Δatp6::ARG8m mtDNA (Table 1). Primers P3 and P4 were used for further validation. (D) Southern blot. mtDNA from C. glabrata Δura3 Δarg8, mtDNA primary transformants, and a homoplasmic Δatp6::ARG8m deletion strain were digested with SwaI or TaqI. DNA fragments were separated in 1% agarose and transferred to nitrocellulose. Membranes were hybridized with digoxigenin-labeled ATP6 and ARG8m probes. The relevant SwaI and TaqI restriction sites and positions of the probes (bar labeled with an asterisk) are shown in panel B.
Oligomycin and anaerobic culture decreases the loss ratio of mtDNA(Δatp6::ARG8m).
Various media were used to culture mtDNA transformants through 20 generations. The ability to grow on MM was lost by 99.6% of colonies from YPD, 98.0% from SM, and 61.0% from MM (Table 2), suggesting that the transformed cells selectively lost viability from growth on MM under normal aerobic conditions, even with media lacking arginine. These results meant that it was difficult to obtain homoplasmic cells that could grow only on MM plates but not on YPG plates. However, in order to determine the physiological characteristics of ATP6 deficiency, homoplasmic cells are necessary. Homoplasmic cells can be obtained by maintaining the transformed mtDNA(Δatp6::ARG8m) while decreasing the copy number of wild-type mtDNA(ATP6). Three independent strategies were attempted to obtain homoplasmic Δatp6::ARG8m cells: (i) aerobic growth in the presence of oligomycin, (ii) anaerobic growth, and (iii) anaerobic growth in the presence of oligomycin. Based on our previous work, the addition of sublethal concentrations of oligomycin decreases the intracellular ATP level by 35.7% (33). Addition of a sublethal concentration of oligomycin, a specific inhibitor of FoF1-ATPase (10), during aerobic growth decreased the loss ratio to 12.0% (t10) and 6.2% (t20) (Table 2). The loss ratio was reduced to 14.0% (t10) and 7.1% (t20) under anaerobic growth conditions. By combining oligomycin treatment with anaerobic growth, the loss ratio was further reduced to 6.1% (t10) and 3.4% (t20). The addition of oligomycin to anaerobic cultures resulted in growth decreases of 22.7% (MM-oligomycin), 43.4% (MM, anaerobic), and 17.6% (MM-oligomycin, anaerobic) relative to that of a 24-h aerobic MM culture. Among the three culture conditions, oligomycin strongly inhibited cell growth but had no obvious effect on mitochondrial biogenesis, and therefore anaerobic growth was employed in later experiments.
Mitochondrial copy number and ATP6/ARG8m ratio decrease under anaerobic growth conditions.
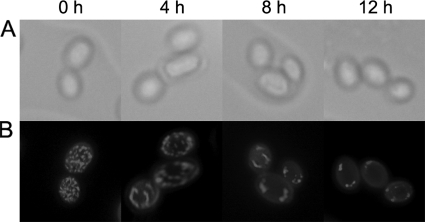
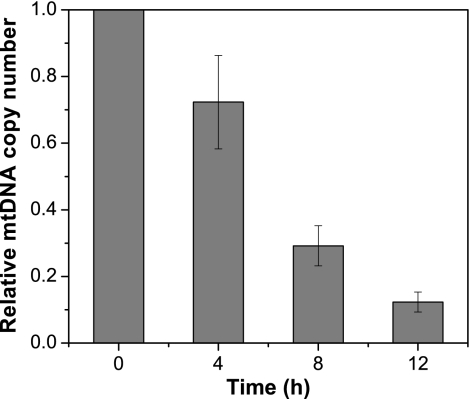
Cells grown under anaerobic conditions were visualized with MitoTracker Green FM staining (46) (Fig. 3), which showed that the copy number of mitochondria was significantly reduced with increasing lengths of anaerobic growth. However, no inhibition of mitochondrial biogenesis was observed with the addition of oligomycin. We propose that the likely decrease in the number of mitochondria, resulting from repressed mitochondrial biogenesis under anaerobic growth conditions, increases the occurrence of homoplasmic cells. The relative mtDNA copy number (RCN) was determined by qPCR (Fig. 4), showing that the RCN decreased with increased periods of anaerobic growth.
Fig. 3.
Mitochondrial biogenesis under anaerobiosis. To visualize mitochondria, cells were stained with MitoTracker Green FM. Discrete green staining creates fluorescence from mitochondria. Phase-contrast (A) and fluorescence (B) images of Δatp6 heteroplasmic cells from 0 h, 4 h, 8 h, and 12 h of anaerobic growth in MM are shown.
