Abstract
HIV-1 drug resistance mutations have been detected at low frequencies after single-dose nevirapine (sdNVP) for prevention of mother-to-child transmission (PMTCT). We investigated the relationship between these “minor variant” NVP-resistant viruses and clinical outcome with NVP-containing antiretroviral therapy (ART). An allele-specific quantitative PCR (ASPCR) assay was used to quantify the pre-ART frequency of K103N and Y181C in 26 women who had received sdNVP. The cohort was composed of 7 patients who experienced virologic failure and 19 control patients who maintained virologic suppression on NVP-containing ART; all were negative for resistance by standard genotyping. NVP resistance mutations were found in 17 of 26 (65%) patients using ASPCR. The frequency of NVP-resistant viruses ranged from 0.1% to 4.11%. Receiver operating characteristics (ROC) analysis identified a clinical threshold frequency of 0.19% for the ASPCR assay. Application of this threshold demonstrated minor variant resistance in 6 of 7 patients (86%) who failed treatment compared to 6 of 19 patients (32%) who were successful (OR = 13; 95% CI 1.27–133). ASPCR provides a means of detecting minor variant drug-resistant viruses that may impact subsequent treatment response. These data suggest a clinical role for highly sensitive assays to detect and quantify resistant viruses at low frequencies.
Introduction
The prevention of mother-to-child transmission (PMTCT) of HIV remains a critical issue in resource-limited settings as they strive to prevent a new generation of infants born with HIV; approximately 420,000 children were infected with HIV in 2007 alone.1 The administration of a single dose of nevirapine (sdNVP), a nonnucleoside reverse transcriptase inhibitor (NNRTI), to the mother during labor has been demonstrated to be a safe and effective means of reducing vertical transmission of HIV.2–6 This approach is an integral component of PMTCT in many developing nations and is particularly valuable when HIV-positive mothers are not identified early enough to receive more effective chemoprophylaxis regimens for PMTCT.
In this setting, the emergence of NVP-resistant viruses in both mothers and infants who received sdNVP administration is now well documented,7–10 and there is evidence that subsequent antiretroviral treatment failure may be related to drug resistance resulting from this prior nevirapine exposure. For example, the PHPT-2 study in Thailand showed that patients who received sdNVP experienced greater virologic failure in the course of postpartum antiretroviral therapy than those who had been in the arm receiving placebo.11 In addition, a study from Botswana showed that women who received sdNVP and delayed initiation of NVP-containing ART until 6 months postpartum had improved clinical outcome compared to those who started therapy earlier.12 This was thought to be related to the outgrowth over time of wild-type virus in the absence of NVP at the expense of drug-resistant virus as has been described for other HIV drug resistance mutations.13
It is commonly believed that the standard commercial drug resistance genotyping platforms have a limit of detection of approximately 20%, i.e., drug-resistant viruses must comprise at least 20% of the total viral population for the resistance mutation to be detected.14 Several studies using more sensitive techniques have demonstrated the presence of NVP resistance mutations at frequencies well below this threshold of detection in both plasma and peripheral blood cells (proviral DNA) following sdNVP exposure.15–19 The role of such “minor variants” in clinical outcome is evolving, as studies of drug-resistant minor variants in the treatment-naive population, both in chronically infected individuals and in those receiving PMTCT, suggest that they may in fact contribute to clinical failure.20–23 In this retrospective study, we sought to determine if the presence of drug-resistant mutations at low frequency in the plasma of mothers who received sdNVP for PMTCT in Botswana was predictive of the clinical outcome with subsequent treatment with NVP-containing antiretroviral treatment (ART).
Materials and Methods
Clinical samples
The Mashi PMTCT trial in Botswana involved pregnant women who received zidovudine beginning at 34 weeks of gestation or zidovudine (beginning at 34 weeks) plus randomization for sdNVP or placebo during labor. Women enrolled in the original Mashi study design and who subsequently had an AIDS-defining illness or a CD4 count below 200 cells/μl were started on NVP-containing ART per the national guidelines in Botswana. The investigators received samples from seven patients who received sdNVP for PMTCT, had no drug resistance mutations detected in their pre-ART plasma sample, and failed NVP-containing ART (virologic failure defined as detectable plasma HIV-1 viral load of >400 copies/ml after 6 months of treatment). Another 19 patients were identified from the same cohort. These women also received sdNVP for PMTCT but successfully maintained virologic control of HIV-1 when subsequently treated with NVP-containing ART. The study was approved by the Human Subjects Committee at the Harvard School of Public Health.
