Abstract
All living organisms respond to changes in their internal and external environment for their survival and existence. Signaling is primarily achieved through reversible phosphorylation of proteins in both prokaryotes and eukaryotes. A change in the phosphorylation state of a protein alters its function to enable the control of cellular responses. A number of serine/threonine kinases regulate the cellular responses of eukaryotes. Although common in eukaryotes, serine/threonine kinases have only recently been identified in prokaryotes. We have described that the human pathogen Group B Streptococcus (GBS, Streptococcus agalactiae) encodes a single membrane-associated, serine/threonine kinase (Stk1) that is important for virulence of this bacterium. In this study, we used a combination of phosphopeptide enrichment and mass spectrometry to enrich and identify serine (S) and threonine (T) phosphopeptides of GBS. A comparison of S/T phosphopeptides identified from the Stk1 expressing strains to the isogenic stk1 mutant indicates that 10 proteins are potential substrates of the GBS Stk1 enzyme. Some of these proteins are phosphorylated by Stk1 in vitro and a site-directed substitution of the phosphorylated threonine to an alanine abolished phosphorylation of an Stk1 substrate. Collectively, these studies provide a novel approach to identify serine/threonine kinase substrates for insight into their signaling in human pathogens like GBS.
Keywords: Bacteria, Streptococcus agalactiae, serine/threonine kinase, substrates, phosphorylation, neutral loss, mass spectrometry
Introduction
To survive, all living organisms must detect and process information from their environment. Signaling responses of living organisms to changes in their external environment is primarily achieved through reversible phosphorylation of proteins. This mechanism enables organisms to appropriately regulate protein function and cellular processes in response to the dynamic internal and external environment. Approximately 30% of all cellular proteins are estimated to be phosphorylated in eukaryotes.1 Multiple amino acid residues can undergo phosphorylation and affect protein function. Protein phosphorylation in eukaryotes commonly occurs on serine, threonine and tyrosine amino acids at an estimated ratio of 1000:100:1.1 These events require the function of serine/threonine and tyrosine kinases and their cognate phosphatases, and as many as 500 are present in the human genome.2 Deviation from normal phosphorylation events can lead to changes in cellular function and disease.3
In contrast to the serine/threonine and tyrosine kinase cascade systems in eukaryotes, signal transduction in prokaryotes is primarily accomplished by two-component systems (TCS). A typical TCS comprises a membrane-associated histidine kinase that is responsive to external signals, and phosphorylates its cognate response regulator at a conserved, active site aspartate residue.4–6 Most often, aspartate phosphorylation of the response regulator alters its DNA binding affinity, resulting in changes in gene expression (for reviews, see refs 5–7). The change in gene expression enables the organism to adapt and respond to the environmental signal.
Because a number of prokaryotes also encode eukaryotic-like serine/threonine protein kinases (STK) and their cognate phosphatases (STP) (for reviews, see refs 8 and 9), it is apparent that these organisms do not solely rely on TCS for signal transduction. Bacterial species such as Myxococcus, Mycobacteria, Cornyebacterium and Bacillus encode multiple STKs, while other bacterial species like Yersinia and Streptococci encode only a single STK.10–18 Interestingly, STK encoded by Mycobacteria (PknG) and Yersinia (YpkA) are secreted to regulate host cell functions.19–21 In contrast, STKs encoded by Gram-positive human pathogens such as Streptococci and Listeria are associated with the bacterial membrane.17 Further, the catalytic domain necessary for kinase activity (ATP binding and phosphotransfer) in Streptococci is intracellular.17,18 Consequently, these membrane-associated STKs can phosphorylate and regulate the function of specific bacterial proteins. Although signaling mediated by STKs in prokaryotes is not completely understood, they are important for virulence of human pathogens such as Yersinia, Pseudomonas, Mycobacterium, Streptococcus and Enterococcus species.13,17–20,22–27 Thus, it is essential to understand signaling mediated by these kinases to comprehensively understand the role of prokaryotic STK in bacterial infections.
A major limitation in understanding serine/threonine kinase signaling in prokaryotes and in bacterial disease pathogenesis is the difficulty in identification of their targets. Many strategies are currently employed to identify phosphorylated proteins and to discern phosphoproteomes.1,28,29 One of these strategies utilizes a mass spectrometric approach that is based on screening for the neutral loss of phosphate during collision induced dissociation (CID).1,28–30 As serine/threonine phosphopeptides readily lose phosphate groups during CID, tandem mass spectrometry (MS/MS) combined with neutral loss of H3PO4 (98 Da) has gained popularity in the identification of these phosphopeptides (see reviews31,32). Recently, this approach revealed that 78 proteins of Bacillus subtilis and 79 proteins in Escherichia coli are phosphorylated at serine, threonine and tyrosine residues.33,34 Because this method utilizes bacterial cells that are grown in normal laboratory media and are not treated or manipulated prior to harvesting, they represent in vivo conditions.33,34 Consequently, this approach has become widely utilized in the identification of phosphopeptides in both prokaryotic and eukaryotic organisms.
Our interest is to understand signaling responses of the human pathogen Group B Streptococcus (GBS, Streptococcus agalactiae) during its lifecycle as a commensal and as an invasive pathogen. While GBS commonly reside as commensal organisms in the genital and gastrointestinal tracks of healthy adult women, they are a significant cause of invasive infections in human newborns and in diabetic, immuno-compromised and elderly adults.35 Previous studies from our laboratory have shown that GBS encodes a single, membrane-associated, serine/threonine kinase known as Stk1 and its cognate phosphatase called Stp1 that are expressed during growth in laboratory media.17,36 Importantly, GBS strains deficient in Stk1 expression demonstrate decreased virulence in sepsis models of infection.17,37 Furthermore, these strains are unable to sustain de novo purine biosynthesis and demonstrate altered toxin expression.36,37 With the use of genetic and biochemical analyses, we discovered that Stk1 and a response regulator (RR) CovR coregulate toxin expression in GBS.37 Recently, we showed that Stk1 phosphorylates CovR at a threonine residue in position 65, and the consequent impact on promoter DNA binding and gene expression was also described.38
Because the GBS stk1 mutants exhibit pleiotropic phenotypes such as altered cell segregation apart from changes in toxin expression,17,37 we hypothesized that Stk1 may have additional targets other than CovR that are responsible for this phenotype. The purpose of this study was to identify proteins that are phosphorylated at serine and/or threonine residues during growth of GBS in rich media (hereafter referred to as in vivo). We used a strategy that employed a phosphopeptide enrichment procedure followed by mass spectrometry. To identify Stk1 specific targets under these in vivo conditions, we enriched and identified phosphopeptides that were unique to the Stk1 expressing GBS strains and confirmed that these were not present in the isogenic stk1 mutant. With these methods, we identified 10 phosphopeptides/proteins that were unique to the Stk1 expressing GBS strains. Therefore, we hypothesize that these 10 proteins are likely substrates of the GBS Stk1 enzyme. In vitro phosphorylation on selected targets and a site-directed substitution mutant support the data obtained from the phosphopeptide enrichment and mass spectrometric methods.
Experimental Procedures
Reagents
All chemicals and reagents used in this work were of mass spectrometric (MS) quality and were purchased from Sigma-Aldrich (St. Louis, MO), unless stated otherwise.
Bacterial Strains and Growth Conditions
The bacterial strains, plasmids and primers used in this study are listed in Table 1. GBS (S. agalactiae) strains were grown in 100 mL of Todd Hewitt Broth (THB, Difco Laboratories) at 37 °C in 5% CO2 to an optical density (OD600nm) of 0.6. Routine cultures of E. coli were performed in Luria–Bertani broth (LB, Difco Laboratories) at 37 °C. A909 is the wild-type (WT) GBS strain belonging to the capsular serotype Ia;39 LR114 was derived from A909 and is deficient in expression of both Stp1 and Stk1 (for details, see ref 17). Strain LR120 was derived from LR114 and is restored (or complemented) for expression of both Stp1 and Stk1 due to the presence of the plasmid pLR10.17 Strain LR119 was also derived from LR114 but is restored only for Stk1 expression due to complementation by the plasmid pLR9.17 To retain the complementing plasmids during growth of the strains LR119 and LR120, chloramphenicol was added to the growth media (THB) at a final concentration of 5 μg mL−1. For enhanced phosphopeptide stability during protein isolation,40,41 the serine/threonine (S/T) phosphatase inhibitor sodium fluoride (NaF) was added at a final concentration of 10 mM to the bacterial cultures when the optical density (OD600) reached 0.6 and incubated for 30 min at room temperature (RT). Subsequently, the cells were centrifuged and washed twice in ice-cold 20 mM Tris-HCl, pH 7.5 containing the phosphatase inhibitors (10 mM NaF, 10 mM Na pyrophosphate and 50 mM β-glycerophosphate). Cell pellets were then processed as described below.