Fig. 4.
Effect of anaerobic growth on the mitochondrial copy number. mtDNA copy number is expressed as the relative value for cells during aerobic growth at the indicated time point. The approximate value of the mtDNA copy number shown here decreased with time during anaerobic growth. mtDNA copy number is a highly approximate value of the number of mitochondria and is not exact because of mtDNA concatemers (52) and mitochondria without mtDNA (48). Error bars indicate standard deviations.
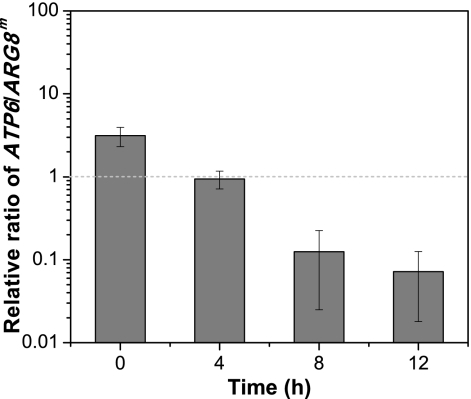
The copy numbers of ATP6 and ARG8m, representing mtDNA(ATP6) and mtDNA(Δatp6::ARG8m), were determined by qPCR. These results showed that the ratio of ATP6 to ARG8m decreases sharply under anaerobic growth conditions (Fig. 5). The decreases in mtDNA(ATP6) copy number concurrent with increases of mtDNA(Δatp6::ARG8m) during anaerobic growth make an attractive protocol for obtaining homoplasmic Δatp6::ARG8m cells.
Fig. 5.
Elimination of mtDNA heteroplasmy. The relative ratio of mtDNA(ATP6) and mtDNA(Δatp6::ARG8m) could be determined by qPCR of ATP6 and ARG8m because the two genes were unique in the two kinds of mtDNA molecules. Error bars indicate standard deviations.
Complete elimination of mtDNA(ATP6).
Based on the above results, anaerobic growth appeared to be an ideal route for screening the two important prerequisites for cells with homoplasmic mtDNA: maintaining mtDNA(Δatp6::ARG8m) and decreasing the mtDNA(ATP6) copy number. By PCR screening of purified mtDNA and selecting for growth on MM but not YPG, we isolated a single colony from an anaerobic broth culture that was confirmed as a Δatp6::ARG8m homoplasmic strain. This strain was confirmed by Southern blotting and by PCR with diagnostic primers, using purified mtDNA as a template, which avoided interference caused by nDNA or plasmids. The results demonstrated that the target gene was completely deleted (Fig. 2C), and the strain could no longer grow on YPG (data not shown).
ROS production in ATP6, ATP6/atp6, and atp6 strains.
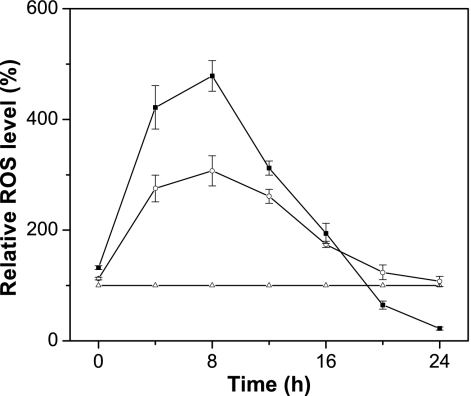
ROS production was measured using the ROS-sensitive fluorescent dye DCFH-DA in the original CCTCC M202019 (ATP6) strain, the ATP6 partial deletion ATP6/atp6 strain, and the homoplasmic Δatp6 strain. To minimize the ROS production, all three C. glabrata strains were grown in YPD under anaerobic conditions for 2 h before growth under aerobic conditions. After 8 h under aerobic conditions, ROS production was 207.2% in the ATP6/atp6 strain and 378.6% in the atp6 strain, which were higher than that in the ATP6 strain. An increase in aerobic culturing rapidly decreased ROS production in the ATP6/atp6 and atp6 strains. At 20 h, the ROS levels in the atp6 strain were equal to or lower than those in the ATP6 strain and the ROS level in the ATP6/atp6 strain was approximately the same as that in the ATP6 strain, while after 24 h of aerobic growth, the ROS level in the atp6 strain was only 22.3% of that in the ATP6 strain (Fig. 6).
Fig. 6.