Measurement of plasma HIV-1 RNA and HIV-1 genotyping
The HIV-1 RNA viral load was quantified using the automated COBAS Amplicor/AmpliPrep HIV-1 Monitor Test V1.5 (Roche Diagnostics) as per the manufacturer's instructions (lower limit of detection 400 copies/ ml). All 26 of the pre-ART baseline samples included in this study were tested for drug resistance using the ViroSeq 2.0 HIV-1 Genotyping assay (Celera Diagnostics) and no drug resistance was found in any of the samples. The reverse transcriptase (RT) amino acid mutations defined as nevirapine resistance mutations per IAS-USA reference of HIV-1 drug resistance mutations are 100I, 103N, 106A/M, 108I, 181C/I, 188L/C/H, and 190A.24 ViroSeq 2.0 was also applied to all seven failure samples.
HIV-1C reverse transcriptase: isolation, PCR amplification, cloning, and sequencing
HIV RNA was isolated from 140 μl of plasma using QIAamp viral RNA Mini Kit (Qiagen) per the manufacturer's protocol. Viral cDNA was generated from 8 μl vRNA using Superscript III First Strand Synthesis for reverse transcriptase polymerase chain reaction (RT-PCR) (Invitrogen) per the manufacturer's protocol and then amplified in two independent nested PCR reactions. Three samples with limited first round amplification when viewed on gel electrophoresis were reisolated from 280 μl of plasma. Five microliters of cDNA was used in the first round PCR with the primers RT-18 (5′-GGA AAC CAA AAA TGA TAG GGG GAA TTG GAG G-3’) and NE1 (5′-CCT ACT AAC TTC TGT ATG TCA TTG ACA GTC CAG CT-3′) in the protocol described previously,25 generating a 957 base pair amplicon comprising HIV-1 nucleotides 2377–3334 (HXB2 numbering).
Then 2.0 μl of the first round PCR was subjected to a second PCR with the primers RTgen2F (5′-AGG TTA AAC AAT GGC CAT TGA CAG AAG-3′) and Zim27R2 (5′-GGA GTT CAT ACC CCA TCC AAA-3′). The 50 μl reaction contained 38.5 μl of water, 5 μl of 10× reaction buffer, 2 μl of dNTPs, 1 μl of each primer Zim27R2 and RTgen2F (10 μM concentration each), 0.5 μl of FastStart enzyme, in addition to the 2.0 μl of first round template. The cycling parameters for the PCR were 94°C for 5 min followed by 35 cycles of 94°C for 20 s, 59°C for 20 s, and 72°C for 1 min, with a final extension at 72°C for 7 min. This generated a 644 base pair amplicon comprising nucleotides 2608–3252 (HXB2 numbering).
For each patient sample, duplicate first and second round PCR amplifications were performed and the resultant amplicon containing HIV-1 pol was cloned by TOPO T/A cloning into the pCR2.1 plasmid vector (Invitrogen) per the manufacturer's protocol. For each sample, 24 individual transformed bacterial colonies were cultured and plasmid DNA was isolated by QIAprep Spin Miniprep kit (Qiagen). Plasmid DNA was submitted to a core facility for sequencing from the -20 M13 primer in pCR2.1. Sequences were manually edited and aligned by Clustal W in BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). They were screened for drug resistance mutations using the HIVdb Program from the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu).
Allele-specific PCR (ASPCR)
ASPCR is a nested PCR assay combining a standard first round PCR and a quantitative, second round PCR with allele-specific primers. The first round primers and reaction conditions were as described above. The first round PCR generated a 957-bp amplicon that was gel purified on 1% agarose followed by QiaQuick gel extraction per the manufacturer's protocol (Qiagen).
This first round PCR amplicon (107 copies as measured by spectrophotometry) served as the template for the second round, quantitative PCR step. The forward primer used in this quantitative step was constant for all reactions to detect resistance mutations at the 103 position (2090F-5′-AAG TGG AGA AAA TTA GTA GAT TTC AGG GA-3′) and for all reactions to detect resistance mutations at the 181 position (2298F-5′-CAC CAG GGA TTA GAT ATC AAT ATA ATG TG-3′). Different sets of primers were created to match nucleotide polymorphisms in the reverse primer binding sites for both the K103N and Y181C drug resistance mutations.