Table 1.
Strains and Plasmids, and Primers Used in This Study
| strains | description | reference |
|---|---|---|
| Group B Streptococcus | ||
| A909 | Wild type (WT) Serotype Ia | 39 |
| LR114 | GBS A909, Stp1−Stk1− | 17 |
| LR119 | LR114/pLR9, Stk1+ | 17 |
| LR120 | LR114/pLR10, Stp1+ Stk1+ | 17 |
| E. coli | ||
| α-Select | E. coli strain for cloning | Bioline |
| BL21DE3 | E. coli strain for protein expression | Novagen |
| Origami | E. coli strain for protein expression | Novagen |
| plasmids | description | reference |
| pLR9 | Stk1+ | 17 |
| pLR10 | Stp1+ Stk1+ | 17 |
| pET32CK | pET32a derivative with trx removed\ApR; cloning vector | 43 |
| pAS5 | pET32CK encoding SAK_2030-His6 fusion protein | This study |
| pAS20 | pET32CK encoding SAK_0373-His6 fusion protein | This study |
| pAS21 | pET32CK encoding SAK_0586-His6 fusion protein | This study |
| pAS24 | pET32CK encoding SAK_0581-His6 fusion protein | This study |
| pAS5mut | pET32CK encoding site directed T4A/T7A mutant SAK_2030-His6 fusion protein | This study |
| Primers | ||
| primer name | sequence | |
| CHP2030_5 | 5′CATGCCATGGGATTTACAGATGAAACTGTTCGT 3′ | |
| CHP2030_3 | 5′CCGCTCGAGCAAATCTATTCCATTTCCTTGT 3′ | |
| ftsZ_5 | 5′CATGCCATGGTATTTTCATTTGATACAGCATC 3′ | |
| ftsZ_3 | 5′CCGCTCGAGACGGTTTTTAAAGAATGGAGGTG 3′ | |
| divIVA-0373_5 | 5′CATGCCATGGCAAGTATTATTTATAGCCC 3′ | |
| divIVA-0373_3 | 5′CCGCTCGAGTTCTCTAATTTGTCTTCCAA 3′ | |
| CHP-0586_5 | 5′CATGCCATGGCACTTACAGCACTTGAAATTA 3′ | |
| CHP-0586_3 | 5′CCGCTCGAGATCTTCAATATTTAATTTAAAAG 3′ | |
| pAS5mut upper | 5′CCCATGGGATTTGCAGATGAAGCTGTTCGTTTTAGATTAGATTAG 3′ | |
| pAS5mut lower | 5′CTAATCTAATCTAAAACGAACAGCTTCATCTGCAAATCCCATGGG 3′ | |
Isolation of Proteins and Tryptic Peptides
Total protein was isolated from the cell pellets using the QIAzol reagent (Qiagen) following the manufacturer instructions. Briefly, the cell suspensions (final volume 500 μL) were lysed using a FastPrep FP101 bead beater (Bio 101), with 3 × 30 s bursts at a power setting of 6. Unlysed cells were removed by centrifugation. Contaminating RNA and DNA were eliminated from the cell supernatants using chloroform extraction and ethanol precipitation, respectively. Finally, the proteins were precipitated using isopropanol, and these protein pellets were washed 3× in 0.3 M guanidine hydrochloride in 95% ethanol to assist in protein denaturation. Protein pellets were dried using a Speedvac, or alternatively were lyophilized. Following drying, the samples were reduced in the presence of 50 mM ammonium bicarbonate, 5 mM DTT and 8 M urea, then alkylated in the dark with 15 mM iodoacetamide for 30 min at RT. Total protein was quantified using BCA (Pierce, Rockford, IL), with the samples diluted to 1 M urea in 50 mM ammonium bicarbonate. The protein samples were then digested overnight (O/N) using sequencing grade trypsin at a ratio of 1:20 (trypsin/total protein) at 37 °C in 1 mM CaCl2. Peptides were separated by ultrafiltration using a 5000 Da cutoff membrane filtering device (Amicon Ultra-4 or “Ultrafree-CL”, Millipore, Billarica, MA) and peptide concentration was determined using BCA as described above. Approximately 1.5 mg of peptides from each GBS strain was acidified to pH ≤ 3.5 using trifluoroacetic acid (TFA), desalted by reverse phase chromatography on C18 column cartridges (Waters, Milford, MA), and eluted with 40% acetonitrile (ACN) in 0.1% TFA. Pure peptides were dried in a Speedvac for phosphomodification and enrichment procedure (see below).
Phosphopeptide Modification and Enrichment
Phosphopeptide modification and enrichment procedures were performed as described previously with a few modifications.42 Briefly, the peptide samples (1 mg) that contained both phosphopeptides and nonphosphopeptides were incubated at 12 °C for 90 min in 100 μL of methanolic HCl. Methanolic HCl was prepared by adding 80 μL of acetyl chloride to 500 μL of anhydrous methanol. Following methyl esterification, the solvent was evaporated using a Speedvac, and peptide methyl esters were resuspended in 16 μL solution of water/methanol/ACN at a ratio of 1:1:2. Subsequently, each sample was incubated with 25 μL of solution (pH 5.6) containing 100 mM imidazole, 100 mM N-(3-dimethylaminopropyl)N′-ethylcarbo-diimide hydrochloride (EDC), 100 mM 2-(N-morpholino) ethane-sulfonic acid (MES), pH 5.8, and 2 M cystamine for 15 h at RT with vigorous shaking. The reaction was stopped by the addition of 1 mL of 0.025% TFA and the peptides were subsequently bound to a C18 column (Waters, Milford, MA). The column was washed with 0.1% TFA and the bound peptides were treated with 10 mM tris-carboxyethyl phosphine (TCEP) in sodium phosphate buffer, pH 6.2, for 10 min to generate free thiol group. The column was washed with 0.1% TFA to remove TCEP and the phosphate buffer. The peptides were then eluted with 6 mL of 80% ACN in 0.1% TFA into 15 mL tubes that contained 180 μL of 1 M MES, pH 6.2. ACN was partially removed using the Speedvac to bring its final concentration to 30–40% and the final volume of the peptides to ~200 μL.
The modified peptide sample was then incubated with 10 mg of maleimide-functionalized glass beads in microcolumns containing a 10 μm pore filter (MoBioTec, Germany) in a rocking mixer at RT for 1 h. Maleimide-functionalized glass beads were prepared by a 2 h reaction between 4 equiv of 3-maleimidopropionoic acid, 1-Hydroxy-benzotriazole hydrate (HOBt) and 1,3-Diisopropylcarbodiimide (DICI) and 1 equiv of aminopropyl controlled pore glass (AMP-CPG) beads (Millipore, Billerica, MA) as described.42 The peptide bound beads were washed sequentially with 3 M NaCl, water, methanol and 80% ACN to remove nonspecifically bound peptides. The beads were then resuspended in 200 μL of water/ACN (1:1), transferred to a glass HPLC tube (Waters, Milford, MA) and incubated for 1.5 h with agitation at room temperature in 2.5% TFA. Subsequently, the beads containing the peptides were dried in a Speedvac, and the phosphopeptides were eluted using 200 μL of 80% ACN in 0.1% TFA. Recovered peptides were dried again in a Speedvac, and reconstituted in 8 μL of 0.1% formic acid for LC-MSn analysis.
Phosphopeptide Identification by Liquid Chromatography and Tandem Mass Spectrometry (LC-MS/MS)
The enriched phosphopeptides were analyzed using LC-MSn following the previously described procedure.42 Chromatographic separation of peptides was achieved using an Agilent Series 1100 LC system (Agilent Technologies) equipped with a reverse phase C18 column, a solvent delivery system and an autosampler as described.42 The peptide mixtures were separated over a 12–33% gradient of ACN, followed by detection, isolation, and fragmentation on a quadropole linear ion trap mass spectrometer (LTQ, Thermo Fisher, San Jose, CA). Data-dependent MSn spectra were acquired as follows. First, all eluted peptides were recorded in the MS mode. Subsequently, the five most intense MS ions were selected for product ion spectrum (MS2). An MS3 spectrum of the peaks in MS2 depicting neutral loss of phosphoric acid was automatically selected for the third scan event. All spectra were searched against the GBS A909 TIGR genome database http://cmr.jcvi.org/tigrscripts/CMR/GenomePage.cgi?org=gbsa909 using SEQUEST.
The MS2 spectra were searched to allow variable modifications of serine, threonine and tyrosine (+79.9663 Da) and static modifications of carboxyaminomethylation on cysteine (+57.0214 Da), as well as methylation of all carboxylate groups (+14.0156 Da). Further, the search results were also subjected to statistical filtering using PeptideProphet (V3.0). Peptides with first filtered with a p-value of at least 0.6. All spectra were manually validated for dominant fragment peaks corresponding to the neutral loss of phosphoric acid. For interpretation of the MS2 spectra, the same static modification described above was used, but variable modifications of −18 Da for S and T were permitted for the detection of neutral loss of phosphoric acid at these sites. These peptides had a PeptideProphet value of 0.9 or greater for these spectra.