ROS production in different kinds of yeast cells. ▵, C. glabrata with only mtDNA(ATP6); □, C. glabrata with both mtDNA(ATP6) and mtDNA(Δatp6::ARG8m); ▪, C. glabrata with only mtDNA(Δatp6::ARG8m). Error bars indicate standard deviations.
DISCUSSION
In this work, the ATP6 gene was deleted from the mtDNA of C. glabrata using DNA fragments containing a recoded ARG8m mitochondrial genetic marker flanked by arms homologous to the target gene, delivered into mitochondria by biolistic transformation. The transformants, identified by arginine auxotrophy, are unable to spontaneously discard the original mtDNA, even in the absence of arginine. In addition, some transformants selectively lost the ARG8m-containing mtDNA under aerobic conditions. The relatively stable heteroplasmy hindered the isolation of Δatp6::ARG8m homoplasmic mtDNA cells, preventing the clear characterization of physiological processes associated with mtDNA genes. Quantitative PCR (qPCR) assays revealed that the ratio of original to transformed mtDNA varied with different periods and conditions of growth. Further investigation showed that the cells with mtDNA(Δatp6::ARG8m) produced more ROS. Therefore, using these alterations in the growth conditions of the mtDNA heteroplasmy strain, we obtained a homoplasmic mutant deficient in ATP6. The mechanisms and strategies derived here should facilitate the efficient and convenient genetic manipulation of haploid yeast mtDNA and provide an alternative route for working with mtDNA in other eukaryotic microorganisms.
Heteroplasmy in eukaryotic cells.
The coexistence of different mtDNA molecules in a single eukaryotic cell, termed mtDNA heteroplasmy, can be observed from yeasts to higher organisms. Under natural conditions, mtDNA heteroplasmy arises from one of four sources: (i) partial damage to mtDNA from agents such as ionizing radiation and environmental toxins (17), (ii) mating of diploid fungal cells bearing different kinds of mtDNA (3, 27), (iii) some types of sexual reproduction in higher organisms (7, 53), and (iv) rare spontaneous heteroplasmy (50). Some plant and animal cells maintain a heteroplasmic state for extended periods (24, 64). However, heteroplasmic S. cerevisiae cells are not stable under most conditions (31) because they rapidly become homoplasmic through mitotic segregation (5), during which new buds receive relatively few copies of mtDNA from mother cells because of highly asymmetric S. cerevisiae budding (69). In mice, single cells with different mtDNAs also eliminate specific mtDNA in order to become homoplasmic (23, 53). The variation in mtDNA ratios during the elimination of the heteroplasmic state can be regarded as a typical mtDNA heteroplasmic dynamic process. Further investigation of heteroplasmic dynamics will contribute to our understanding of numerous mtDNA-associated physiological processes.
C. glabrata mtDNA transformants as a model for characterization of mtDNA heteroplasmy.
Many conserved nuclear factors control mtDNA integrity and transmission. Increased understanding of the problem in yeast may shed light on the relevant nuclear genes in higher eukaryotes. However, relatively stable and easily identifiable models are rare (31) and have not been extensively characterized (14) because in S. cerevisiae, the most widely investigated mtDNA transformation model, pure recombinant clones result from uncontrollable homologous recombination among the initial heteroplasmic cells (26). Although the rapid elimination of heteroplasmy facilitates the screening of homoplasmic mtDNA transformants, it hinders investigations on heteroplasmy dynamics, for example, determining the mechanism by which specific mitochondria with different mtDNAs are eliminated. Most naturally occurring heteroplasmic examples are small nucleotide polymorphisms or fragment length polymorphisms, which are difficult to identify (7, 50).
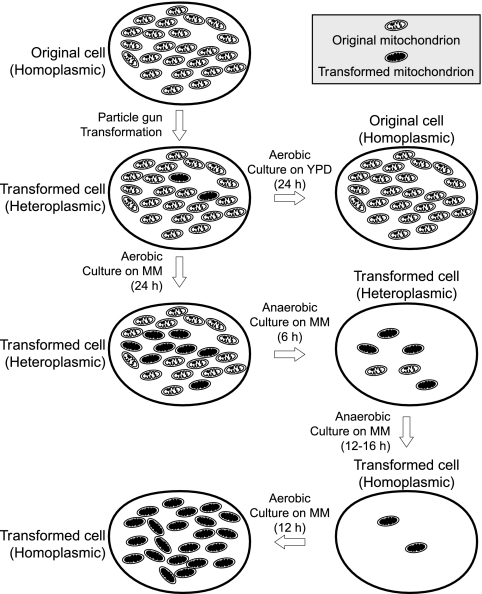
In this work, C. glabrata transformed with mtDNA was used as a model with easily identifiable phenotypes for studies on heteroplasmy dynamics and the relatively stable heteroplasmy. Heteroplasmy dynamics in C. glabrata are illustrated in Fig. 7, which describes the ratio of mtDNA(ATP6) to mtDNA(Δatp6::ARG8m) under various growth conditions. Based on our current hypothesis for heteroplasmy elimination, the phenomenon appears to be determined by three principal processes: (i) mtDNA instability, (ii) the selective loss of specific mtDNA, and (iii) mitochondrial biogenesis.