Expression of Stk1 Targets for in Vitro Phosphorylation
The genes for selected targets from Table 2A were cloned into the expression vector pET32ck43 to generate C-terminal His-tagged fusions. The primers and E. coli host strains used for construction of recombinant plasmids and expression of proteins are listed in Table 1. Briefly, the target genes were PCR amplified using Pfu Turbo DNA Polymerase (Stratagene) and A909 genomic DNA was used as the template. All PCR products were digested with the enzymes for which restriction sites were engineered in the primers and were cloned in frame into the multiple cloning site (MCS) of pET32CK to obtain C-terminal His6 fusion proteins. Ligation products were cloned in E. coli α-select (Bioline, Taunton, MA) and all recombinant plasmids were sequenced. Subsequently, the plasmids were transformed into the E. coli strain Origami B (Novagen). Cultures were induced with 1 mM IPTG at OD600 = 0.6 for at least 3 h. Cells were harvested by centrifugation, and His-tagged fusion proteins were purified from cell free extracts using nickel affinity chromatography as described by the manufacturer (Qiagen). Subsequent in vitro phosphorylation of these substrates in the presence and absence of Stk1 was performed as described previously.37
Table 2 a.
| strain |
||||||
|---|---|---|---|---|---|---|
| locus | phosphopeptide | protein description | A909 Stk1+ Stp1+ | LR119b pStk1+ Stp1− | LR120b pStk1+ pStp1+ | LR114 Stk1− Stp1− |
| (A) S/T Phosphopeptides Unique to GBS Expressing Stk1 | ||||||
| Cell Division | ||||||
| SAK_0373 | NS*GTAMYNQKPIAQSATNFDIL | DivIVA domain | × | ✓ | × | × |
| NSGT*AMYNQKPIAQSATNFDIL | × | ✓ | × | × | ||
| NS*GTAMYNQK | × | ✓ | × | × | ||
| SAK_0586 | LPVDDTESFDAT*R | DivIVA | ✓ | ✓ | ✓ | × |
| VLDEDDALPVVDDTESFDAT*R | ✓ | ✓ | ✓ | × | ||
| SAK_0581 | VSFDT*ASVQGAVIK | FtsZ | × | ✓ | ✓ | × |
| TNQVSGFT*TSAPTNQAPSER | × | ✓ | × | × | ||
| DNISRPTEGELDS*K | × | ✓ | × | × | ||
| GNFDMT*ESR | × | ✓ | ✓ | × | ||
| GNFDMTES*R | × | ✓ | ✓ | × | ||
| Regulatory Functions | ||||||
| SAK_0389 | TLPKVTSTVSSLT*TEQLLR | Serine/threonine kinase, Stk1 | × | ✓ | ✓ | × |
| Unknown Functions | ||||||
| SAK_0375 | SSDFANLDT*ASLDDFIK | Hypothetical protein | × | ✓ | ✓ | × |
| SAK_0583 | TGQET*SFDFDMK | Hypothetical protein | × | ✓ | ✓ | × |
| SAK_0865 | GKFES*GELTTEDIVSAVK | Hypothetical protein | × | ✓ | ✓ | × |
| SAK_1559 | VSGQTILDQET*K | Hypothetical protein | ✓ | ✓ | ✓ | × |
| SAK_1628 | TDVATAVPNQET*EEIFLVR | Hypothetical protein | × | ✓ | ✓ | × |
| SAK_2030 | GFTDET*VR | Hypothetical protein | ✓ | ✓ | ✓ | × |
| GFT*DETVR | × | ✓ | × | × | ||
| (B) S/T Phosphopeptides Not Specific to Stk1 | ||||||
| Metabolic Functions | ||||||
| SAK_1010 | ASAGVMISAS*HNPALDNGIK | Phospho-glucosamine mutase, GlmM | ✓ | ✓ | ✓ | ✓ |
| SAK_1155 | AGIMVTAS*HNPAPFNGYK | Phospho-glucomutase/mannomutase | ✓ | ✓ | ✓ | ✓ |
| SAK_1682 | GGMT*SHAAVVAR | Pyruvate phosphate dikinase, PpdK | × | ✓ | × | ✓ |
Peptides listed showed neutral loss of phosphoric acid in the MS analysis. A909 represents wild-type GBS and LR114 represents the Stk1 and Stp1 deficient strain.
The complementing plasmid in LR119 and LR120 restore Stk1 and both Stp1 and Stk1 expression, respectively. The threonine or serine amino acid that is phosphorylated on the peptide is indicated as T* or S*, respectively. A ‘✓’ indicates that the phosphopeptide was identified in the mass spectrometric analysis from the corresponding strain. An ‘×’ indicates that the phosphopeptide was absent in the mass spectrometric analysis from the corresponding strain. SAK numbers correspond to the ORF of the gene in the genome sequence.62
Construction of Site-Directed Mutants
Site-directed substitution of the phosphorylated threonine to an alanine at positions 4 and 7 in SAK_2030 was performed to generate T4A/T7A SAK_2030. Inside-out PCR on the plasmid pAS5 that encodes SAK_2030 was performed using the plasmid as template and the corresponding mutagenic primers (Table 1). The QuickChange II Site-Directed Mutagenesis Kit from Stratagene was used to generate the mutant as per manufacturer’s instructions. The plasmid was sequenced to confirm the presence of the threonine to alanine substitutions and the absence of other mutations in the construct. Protein expression, purification and in vitro phosphorylation reactions were performed as described previously.37
Results
Enrichment and Isolation of Phosphopeptides from GBS
The human pathogen GBS encodes a single eukaryotic-like serine/threonine kinase Stk1 and its cognate phosphatase Stp1 (see Figure 1A for the operon structure and Rajagopal et al.17 for homology alignments). Stk1 is a 69 kDa membrane-associated serine/threonine protein kinase (STK) that has an N-terminal catalytic domain with the conserved Hanks domains.17 Stp1 is the cognate phosphatase of Stk1 and belongs to the PP2C family of protein phosphatases.17 We have previously shown that GBS deficient in Stk1 expression are attenuated for virulence in sepsis models of infection and demonstrate altered cell segregation and toxin expression.17,37 The purpose of this study was to identify potential in vivo substrates of the GBS Stk1 enzyme and their target phosphorylation sites.
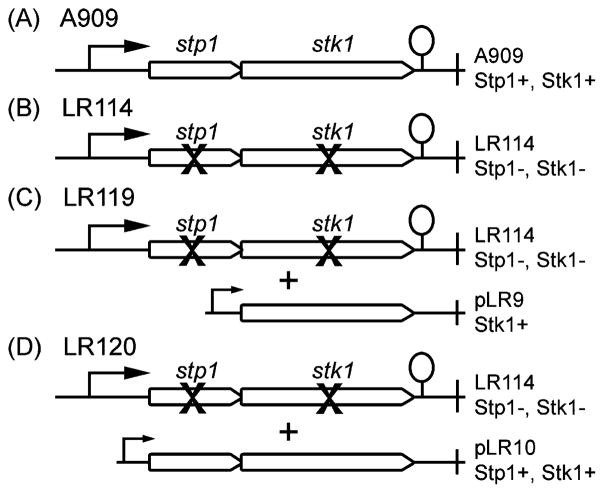
Figure 1.
Schematic representation of the serine/threonine kinase Stk1 and phosphatase Stp1 operon in GBS. (A) In wild-type GBS A909, genes stp1 and stk1 encode the serine/threonine phosphatase and serine/threonine kinase, respectively. (B) Mutant LR114 is deficient in expression of stp1 and stk1. (C) Partially complemented strain LR119 was derived from LR114 and contains the plasmid pLR9, which restores expression Stk1. (D) Fully complemented strain LR120 also derived from LR114 and contains the plasmid pLR10 that restores expression of stp1 and stk1.
Given that phosphoproteins are normally present in low abundance, we utilized a phosphopeptide enrichment approach42 for identification of Stk1 substrates. While there are multiple approaches for phosphopeptide enrichment, we utilized the chemical approach based on reversible covalent modification of the phosphate moietydue to its high specificity.42,44 This enrichment procedure enhances the success of identification of the phosphorylated proteins in subsequent mass spectrometric analysis.42 To identify the Stk1 target proteins that are phosphorylated in vivo, we enriched, isolated and identified phosphopeptides from four GBS strains. These included the wild-type GBS serotype Ia strain known as A90945 and an isogenic mutant (LR114) that is defective in expression of both the kinase Stk1 and its cognate phosphatase Stp1 (see panels A and B in Figure 1 and ref 17). We reasoned that a comparison of phosphorylated peptides from the WT strain (A909) to those present in the mutant LR114 would enable us to identify phosphopeptides that are unique to the WT and, hence, are likely substrates of the kinase Stk1. Because phosphorylation is reversible and general inhibitors such as NaF and sodium pyrophosphate were reported to only partially inhibit the function of bacterial PP2C serine/threonine phosphatases,46 we also included two complemented strains LR119 and LR120 in these analyses. LR119 is restored for Stk1 expression from a constitutive promoter on a multi copy plasmid and strain LR120 that is restored for both Stk1 and Stp1 expression (see panels C and D in Figure 1, also see Table 1, and refs 17 and 37). As Stp1 mediated dephosphorylation should not occur in LR119, we predicted that inclusion of this strain would enable us to identify most, if not all, Stk1 targets during GBS growth in laboratory media.
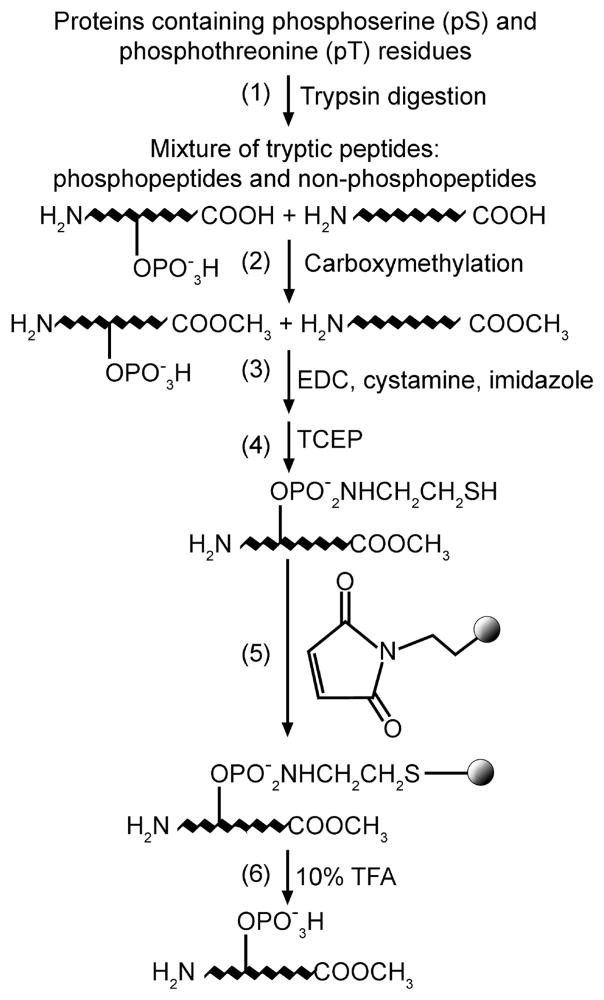
Phosphopeptides were isolated from A909, LR114, LR119 and LR120 in parallel as described in the Experimental Procedures (also see Figure 2). Briefly, equal amounts of tryptic peptides from each GBS strain (containing both phosphopeptides and nonphosphopeptides) were methylated to protect the caboxylate groups as described.42,47 Phosphoramidate bonds were then generated between the O-phosphoesters and cystamine in a one-pot reaction containing carbodiimide (EDC), imidazole and cystamine. A reducing agent such as tris(2-carboxyethyl) phosphine (TCEP) was employed to generate free thiol groups on the modified phosphopeptides. The derivatized phosphopeptides were captured on a solid phase through the reaction between thiol groups and maleimide immobilized on glass beads. After stringent washes to remove nonspecifically bound peptides, phosphopeptides were recovered by cleavage of the phosphoramidate bonds under mild acidic conditions. To increase the confidence of the generated data, the experiment was repeated three times for each GBS strain using independent biological samples. In addition, prior to each experiment, the phosphopeptide enrichment procedure and MSn analysis were validated using the standard phosphopeptide Angiotensin II as described previously.42
Figure 2.
Flowchart represents the phosphopeptide enrichment method. Total proteins were isolated from GBS strains that express the serine/threonine kinase (Stk1). Controls included strains that did not express Stk1. Tryptic peptides from each GBS strain were selectively enriched for phosphopeptides using a combination of phosphoramidate chemistry and solid phase capture. Phosphopeptides were identified using mass spectrometry (see Experimental Procedures for details).
Mass Spectrometric Analysis and Identification of GBS Phosphopeptides
Phosphopeptides isolated from the GBS strains were analyzed using multistage mass spectrometry (MSn) as described previously.42 Briefly, the top five most intense ions were selected from the MS mode for product ion spectrum and were validated for the neutral loss of phosphoric acid (see Experimental Procedures). Neutral loss of phosphoric acid (H3PO4) was measured as the loss of 98 Da (−98 Da) for singly charged ions, − 49 Da for doubly charged ions, and −33 Da for triply charged ions. The occurrence of a neutral loss of phosphoric acid is a strong indication that the peptide is a serine/threonine containing phosphopeptide.42 Subsequently, a third scan event was automatically performed to generate an MS3 spectrum of the MS2 ions showing the neutral loss (see Figures 3 and 4 for the MS2 and MS3 spectra of two threonine phosphopeptides). To determine that a phosphopeptide may be specific to the kinase activity of Stk1, the following criteria were used: (i) all peptides should have a Peptide Prophet probability of at least 0.6, cross correlation score (Xcorr) of at least 3.0 and top Sp Rank; (ii) the MS2 ion spectrum of the phosphopeptide should reveal neutral loss of phosphoric acid; (iii) MS2 and its corresponding MS3 spectra should reveal the same phosphopeptide sequence; and (iv) both the MS2 and MS3 ion spectra for the phosphopeptide should not be present in the Stk1 deficient strain LR114. We considered phosphopeptides that cannot be identified by MS2 or MS3 in the Stk1 deficient strain LR114 as absent. These criteria were established because the mass accuracy of the LTQ instrument (m/z in the MS1 measurements) can be misleading to confirm the absence of the respective phosphopeptide ion.
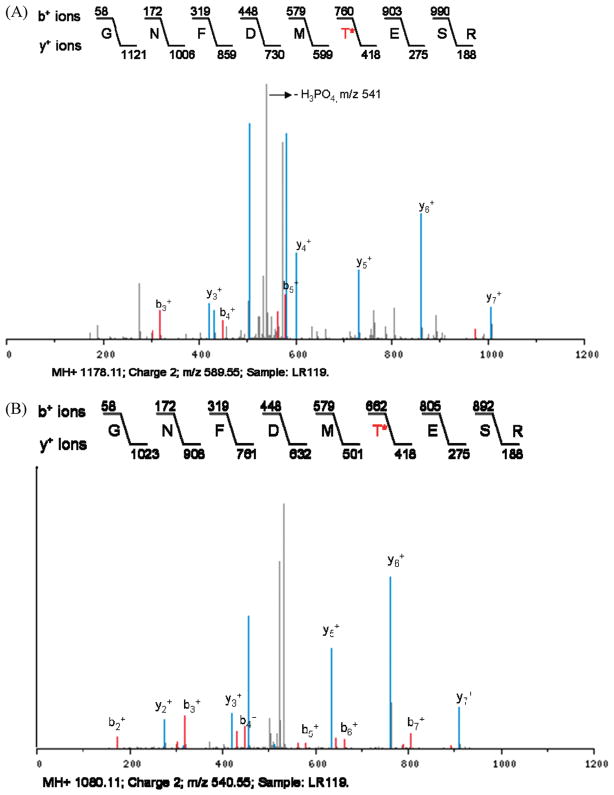
Figure 3.
MS2 and MS3 spectra of a threonine phosphorylated peptide corresponding to SAK_0581, the cell division protein FtsZ. (A) In the MS2 spectrum, the neutral loss of phosphoric acid from a threonine phosphopeptide is observed. The m/z of the doubly charged precursor ion is 589.55. Note that the peak depicting neutral loss of phosphoric acid (H3PO4) has an m/z of 541 due to the loss of 49 Da from the double charged peptide. LR119 corresponds to the strain or sample from which the peptide was identified. Asterisk (*) indicates the phosphorylated threonine in the peptide. (B) The MS3 spectrum of the corresponding MS2 ion from Figure panel A is shown. Note that the m/z of the same peptide is 540.55.