Fig. 7.
Process of mtDNA transformation and heteroplasmy dynamics. The original mtDNA contained all essential genes for oxidative phosphorylation, which supplied the cell with more ATP and facilitated growth. The transformed mtDNA lacked the ATP6 gene, which is essential for oxidative phosphorylation, but contained the ARG8m gene, which is essential for cell growth on medium without arginine.
Role of mtDNA instability in heteroplasmy.
The stability of mtDNA plays a significant role in the emergence of heteroplasmy. In yeast, mtDNA encodes the subunits of oxidative phosphorylation complexes that are essential for cellular respiration and ATP production, so maintenance of mtDNA integrity is essential for energy supply and robust cell growth (6, 13). Most deficiencies in FoF1-ATPase subunit genes, such as ATP1, ATP3 (21), ATP5 (45), ATP6 (48), ATP14 (22), and ATP15 (11), lead to mtDNA instability. Deficiency in the FoF1-ATP synthase induces a series of physiological changes, including accumulation of ROS (16, 43, 58), decreases in ATP synthesis, and alterations in mitochondrial crista morphology (42). A higher ROS level directly increases mtDNA instability (38, 58). Decreases in ATP synthesis may also influence mtDNA stability through effects on replication and ATP requirements for mtDNA maintenance. Deficiency in FoF1-ATP synthase subunits might inhibit complex assembly, resulting in alterations in mitochondrial crista morphology, all of which affect mtDNA stability (42). Cells with wild-type mtDNA(ATP6) have normal physiological ATP biosynthesis during aerobic growth. Arginine released from other cells may supply Δarg8 cells, because these cells grow well, even in MM with 10 mg·liter−1 of arginine. Thus, cells with only mtDNA(ATP6) could grow in liquid YPD or MM but not on MM plates.
Role of ROS production in mtDNA heteroplasmy.
Loss of mtDNA is common in yeast cells lacking the genes for FoF1-ATP synthase subunits. However, the detailed mechanism of this loss is not well elucidated (14). In this work, strains with partial and full deletions of ATP6 showed significantly different ROS levels after aerobic culture (Fig. 6). After 2 h of anaerobic growth, the ROS level was minimal for the three strains, C. glabrata Δarg8 Δura3 [with only mtDNA(ATP6)], C. glabrata with both mtDNA(ATP6) and mtDNA(Δatp6::ARG8m), and C. glabrata with only mtDNA(Δatp6::ARG8m). When these strains were aerobically cultured, ROS levels in the latter two sharply increased (Fig. 6), suggesting that mitochondria of the mtDNA(Δatp6::ARG8m) produce more ROS. ROS are respiratory by-products that damage mtDNA and other cellular components (8). The time course of C. glabrata with both mtDNA(ATP6) and mtDNA(Δatp6::ARG8m) initially showed sharply decreased ROS levels, which eventually reached the level in C. glabrata with only mtDNA(ATP6), suggesting that mtDNA(Δatp6::ARG8m) was gradually lost, leaving only mtDNA(ATP6). Furthermore, the time course of C. glabrata with only mtDNA(Δatp6::ARG8m) suggested that mtDNA(Δatp6::ARG8m) was almost completely lost after 20 h. Several key enzymes of the respiratory chain are encoded by the C. glabrata mtDNA, including Cox1p, Cox2p, and Cox3p. Therefore, complete loss of mtDNA would interrupt the respiratory chain (4), resulting in a lower ROS level than in C. glabrata with only mtDNA(ATP6).
Selective loss of mtDNA.