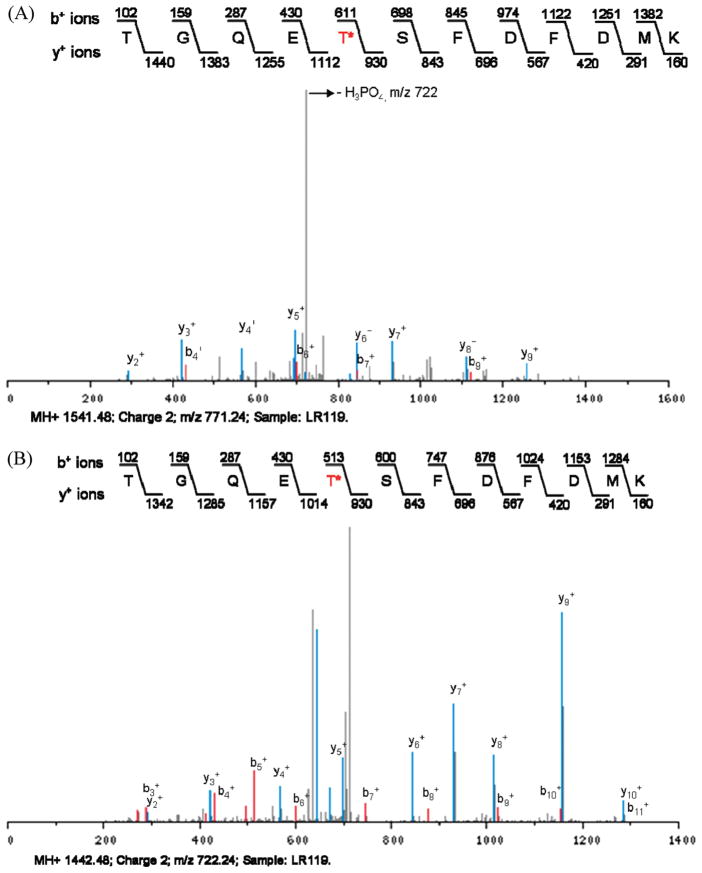
Figure 4.
MS2 and MS3 spectra of a threonine phosphorylated peptide corresponding to SAK_0583, a protein of unknown function in GBS. (A) In the MS2 spectrum, the neutral loss of phosphoric acid from a threonine phosphopeptide is observed. The m/z of the doubly charged precursor ion is 771.2. Note that the peak depicting neutral loss of phosphoric acid (H3PO4) has an m/z of 722 due to the loss of 49 Da from the double charged peptide. LR119 corresponds to the strain or sample from which the peptide was identified. Asterisk (*) indicates the phosphorylated threonine in the peptide. (B) The MS3 spectrum of the corresponding MS2 ion from Figure panel A is shown. Note that the m/z of the same peptide is 722.2.
Phosphopeptides selected using the above criteria were considered specifically phosphorylated by Stk1 and are listed in Table 2A. These phosphopeptides were identified in at least two of the three independent experiments from any one of the Stk1 expressing GBS strains (A909, LR119, and LR120, Table 2A). Since these peptides were not identified as phosphopeptides in the stk1 mutant LR114, the likelihood that their phosphorylation is Stk1-dependent is compelling. Consistent with its ability to autophosphorylate in vitro,17,36,37 we identified phosphopeptides corresponding to the Stk1 protein (Table 2A). These data confirm that Stk1 autophosphorylates in GBS in vivo and also validates our analyses. Three out of the 10 phosphopeptides unique to the Stk1 expressing strains were FtsZ, DivIVA and a protein with the conserved DivIVA domain (Table 2A). Orthologs of DivIVA and FtsZ regulate cell division and cell separation in a number of bacteria such as B. subtilis, Enterococcus faecalis, Streptococus pneumoniae and Mycobacterium tuberculosis.48–54 The identification of phosphopeptides corresponding to these proteins suggests that phosphorylation of these substrates affects cell division/cell segregation of GBS17 similar to their role in M. tuberculosis.48,49 In particular, we noted that threonine phosphorylation of DivIVA is observed in the WT, absent in the Stk1 deficient strain LR114 and is restored in the complemented strain LR120 (see Table 2A). As abnormal chaining was observed in LR114 and not in the WT or LR120,17 together, these data suggest that reversible phosphorylation of DivIVA is important for normal cell segregation/separation of GBS. Likewise, phosphorylation of two proteins of unknown function, namely, SAK_2030 and SAK_1559, is observed in the WT, absent in the Stk1 deficient strain LR114 and is restored in the complemented strain LR120 (see Table 2A). These data provide the foundation for future studies on the role of their phosphorylation to GBS growth, cell division and virulence.
As phosphorylation of the DivIVA domain protein is only seen in the LR119 strain (that is deficient for Stp1, Table 2A), we speculate that the smaller size and altered colony morphology of this strain (see ref 17) may in part be due to increased phosphorylation of the DivIVA domain protein. Other phosphopeptides that were unique to the Stk1 expressing strain/s correspond to 4 proteins whose functions in GBS are as yet unknown (see LR119 and LR120 in Table 2A). As expected, phosphopeptides unique to Stk1 were identified in GBS strains that constitutively expressed Stk1 (LR119) or both Stk1 and Stp1 (LR120, see Table 2A). We employed the use of the complemented strains that constitutively express Stk1 because the external signals for Stk1 activation in vivo are not known and construction of a GBS strain lacking only Stp1 was unsuccessful.17 One possibility for the increased identification of phosphopeptides from the complemented strains is that constitutive expression of Stk1 from the plasmid17 increased phosphorylation of the substrates and enabled their identification. It is also likely that the nonspecific phosphatase inhibitors used in this study may not effectively inhibit all phosphatase activity in GBS, particularly in the WT strain. Support for this hypothesis is provided by observations that the lack of specific inhibitors to the PP2C family of serine/threonine phosphatases has limited the identification of their in vivo substrates.55,56 Only recently, a few specific inhibitors were identified to a eukaryotic PP2C enzyme using virtual screening.55 Furthermore, general inhibitors such as NaF and sodium pyrophosphate only partially inhibit the function of bacterial PP2C serine/threonine phosphatases.46 Therefore, it is likely that the number of phosphopeptides identified in the wild-type GBS strain is fewer due to incomplete inhibition of Stp1. These data also suggest that phosphorylation of kinase substrates during growth of wild-type GBS is dynamically reversible. Importantly, as the strain LR120 is similar to the wild-type for growth, cell division and toxin phenotypes,17,37 we predict that the phosphopeptides identified in this strain represent in vivo substrates of Stk1 in GBS.
Listed in Table 2B are phosphopeptides that were identified in both the Stk1 expressing strains (A909, LR119, and LR120) as well as in the Stk1-deficient mutant (LR114). The identification of these phosphopeptides in strains that are proficient and deficient in Stk1 expression suggests their phosphorylation is independent of Stk1. It is likely that these proteins either autophosphorylate at serine or threonine or that their phosphorylation is mediated by other kinases not yet characterized in GBS.
Although eukaryotic serine/threonine kinases most often recognize a short conserved motif sequence in their target substrates,57,58 we were unable to identify a consensus recognition motif as the total number of phosphopeptides identified in this study is below the limit of detection for accurate motif prediction.
Validation of the Potential Stk1 Targets Using in Vitro Phosphorylation
In vitro phosphorylation assays were performed on selected Stk1 substrates to validate the MS data. Selection for substrates was based on the criteria that the phosphopeptides were present in a Stk1-expressing strain (A909, LR119, LR120) and demonstrated neutral loss of phosphate in MSn analyses. We derived C-terminal, His-tag fusions for these peptides, and recombinant proteins were purified as described in the Experimental Procedures. Subsequently, in vitro phosphorylation assays were performed using kinase buffer containing [γ-32P]-ATP in the presence and absence of a recombinant Stk1 fusion protein as described previously.17,37 We have previously shown that the Stk1 fusion protein autophosphorylates in vitro in the presence of [γ-32P]-ATP.17
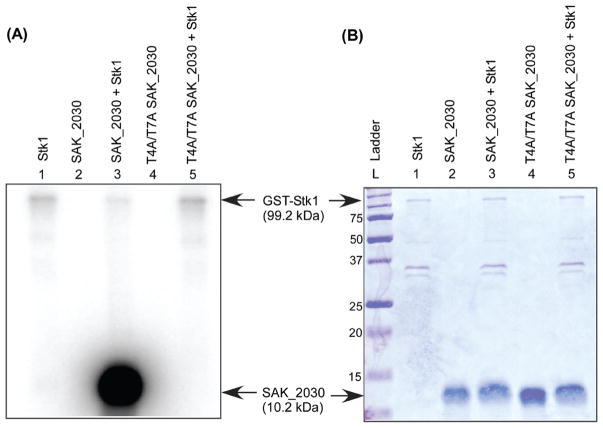
The results shown in Figure 5A indicate that Stk1 autophosphorylates in vitro (see lanes 1, 3, and 5) and can phosphorylate the hypothetical protein SAK_2030 (Figure 5A, lane 3). Figure 5B shows the corresponding Coomassie stained gel. Autophosphorylation was not observed with SAK_2030 alone (see Figure 5A, lane 2), indicating that phosphorylation of this protein requires Stk1. Furthermore, site-directed substitutions of the phosphorylated threonine at positions 4 and 7 to alanine (T4A/T7A) in SAK_2030 abolished phosphorylation by Stk1 (Figure 5A, lane 5). These data confirm that the threonine residues in positions 4 and 7 are critical for Stk1 phosphorylation of SAK_2030 and, thus, validate the phosphopeptide enrichment and MSn analysis.
Figure 5.
In vitro phosphorylation of SAK_2030 by Stk1. In vitro phosphorylation assays were performed as described in the Experimental Procedures. Panel A shows the phosphoimage and panel B shows the corresponding Coomassie stained gel. Autophosphorylation of Stk1 is evident in all lanes to which it was added (lanes 1, 3 and 5). The protein ladder “L” is labeled with the kDa sizes. Note that Stk1 phosphorylates the wild-type SAK_2030 (“2030”) protein but not the site-directed T4A/T7A SAK_2030.
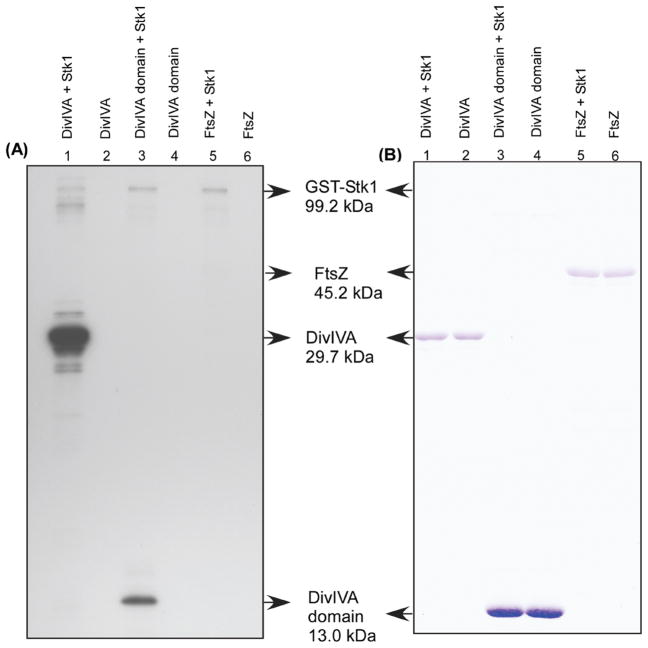
We also show that the putative Stk1 substrates SAK_0586 (DivIVA586), SAK_0373 (DivIVA373) and FtsZ do not autophosphorylate in vitro (see lanes 2, 4, and 6 in Figure 6A), indicating that their phosphorylation is dependent on protein kinase/s. Consistent with this hypothesis, phosphorylation of DivIVA domain protein 373 and the DivIVA586 is observed in the presence of Stk1 (see lanes 1 and 3 in Figure 6A). Notably, phosphorylation of FtsZ was not observed even in the presence of Stk1 (Figure 6A, lane 5). Phosphorylation of FtsZ was also not observed with increasing concentrations of the Stk1 protein (data not shown). This suggests that Stk1-mediated phosphorylation of FtsZ may require the presence of other factors not present in the in vitro phosphorylation assays. Alternatively, it is possible that phosphorylation of FtsZ does not require Stk1 but that stability of phosphorylated FtsZ (FtsZ~P) is enhanced in the presence of Stk1 in GBS. Collectively, this study demonstrates that the phosphopeptide enrichment-mass spectrometric approach is successful for the identification of phosphopeptides and serine/threonine kinase substrates in human bacterial pathogens such as GBS. Consequently, these studies provide a useful tool for further studies and insight into serine/threonine kinase signaling in prokaryotes.
Figure 6.
In vitro phosphorylation of Stk1 substrates. Phosphorylation assays were performed as described in the Experimental Procedures. Panel A shows the phosphoimage and panel B shows the corresponding Coomassie stained gel. Although autophosphorylation of Stk1 is evident in all lanes to which it was added, a Coomassie band corresponding to Stk1 is not seen due to its lower concentration. Note that Stk1 phosphorylated DivIVA 586 and DivIVA373 and did not phosphorylate FtsZ. Phosphorylation of FtsZ was not observed on prolonged exposure or if more Stk1 was added (data not shown).
Discussion
In this work, we used an in vivo proteomic-mass spectrometry approach, including a critical phosphopeptide-enrichment step, to identify targets of the unique serine/threonine protein kinase in GBS. These in vivo data are supported by in vitro assays, raising the level of confidence in identifying kinase substrates. In this phosphoproteome screening, we identified 21 phosphopeptides, belonging to 13 unique proteins that were phosphorylated at serine or threonine residues.
The number of phosphopeptides identified in GBS is few compared to the serine/threonine and tyrosine phosphoproteins of B. subtilis, E. coli and recently in Lactococcus lactis.33,34,59 However, a large number of the phosphoproteins identified in B. subtilis, E. coli and L. lactis are involved in carbohydrate metabolism.59 We speculate that differences in growth media may contribute to the discrepancy in the total number of phosphoproteins identified in GBS to these other bacteria. It is also noteworthy that the volume of bacterial cell culture used for phosphopeptide enrichment in B. subtilis, E. coli and L. lactis ranged from 4 to 12 L;33,34,59 these represent approximately 40- to 120-fold greater bacterial colony forming units (cfu) compared to our studies in GBS. Because of the potential of GBS as both a neonatal and adult human pathogen and constraints in its cell lysis, we have refrained from using such large cultures of GBS in our study. Furthermore, differences in phosphopeptide enrichment techniques such as IMAC (TiO233,34,59) versus modification of the phosphate using phosphoramidate chemistry (this study) may also contribute to differences in discerning bacterial phosphoproteomes. However, compared to the previous studies on phosphoproteomes of B. subtilis, E. coli and L. lactis that have only used the wild-type strain,33,34,59 we have examined S/T phosphorylation in wild-type and its isogenic kinase and phosphatase mutant and have also confirmed phosphorylation in two different complemented strains.
A comparison of S/T phosphopeptides between the GBS strains led us to identify 10 proteins from three biologically independent phosphopeptide enrichments that were unique to the Stk1 expressing strains (see Table 2A) and, hence, are suggested as targets of Stk1 phosphorylation. The functional descriptions of these proteins are indicated and include the serine/threonine kinase Stk1, three proteins that regulate bacterial cell division (such as DivIVA and FtsZ), and six proteins whose functions are as yet unknown. The identification of proteins predicted to control cell division/separation as targets of Stk1 are consistent with the altered cell segregation phenotypes of the stk1 mutants described previously.17 Threonine phosphorylation of DivIVA was seen in the WT and complemented LR120 strain and not in the Stk1 deficient strain LR114. Therefore, we predict that reversible phosphorylation of this protein is required for normal cell separation of GBS. It is noteworthy that orthologs of DivIVA (Wag31) and FtsZ were previously identified as substrates of the serine/threonine kinases PknA and PknB from M. tuberculosis.48,49 Further, Kang et al. demonstrated that both overexpression and depletion of Wag31 can alter cell shape in M. tuberculosis.49 Previous studies have also established that the GTPase activity of FtsZ is impaired when FtsZ is phosphorylated by the Mycobacterial STK PknA.48 As LR120 resembles the wild-type strain for growth, cell division and toxin expression phenotypes,17,37 we predict that phosphopeptides identified in this strain represent in vivo Stk1 substrates of GBS. Although Stk1 did not directly phosphorylate FtsZ in vitro, it is likely that the in vitro phosphorylation assays are not ideal for FtsZ phosphorylation and may require the presence of other factors. Alternatively, it is possible that the stability of phospho-FtsZ (FtsZ~P) is enhanced by the presence of Stk1 in vivo. Because the Stk1 expressing strain LR119 lacking Stp1 demonstrated smaller cell size and chaining,17 and phosphopeptides corresponding to DivIVA domain protein were observed only in LR119, we speculate that this phenotype may in part be linked to altered activity of the DivIVA domain protein. Further studies are necessary to establish the relevance of phosphorylation of proteins identified only in LR119 to GBS growth and pathogenesis.
We observed that GBS mutants deficient for SAK_2030 expression did not demonstrate changes in either toxin expression or altered cell segregation (data not shown). Therefore, the contribution of SAK_2030 to growth and virulence of GBS is not yet known and is under investigation in our laboratory. Importantly, the role of phosphorylation of DivIVA, SAK_1559 and other substrates identified in the study from WT and the LR120 strain remains to be elucidated and is essential for a deeper understanding of serine/threonine kinase signaling in GBS.