Host cells that were Δarg8 could not synthesize arginine, making the arginine-synthesizing ability of mtDNA(Δatp6::ARG8m) essential and forcing the maintenance of mtDNA(Δatp6::ARG8m). Therefore, although a large number of cells selectively lost mtDNA(Δatp6::ARG8m) under aerobic conditions, a group maintained mtDNA(Δatp6::ARG8m). Only 0.4% of cells retained the mtDNA(Δatp6::ARG8m) after 20 generations of aerobic growth in YPD; on the other hand, 92.9% retained the mtDNA(Δatp6::ARG8m) after 20 generations of anaerobic growth in MM (Table 2). The reason for these differences likely results from mitochondria with either mtDNA(ATP6) or mtDNA(Δatp6::ARG8m) failing to generate ATP from the oxidative phosphorylation during anaerobic growth (68). Thus, the selective advantage resulting from efficient ATP synthesis was eliminated, making the requirement for arginine synthesis the dominant selective advantage. This increases the viability of cells with mtDNA(Δatp6::ARG8m) during anaerobic growth.
Repression of mitochondrial biogenesis for screening homoplasmic cells.
Mitochondrial biogenesis is a complex physiological process involving a series of genes (19, 25) that play significant roles in the maintenance of mitochondrial morphology and function (3). Novel proteins and pathways that control mitochondrial biogenesis continue to be discovered, indicating that the mechanisms governing the behavior of this organelle are still not completely understood (41). In this work, we found that the mtDNA copy number in yeast cells can be decreased to a very low level during anaerobic growth (Fig. 3). This repression of mitochondrial biogenesis resulted in changes in the ratio of the two kinds of mtDNA used in this study. The decrease in mitochondrial number resulted in an increased frequency of occurrence of homoplasmic cells (Fig. 5).
In general, anaerobic culture had two effects on mtDNA maintenance. Under anaerobic conditions, integral FoF1-ATPase did not affect the ATP supply, because ATP was generated by substrate phosphorylation. Anaerobic growth inhibited mitochondrial biogenesis, thus decreasing the mtDNA copy number. Repression of mitochondrial biogenesis under anaerobic conditions further decreased the copy number of mtDNA(ATP6). Since the distribution of mitochondria during S. cerevisiae cell division is asymmetric, this led to an increased loss of mtDNA(ATP6) in some heteroplasmic cells (3).
In conclusion, we describe a reliable method for screening of homoplasmic mtDNA transformants as well as the primary mechanisms for the maintenance or elimination of mtDNA heteroplasmy using mitochondrial transformed C. glabrata cells. Furthermore, our results reveal a method for the controlled elimination of mtDNA heteroplasmy, making it an attractive model for further research concerning how specific mtDNA or mitochondria with specific mtDNA are selectively removed from the heteroplasmic cell.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Thomas D. Fox for the kind donation of pDS24 and continuous technical support, to Nathalie Bonnefoy and Malgorzata Rak for constructive discussion, and to Xiaowei Niu for help with biolistic transformation.
This work was supported by grants from the National Science Fund for Distinguished Young Scholars (grant no. 20625619), the Key Program of the National Natural Science Foundation of China (grant no. 20836003), the National Basic Research Program of China (973) (grant no. 2007CB714306), the National High-Tech Research and Development Program of China (863) (grant no. 2007AA100402), and the National Natural Science Foundation of China, General Program (grant no. 20706025).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 5 March 2010.
REFERENCES
- 1.Adrio J. L., Demain A. L. 2006. Genetic improvement of processes yielding microbial products. FEMS Microbiol. Rev. 30:187–214 [DOI] [PubMed] [Google Scholar]
- 2.Balagurumoorthy P., Adelstein S. J., Kassis A. I. 2008. Method to eliminate linear DNA from mixture containing nicked circular, supercoiled, and linear plasmid DNA. Anal. Biochem. 381:172–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger K. H., Yaffe M. P. 2000. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 8:508–513 [DOI] [PubMed] [Google Scholar]