In this study, we also identified three phosphopeptides corresponding to three unique proteins (PpdK, GlmM and SAK_1155) in GBS strains that were both proficient and deficient in Stk1 expression (see Table 2B). Therefore, we predict that phosphorylation of these proteins does not require Stk1 in GBS. Two of these proteins are phosphorylated at serine and one at threonine. To our knowledge, Stk1 is the only eukaryotic-type serine/threonine kinase in GBS; this conclusion is based on the genome sequence of GBS60–62 and our studies on Stk1.17,36,37 We speculate that phosphorylation of these proteins may be mediated by other kinases such as SAK_1204 (homoserine kinase, ThrB). It is interesting that homologues of GlmM and SAK_1155 were previously reported to be substrates of the serine/threonine kinase StkP in S. pneumoniae.63 One possibility for these differing conclusions might be that the serine/threonine kinase may not be essential but can enhance phosphorylation of these targets. Alternatively, it is likely that substrates for this family of kinases are specific to the host organism. Support for this hypothesis stems from the differing phenotypes of serine/threonine kinase mutants in various bacterial pathogens. For example, serine/threonine kinase homologues regulate type VI secretion in Pseudomonas aeruginosa,64 antimicrobial peptide resistance of E. faecalis,27 competence of S. penumoniae and cell segregation and toxin expression in GBS.37
Surprisingly, the mass spectrometric analyses failed to identify a phosphopeptide corresponding to the previously reported target of Stk1 known as CovR, which regulates toxin expression in GBS.37 In recent studies, we showed that phosphorylation of CovR by Stk1 alters toxin expression during GBS growth in rich media.37 On the basis of our recent observations,38 we expected that a CovR phosphopeptide with phosphothreonine at position 65 could be identified in the Stk1-expressing strains. One possibility for the absence of this peptide in the mass spectrometric analyses is our use of trypsin for cleavage of proteins to peptides. Trypsin cleavage of CovR results in a relatively large peptide (27 amino acid residues) that may be difficult to detect in the mass spectrometric analyses. Alternatively, because CovR is also phosphorylated by its cognate histidine kinase CovS at the conserved aspartate (D53), and Stk1 and CovS phosphorylation of CovR are mutually exclusive (for details, see ref 38), the number of threonine phosphorylated CovR peptides may be lower due to competition for aspartate phosphorylation by CovS and low molecular weight phosphodonors like acetyl phosphate.38,65,66 Construction of GBS strains lacking CovS is currently in progress in our laboratory, and phosphopeptide enrichment analysis in this strain will enable us to circumvent CovR phosphorylation by CovS.
Although we had previously described that PurA encoding an adenylosuccinate synthetase and PpaC encoding an inorganic pyrophosphatase are phosphorylated and regulated by Stk1, these conclusions were in part based on growth of GBS in nutrient deficient conditions where the bacteria are forced to utilize their de novo biosynthetic pathways.17,36 Because the current studies were performed with GBS grown in rich media where altered activities of PurA and PpaC were not observed,37 phosphorylation of PurA and PpaC is less likely to occur, and hence, phosphopeptides corresponding to these proteins were not identified in our analyses. Taken together, these data suggest that serine/threonine kinases may preferentially alter substrate specificity depending on their external environment, as can be expected for sensor kinases that regulate adaptive responses of living organisms. Importantly, these studies also show that STK substrates can be identified using phosphopeptide enrichment approaches. A deeper understanding of the role of cognate regulators of the kinases and their targets, identification of the activating signals and the role of reversible phosphorylation on substrate activity is also essential for a complete picture of STK signaling in prokaryotic organisms.
Acknowledgments
This work was supported by funding from the National Institutes of Health, Grant No. RO1 AI070749 to L.R. and National Science Foundation (NSF) CAREER award to W.A.T. We thank Louis Paolella and Thao Tran for technical assistance. We are grateful to Dr. Kellie Burnside for critical reading of the manuscript.
Abbreviations
- STK
serine/threonine kinase
- WT
wild-type
- TCS
two-component system
References
- 1.Raggiaschi R, Gotta S, Terstappen GC. Phosphoproteome analysis. Biosci Rep. 2005;25(1–2):33–44. doi: 10.1007/s10540-005-2846-0. [DOI] [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. Protein kinases--the major drug targets of the twenty-first century. Nat Rev Drug Discovery. 2002;1(4):309–15. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 4.Hoch J, Silhavy T. Two-Component Signal Transduction. American Society for Microbiology; Washington, DC: 1995. [Google Scholar]
- 5.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Foussard M, Cabantous S, Pedelacq J, Guillet V, Tranier S, Mourey L, Birck C, Samama J. The molecular puzzle of two-component signaling cascades. Microbes Infect. 2001;3(5):417–24. doi: 10.1016/s1286-4579(01)01390-9. [DOI] [PubMed] [Google Scholar]
- 7.Calva E, Oropeza R. Two-component signal transduction systems, environmental signals, and virulence. Microb Ecol. 2006;51(2):166–76. doi: 10.1007/s00248-005-0087-1. [DOI] [PubMed] [Google Scholar]
- 8.Deutscher J, Saier MH., Jr Ser/Thr/Tyr protein phosphorylation in bacteria - for long time neglected, now well established. J Mol Microbiol Biotechnol. 2005;9(3–4):125–31. doi: 10.1159/000089641. [DOI] [PubMed] [Google Scholar]
- 9.Kennelly PJ. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol Lett. 2002;206(1):1–8. doi: 10.1111/j.1574-6968.2002.tb10978.x. [DOI] [PubMed] [Google Scholar]
- 10.Inouye S, Jain R, Ueki T, Nariya H, Xu CY, Hsu MY, Fernandez-Luque BA, Munoz-Dorado J, Farez-Vidal E, Inouye M. A large family of eukaryotic-like protein Ser/Thr kinases of Myxococcus xanthus, a developmental bacterium. Microb Comp Genomics. 2000;5(2):103–20. doi: 10.1089/10906590050179783. [DOI] [PubMed] [Google Scholar]
- 11.Av-Gay Y, Everett M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 2000;8(5):238–44. doi: 10.1016/s0966-842x(00)01734-0. [DOI] [PubMed] [Google Scholar]
- 12.Han G, Zhang CC. On the origin of Ser/Thr kinases in a prokaryote. FEMS Microbiol Lett. 2001;200(1):79–84. doi: 10.1111/j.1574-6968.2001.tb10696.x. [DOI] [PubMed] [Google Scholar]
- 13.Hakansson S, Galyov EE, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20(3):593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 14.Galyov EE, Hakansson S, Forsberg A, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature (London) 1993;361(6414):730–2. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- 15.Fiuza M, Canova MJ, Zanella-Cleon I, Becchi M, Cozzone AJ, Mateos LM, Kremer L, Gil JA, Molle V. From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J Biol Chem. 2008;283(26):18099–112. doi: 10.1074/jbc.M802615200. [DOI] [PubMed] [Google Scholar]
- 16.Kennelly PJ, Potts M. Fancy meeting you here! A fresh look at “prokaryotic” protein phosphorylation. J Bacteriol. 1996;178(16):4759–64. doi: 10.1128/jb.178.16.4759-4764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajagopal L, Clancy A, Rubens CE. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J Biol Chem. 2003;278(16):14429–41. doi: 10.1074/jbc.M212747200. [DOI] [PubMed] [Google Scholar]
- 18.Jin H, Pancholi V. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J Mol Biol. 2006;357(5):1351–72. doi: 10.1016/j.jmb.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, Betts JC, Mizrahi V, Smith DA, Stokes RW, Av-Gay Y. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol. 2004;52(6):1691–702. doi: 10.1111/j.1365-2958.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- 20.Koul A, Choidas A, Tyagi AK, Drlica K, Singh Y, Ullrich A. Serine/threonine protein kinases PknF and PknG of Mycobacterium tuberculosis: characterization and localization. Microbiology. 2001;147(Pt. 8):2307–14. doi: 10.1099/00221287-147-8-2307. [DOI] [PubMed] [Google Scholar]
- 21.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004;304(5678):1800–4. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 22.Barz C, Abahji TN, Trulzsch K, Heesemann J. The Yersinia Ser/Thr protein kinase YpkA/YopO directly interacts with the small GTPases RhoA and Rac-1. FEBS Lett. 2000;482(1–2):139–43. doi: 10.1016/s0014-5793(00)02045-7. [DOI] [PubMed] [Google Scholar]
- 23.Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc Natl Acad Sci USA. 2000;97(17):9431–6. doi: 10.1073/pnas.170281997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madec E, Laszkiewicz A, Iwanicki A, Obuchowski M, Seror S. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol Microbiol. 2002;46(2):571–86. doi: 10.1046/j.1365-2958.2002.03178.x. [DOI] [PubMed] [Google Scholar]