- 4.Bialkova A., Subik J. 2006. Biology of the pathogenic yeast Candida glabrata. Folia Microbiol. (Praha) 51:3–20 [DOI] [PubMed] [Google Scholar]
- 5.Bonnefoy N., Remacle C., Fox T. D. 2007. Genetic transformation of Saccharomyces cerevisiae and Chlamydomonas reinhardtii mitochondria. Methods Cell Biol. 80:525–548 [DOI] [PubMed] [Google Scholar]
- 6.Brun S., Dalle F., Saulnier P., Renier G., Bonnin A., Chabasse D., Bouchara J. P. 2005. Biological consequences of petite mutations in Candida glabrata. J. Antimicrob. Chemother. 56:307–314 [DOI] [PubMed] [Google Scholar]
- 7.Burgstaller J. P., Schinogl P., Dinnyes A., Muller M., Steinborn R. 2007. Mitochondrial DNA heteroplasmy in ovine fetuses and sheep cloned by somatic cell nuclear transfer. BMC Dev. Biol. 7:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burhans W. C., Weinberger M. 2007. DNA replication stress, genome instability and aging. Nucleic Acids Res. 35:7545–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butow R. A., Henke R. M., Moran J. V., Belcher S. M., Perlman P. S. 1996. Transformation of Saccharomyces cerevisiae mitochondria using the biolistic gun. Methods Enzymol. 264:265–278 [DOI] [PubMed] [Google Scholar]
- 10.Charton C., Ulaszewski S., Vieira M. R. D., Henoux V., Claisse M. L. 2004. Effects of oligomycins on adenosine triphosphatase activity of mitochondria isolated from the yeasts Saccharomyces cerevisiae and Schwanniomyces castellii. Biochem. Biophys. Res. Commun. 318:67–72 [DOI] [PubMed] [Google Scholar]
- 11.Chen X. J. 2000. Absence of F1-ATPase activity in Kluyveromyces lactis lacking the epsilon subunit. Curr. Genet. 38:1–7 [DOI] [PubMed] [Google Scholar]
- 12.Chen X. J., Clark-Walker G. D. 2000. The petite mutation in yeasts: 50 years on. Int. Rev. Cytol. 194:197–238 [DOI] [PubMed] [Google Scholar]
- 13.Clark-Walker G. D. 2007. The F1-ATPase inhibitor lnh1 (IF1) affects suppression of mtDNA loss-lethality in Kluyveromyces lactis. FEMS Yeast Res. 7:665–674 [DOI] [PubMed] [Google Scholar]
- 14.Contamine V., Picard M. 2000. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64:281–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Defontaine A., Lecocq F. M., Hallet J. N. 1991. A rapid miniprep method for the preparation of yeast mitochondrial DNA. Nucleic Acids Res. 19:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doudican N. A., Song B., Shadel G. S., Doetsch P. W. 2005. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:5196–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Druzhyna N. M., Wilson G. L., LeDoux S. P. 2008. Mitochondrial DNA repair in aging and disease. Mech. Ageing Dev. 129:383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duenas E., Revuelta J. L., del Rey F., De Aldana C. R. V. 1999. Disruption and basic phenotypic analysis of six novel genes from the left arm of chromosome XIV of Saccharomyces cerevisiae. Yeast 15:63–72 [DOI] [PubMed] [Google Scholar]
- 19.Falkenberg M., Larsson N. G., Gustafsson C. M. 2007. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76:679–699 [DOI] [PubMed] [Google Scholar]
- 20.Foury F., Roganti T., Lecrenier N., Purnelle B. 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440:325–331 [DOI] [PubMed] [Google Scholar]
- 21.Francis B. R., White K. H., Thorsness P. E. 2007. Mutations in the Atp1p and Atp3p subunits of yeast ATP synthase differentially affect respiration and fermentation in Saccharomyces cerevisiae. J. Bioenerg. Biomembr. 39:127–144 [DOI] [PubMed] [Google Scholar]
- 22.Goyon V., Fronzes R., Salin B., di-Rago J. P., Velours J., Brethes D. 2008. Yeast cells depleted in atp14p fail to assemble Atp6p within the ATP synthase and exhibit altered mitochondrial cristae morphology. J. Biol. Chem. 283:9749–9758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyllensten U., Wharton D., Josefsson A., Wilson A. C. 1991. Paternal inheritance of mitochondrial-DNA in mice. Nature 352:255–257 [DOI] [PubMed] [Google Scholar]
- 24.Hanson M. R., Folkerts O. 1992. Structure and function of the higher plant mitochondrial genome. Int. Rev. Cytol. 141:129–172 [Google Scholar]
- 25.Hoppins S., Lackner L., Nunnari J. 2007. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76:751–780 [DOI] [PubMed] [Google Scholar]
- 26.Johnston S. A., Anziano P. Q., Shark K., Sanford J. C., Butow R. A. 1988. Mitochondrial transformation in yeast by bombardment with microprojectiles. Science 240:1538–1541 [DOI] [PubMed] [Google Scholar]
- 27.Kang D., Hamasaki N. 2002. Maintenance of mitochondrial DNA integrity: repair and degradation. Curr. Genet. 41:311–322 [DOI] [PubMed] [Google Scholar]
- 28.Kaur R., Domergue R., Zupancic M. L., Cormack B. P. 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8:378–384 [DOI] [PubMed] [Google Scholar]
- 29.Kominsky D. J., Thorsness P. E. 2000. Expression of the Saccharomyces cerevisiae gene YME1 in the petite-negative yeast Schizosaccharomyces pombe converts it to petite-positive. Genetics 154:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koszul R., Malpertuy A., Frangeul L., Bouchier C., Wincker P., Thierry A., Duthoy S., Ferris S., Hennequin C., Dujon B. 2003. The complete mitochondrial genome sequence of the pathogenic yeast Candida (Torulopsis) glabrata. FEBS Lett. 534:39–48 [DOI] [PubMed] [Google Scholar]
- 31.Lewin A. S., Morimoto R., Rabinowitz M. 1979. Stable heterogeneity of mitochondrial DNA in grande and petite strains of S. cerevisiae. Plasmid 2:474–484 [DOI] [PubMed] [Google Scholar]
- 32.Liu L. M., Li Y., Li H. Z., Chen J. 2004. Manipulating the pyruvate dehydrogenase bypass of a multi-vitamin auxotrophic yeast Torulopsis glabrata enhanced pyruvate production. Lett. Appl. Microbiol. 39:199–206 [DOI] [PubMed] [Google Scholar]
- 33.Liu L. M., Li Y., Li H. Z., Chen J. 2006. Significant increase of glycolytic flux in Torulopsis glabrata by inhibition of oxidative phosphorylation. FEMS Yeast Res. 6:1117–1129 [DOI] [PubMed] [Google Scholar]
- 34.Lovett S. T., Kolodner R. D. 1989. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machida K., Tanaka T., Fujita K. I., Taniguchi M. 1998. Farnesol-induced generation of reactive oxygen species via indirect inhibition of the mitochondrial electron transport chain in the yeast Saccharomyces cerevisiae. J. Bacteriol. 180:4460–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBride H. M., Neuspiel M., Wasiak S. 2006. Mitochondria: more than just a powerhouse. Curr. Biol. 16:R551–R560 [DOI] [PubMed] [Google Scholar]
- 37.McMullin T. W., Fox T. D. 1993. COX3 messenger RNA-specific translational activator proteins are associated with the inner mitochondrial-membrane in Saccharomyces cerevisiae. J. Biol. Chem. 268:11737–11741 [PubMed] [Google Scholar]
- 38.Muller F. L., Lustgarten M. S., Jang Y., Richardson A., Van Remmen H. 2007. Trends in oxidative aging theories. Free Radic. Biol. Med. 43:477–503 [DOI] [PubMed] [Google Scholar]
- 39.Muller H., Hennequin C., Gallaud J., Dujon B., Fairhead C. 2008. The asexual yeast Candida glabrata maintains distinct a and alpha haploid mating types. Eukaryot. Cell 7:848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson D. L., Cox M. M. 2004. Lehninger principles of biochemistry W. H. Freeman, New York, NY [Google Scholar]
- 41.Okamoto K., Shaw J. M. 2005. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39:503–536 [DOI] [PubMed] [Google Scholar]
- 42.Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D. M., Brethes D., di Rago J. P., Velours J. 2002. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21:221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perrone G. G., Tan S. X., Dawes I. W. 2008. Reactive oxygen species and yeast apoptosis. Biochim. Biophys. Acta 1783:1354–1368 [DOI] [PubMed] [Google Scholar]
- 44.Polakova S., Blume C., Zarate J. A., Mentel M., Jorck-Ramberg D., Stenderup J., Piskur J. 2009. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. U. S. A. 106:2688–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prescott M., Bush N. C., Nagley P., Devenish R. J. 1994. Properties of yeast cells depleted of the OSCP subunit of mitochondrial ATP synthase by regulated expression of the ATP5 gene. Biochem. Mol. Biol. Int. 34:789–799 [PubMed] [Google Scholar]
- 46.Presley A. D., Fuller K. M., Arriaga E. A. 2003. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. J. Chromatogr. B 793:141–150 [DOI] [PubMed] [Google Scholar]
- 47.Rak M., Tetaud E., Duvezin-Caubet S., Ezkurdia N., Bietenhader M., Rytka J., di Rago J. P. 2007. A yeast model of the neurogenic ataxia retinitis pigmentosa (NARP) T8993G mutation in the mitochondrial ATP synthase-6 gene. J. Biol. Chem. 282:34039–34047 [DOI] [PubMed] [Google Scholar]