- 25.Chaba R, Raje M, Chakraborti PK. Evidence that a eukaryotic-type serine/threonine protein kinase from Mycobacterium tuberculosis regulates morphological changes associated with cell division. Eur J Biochem. 2002;269(4):1078–85. doi: 10.1046/j.1432-1033.2002.02778.x. [DOI] [PubMed] [Google Scholar]
- 26.Echenique J, Kadioglu A, Romao S, Andrew PW, Trombe MC. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect Immun. 2004;72(4):2434–7. doi: 10.1128/IAI.72.4.2434-2437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kristich CJ, Wells CL, Dunny GM. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci USA. 2007;104(9):3508–13. doi: 10.1073/pnas.0608742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinders J, Sickmann A. State-of-the-art in phosphoproteomics. Proteomics. 2005;5(16):4052–61. doi: 10.1002/pmic.200401289. [DOI] [PubMed] [Google Scholar]
- 29.Morandell S, Stasyk T, Grosstessner-Hain K, Roitinger E, Mechtler K, Bonn GK, Huber LA. Phosphoproteomics strategies for the functional analysis of signal transduction. Proteomics. 2006;6(14):4047–56. doi: 10.1002/pmic.200600058. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H, Watts JD, Abersold R. A systemic approach to the analysis of protein phosphorylation. Nat Biotechnol. 2001;19:379–8. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 31.Delom F, Chevet E. Phosphoprotein analysis: from proteins to proteomes. Proteome Sci. 2006;4:15. doi: 10.1186/1477-5956-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goshe MB. Characterizing phosphoproteins and phosphoproteomes using mass spectrometry. Briefings Funct Genomics Proteomics. 2006;4(4):363–76. doi: 10.1093/bfgp/eli007. [DOI] [PubMed] [Google Scholar]
- 33.Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics. 2007;6(4):697–707. doi: 10.1074/mcp.M600464-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7(2):299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Baker CJ, Edwards MW. Group B Streptococcal Infections. In: Remington JS, Klein JO, editors. Infectious Diseases of the Fetus and Newborn Infant. W.B. Saunders; Philadelphia, PA: 1995. pp. 980–1054. [Google Scholar]
- 36.Rajagopal L, Vo A, Silvestroni A, Rubens CE. Regulation of purine biosynthesis by a eukaryotic-type kinase in Streptococcus agalactiae. Mol Microbiol. 2005;56(5):1329–46. doi: 10.1111/j.1365-2958.2005.04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajagopal L, Vo A, Silvestroni A, Rubens CE. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol Microbiol. 2006;62(4):941–57. doi: 10.1111/j.1365-2958.2006.05431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin WJ, Walthers D, Connelly JE, Burnside K, Jewell KA, Kenney LJ, Rajagopal L. Threonine phosphorylation prevents promoter DNA binding of the Group B Streptococcus response regulator CovR. Mol Microbiol. 2009;71(6):1477–95. doi: 10.1111/j.1365-2958.2009.06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madoff LC, Michel JL, Kasper DL. A monoclonal antibody identifies a protective C-protein alpha-antigen epitope in group B streptococci. Infect Immun. 1991;59(1):204–10. doi: 10.1128/iai.59.1.204-210.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner V, Gessner G, Heiland I, Kaminski M, Hawat S, Scheffler K, Mittag M. Analysis of the phosphoproteome of Chlamydomonas reinhardtii provides new insights into various cellular pathways. Eukaryotic Cell. 2006;5(3):457–68. doi: 10.1128/EC.5.3.457-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shu H, Chen S, Bi Q, Mumby M, Brekken DL. Identification of phosphoproteins and their phosphorylation sites in the WEHI-231 B lymphoma cell line. Mol Cell Proteomics. 2004;3(3):279–86. doi: 10.1074/mcp.D300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Bodenmiller B, Mueller LN, Pedrioli PG, Pflieger D, Junger MA, Eng JK, Aebersold R, Tao WA. An integrated chemical, mass spectrometric and computational strategy for (quantitative) phosphoproteomics: application to Drosophila melanogaster Kc167 cells. Mol BioSyst. 2007;3(4):275–86. doi: 10.1039/b617545g. [DOI] [PubMed] [Google Scholar]
- 43.Seepersaud R, Needham RH, Kim CS, Jones AL. Abundance of the {delta} Subunit of RNA Polymerase Is Linked to the Virulence of Streptococcus agalactiae. J Bacteriol. 2006;188(6):2096–105. doi: 10.1128/JB.188.6.2096-2105.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4(3):231–7. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 45.Madoff LC, Hori S, Michel JL, Baker CJ, Kasper DL. Phenotypic diversity in the alpha C protein of group B streptococci. Infect Immun. 1991;59(8):2638–44. doi: 10.1128/iai.59.8.2638-2644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai SM, Le Moual H. PrpZ, a Salmonella enterica serovar Typhi serine/threonine protein phosphatase 2C with dual substrate specificity. Microbiology. 2005;151(Pt. 4):1159–67. doi: 10.1099/mic.0.27585-0. [DOI] [PubMed] [Google Scholar]
- 47.Tao WA, Wollscheid B, O’Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods. 2005;2(8):591–8. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 48.Thakur M, Chakraborti PK. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J Biol Chem. 2006;281(52):40107–13. doi: 10.1074/jbc.M607216200. [DOI] [PubMed] [Google Scholar]
- 49.Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19(14):1692–704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bramkamp M, Emmins R, Weston L, Donovan C, Daniel RA, Errington J. A novel component of the division site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol Microbiol. 2008;70(6):1556–69. doi: 10.1111/j.1365-2958.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- 51.Howard M. A mechanism for polar protein localization in bacteria. J Mol Biol. 2004;335(2):655–63. doi: 10.1016/j.jmb.2003.10.058. [DOI] [PubMed] [Google Scholar]
- 52.Ramirez-Arcos S, Liao M, Marthaler S, Rigden M, Dillon JA. Enterococcus faecalis divIVA: an essential gene involved in cell division, cell growth and chromosome segregation. Microbiology. 2005;151(Pt. 5):1381–93. doi: 10.1099/mic.0.27718-0. [DOI] [PubMed] [Google Scholar]
- 53.Fadda D, Santona A, D’Ulisse V, Ghelardini P, Ennas MG, Whalen MB, Massidda O. Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD-free context. J Bacteriol. 2007;189(4):1288–98. doi: 10.1128/JB.01168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6(11):862–71. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogers JP, Beuscher AEt, Flajolet M, McAvoy T, Nairn AC, Olson AJ, Greengard P. Discovery of protein phosphatase 2C inhibitors by virtual screening. J Med Chem. 2006;49(5):1658–67. doi: 10.1021/jm051033y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCluskey A, Sakoff JA. Small molecule inhibitors of serine/threonine protein phosphatases. Mini Rev Med Chem. 2001;1(1):43–55. doi: 10.2174/1389557013407205. [DOI] [PubMed] [Google Scholar]
- 57.Yang SD, Huang TJ. Identification of -R-X-(X)-S/T-X3-S/T- as consensus sequence motif for autophosphorylation-dependent protein kinase. J Biol Chem. 1994;269(47):29855–9. [PubMed] [Google Scholar]
- 58.Wang Y, Klemke RL. PhosphoBlast, a computational tool for comparing phosphoprotein signatures among large datasets. Mol Cell Proteomics. 2008;7(1):145–62. doi: 10.1074/mcp.M700207-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Soufi B, Gnad F, Jensen PR, Petranovic D, Mann M, Mijakovic I, Macek B. The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics. 2008;8(17):3486–93. doi: 10.1002/pmic.200800069. [DOI] [PubMed] [Google Scholar]
- 60.Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, Paulsen IT, Nelson KE, Margarit I, Read TD, Madoff LC, Wolf AM, Beanan MJ, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Kolonay JF, Madupu R, Lewis MR, Radune D, Fedorova NB, Scanlan D, Khouri H, Mulligan S, Carty HA, Cline RT, Van Aken SE, Gill J, Scarselli M, Mora M, Iacobini ET, Brettoni C, Galli G, Mariani M, Vegni F, Maione D, Rinaudo D, Rappuoli R, Telford JL, Kasper DL, Grandi G, Fraser CM. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci USA. 2002;99(19):12391–6. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, Zouine M, Couve E, Lalioui L, Poyart C, Trieu-Cuot P, Kunst F. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol. 2002;45(6):1499–513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 62.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit Y, Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ”pan-genome. Proc Natl Acad Sci USA. 2005;102(39):13950–5. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novakova L, Saskova L, Pallova P, Janecek J, Novotna J, Ulrych A, Echenique J, Trombe MC, Branny P. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 2005;272(5):1243–54. doi: 10.1111/j.1742-4658.2005.04560.x. [DOI] [PubMed] [Google Scholar]
- 64.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9(7):797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 65.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89(2):718–22. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol. 2007;189(15):5574–81. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]