- 48.Rak M., Tetaud E., Godard F., Sagot I., Salin B., Duvezin-Caubet S., Slonimski P. P., Rytka J., di Rago J. P. 2007. Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J. Biol. Chem. 282:10853–10864 [DOI] [PubMed] [Google Scholar]
- 49.Ryan M. T., Hoogenraad N. J. 2007. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 76:701–722 [DOI] [PubMed] [Google Scholar]
- 50.Sachadyn P., Zhang X. M., Clark L. D., Naviaux R. K., Heber-Katz E. 2008. Naturally occurring mitochondrial DNA heteroplasmy in the MRL mouse. Mitochondrion 8:358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt P., Walker J., Selway L., Stead D., Yin Z., Enjalbert B., Weig M., Brown A. J. 2008. Proteomic analysis of the pH response in the fungal pathogen Candida glabrata. Proteomics 8:534–544 [DOI] [PubMed] [Google Scholar]
- 52.Sedman T., Joers P., Kuusk S., Sedman J. 2005. Helicase Hmi1 stimulates the synthesis of concatemeric mitochondrial DNA molecules in yeast Saccharomyces cerevisiae. Curr. Genet. 47:213–222 [DOI] [PubMed] [Google Scholar]
- 53.Shitara H., Hayashi J., Takahama S., Kaneda H., Yonekawa H. 1998. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 148:851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh K. K. 2004. Mitochondria damage checkpoint in apoptosis and genome stability. FEMS Yeast Res. 5:127–132 [DOI] [PubMed] [Google Scholar]
- 55.Steele D. F., Butler C. A., Fox T. D. 1996. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. U. S. A. 93:5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoyan T., Carbon J. 2004. Inner kinetochore of the pathogenic yeast Candida glabrata. Eukaryot. Cell 3:1154–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strand M. K., Stuart G. R., Longley M. J., Graziewicz M. A., Dominick O. C., Copeland W. C. 2003. POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryot. Cell 2:809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stuart J. A., Brown M. F. 2006. Mitochondrial DNA maintenance and bioenergetics. Biochim. Biophys. Acta 1757:79–89 [DOI] [PubMed] [Google Scholar]
- 59.Subramanian K., Rutvisuttinunt W., Scott W., Myers R. S. 2003. The enzymatic basis of processivity in lambda exonuclease. Nucleic Acids Res. 31:1585–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swayne T. C., Gay A. C., Pon L. A. 2007. Visualization of mitochondria in budding yeast. Methods Cell Biol. 80:591–626 [DOI] [PubMed] [Google Scholar]
- 61.Talla E., Anthouard V., Bouchier C., Frangeul L., Dujon B. 2005. The complete mitochondrial genome of the yeast Kluyveromyces thermotolerans. FEBS Lett. 579:30–40 [DOI] [PubMed] [Google Scholar]
- 62.Taylor S. D., Zhang H., Eaton J. S., Rodeheffer M. S., Lebedeva M. A., O'Rourke T. W., Siede W., Shadel G. S. 2005. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol. Biol. Cell 16:3010–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toogood P. L. 2008. Mitochondrial drugs. Curr. Opin. Chem. Biol. 12:457–463 [DOI] [PubMed] [Google Scholar]
- 64.Wallace D. C. 1992. Diseases of the mitochondrial DNA. Annu. Rev. Biochem. 61:1175–1212 [DOI] [PubMed] [Google Scholar]
- 65.Wang Q. H., He P., Lu D. J., Shen A., Jiang N. 2005. Metabolic engineering of Torulopsis glabrata for improved pyruvate production. Enzyme Microb. Technol. 36:832–839 [Google Scholar]
- 66.Zhou J. W., Dong Z. Y., Liu L. M., Du G. C., Chen J. 2009. A reusable method for construction of non-marker large fragment deletion yeast auxotroph strains: a practice in Torulopsis glabrata. J. Microbiol. Methods 76:70–74 [DOI] [PubMed] [Google Scholar]
- 67.Zhou J. W., Huang L. X., Liu L. M., Chen J. 2009. Enhancement of pyruvate productivity by inducible expression of a F0F1-ATPase inhibitor INH1 in Torulopsis glabrata CCTCC M202019. J. Biotechnol. 144:120–126 [DOI] [PubMed] [Google Scholar]
- 68.Zhou J. W., Liu L. M., Shi Z. P., Du G. C., Chen J. 2009. ATP in current biotechnology: regulation, applications and perspectives. Biotechnol. Adv. 27:94–101 [DOI] [PubMed] [Google Scholar]
- 69.Zinn A. R., Pohlman J. K., Perlman P. S., Butow R. A. 1987. Kinetic and segregational analysis of mitochondrial DNA recombinant in yeast. Plasmid 17:248–256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.