In addition to accounting for naturally occurring nucleotide polymorphisms, the assay was designed to detect and quantify the presence of each of the four possible nucleotides at the drug resistance determinant position thus allowing for the most accurate calculation of the relative frequencies of each mutation in each sample.25 These criteria required the use of a total of 68 different reverse primers: position 103 required 9 primer sets to account for binding site polymorphisms with each primer set including each of the four nucleotides at the 3′ terminus (n = 36) and position 181 required 8 primer sets to account for binding site polymorphisms with each primer set also including each of the four nucleotides at the 3′ terminus (n = 32). Each individual sample had two different sets of four primers used to calculate the allele percentage at both the 103 and 181 positions. The percentage of allele was simply defined as the quantity of the allele in the sample/sum of the four alleles (A,C, T, and G), multiplied by 100. The position 103 binding site polymorphism-specific reverse primers for group I are 5-CCC ACA TCT AGT ACT GTC ACT GAT TCA-3, 5-CCC ACA TCT AGT ACT GTC ACT GAT TCC-3, and 5-CCC ACA TCT AGT ACT GTC ACT GAT TCT-3, 5-CCC ACA TCT AGT ACT GTC ACT GAT TCG-3. The position 181 binding site polymorphism-specific reverse primers for group I are 5-CTA CAT ACA AGT CAT CCA TAT ATT GCA-3, 5-CTA CAT ACA AGT CAT CCA TAT ATT GC C-3, 5-CTA CAT ACA AGT CAT CCA TAT ATT GC T-3, and 5-CTA CAT ACA AGT CAT CCA TAT ATT GC G-3. The reverse primer binding site polymorphisms present in each patient, and thus the polymorphism-specific reverse primer set to be used for ASPCR, were determined by sequencing of cloned RT. Additionally, every reverse primer included an intentional mismatch at the penultimate (–2) position to enhance primer binding specificity.
Each sample had the second round reaction performed in triplicate on the Applied Biosystems Sequence Detector System 7500 in a 25 μl reaction with plasmid controls and standard curves generated for all 68 combinations of resistance mutations and reverse binding site polymorphisms using methods previously described.25 The sensitivity of the ASPCR assay for each resistance mutation in the context of each polymorphic reverse primer binding site was determined by analyzing mixed plasmid control samples prepared to contain from 100% mutation/0% wild type down to 0.01% mutation/99.9% wild type.
Statistics
Statistics were performed by using SigmaStat 3.11 and SigmaPlot 10 (Systat). Wilcoxon rank sum tests were performed to compare the distribution of the data in the failure and control groups with data summarized with medians, interquartile range (IQR), and overall range of data shown. A receiver operating characteristic (ROC) curve was used to assign a clinical cutoff for the ASPCR assay. This curve was calculated by SigmaStat and plots sensitivity (%) versus 100-specificity (100%) for various resistance thresholds. The cutoff determined by the technique detected a level of K103N and Y181C minor variants that provided the optimal sensitivity and specificity of the ASPCR assay for predicting virologic failure.
Results
The sensitivity of the ASPCR assay was determined by mixing different percentages of wild-type/mutant template ranging from 100% mutant template to 0.01%. The r-squared for these serial dilutions for the K103N mutation (ntAAC) was 0.997 and for the other K103N mutation (ntAAT) it was 0.999. Background readings of the assay when 100% wild-type template was entered into the reaction had a mean of 0.028% (SD 0.021) with K103N primers (ntAAC) and 0.010% (SD 0.015) with K103N primers (ntAAT) when tested against five different sets of plasmid PCR products. The Y181C mutation had an r-squared of 0.986 for the serial dilutions with a background false positive of 0.027%. The sensitivity of the assay for this study was conservatively set at 0.10%.
The characteristics of the cases and controls are listed in Table 1. There was a significant shift in the distribution of the baseline pretreatment CD4+ cell counts in the 6 patients with CD4+ data who failed therapy (51 cells/μl, IQR 46–84, range 22–151) than in the 17 patients with CD4+ data who did not experience virologic failure (179 cells/μl, IQR 156–231, range 50–294), p = 0.001. There were no statistically significant differences in the distribution of HIV-1 RNA level at the time of ART initiation with the cases having a median log10 5.31 copies/ml (IQR 4.91–5.88, range of 4.45–5.88) and the controls having log10 4.90 copies/ml (IQR 4.57–5.5, range of 2.94–5.88), respectively, p = 0.19. The time after nevirapine administration until ART initiation in the two groups was 5.5 months (IQR 3.75–8.63, range of 1–10) in the treatment failure group compared to 9.5 months (IQR 7–13.56, range of 0–38.5) in the control group (p = 0.09). The median duration of follow-up was 6 months in the failure group and 31 months in the group without treatment failure.
Table 1.
Clinical Characteristics of the Patients Analyzed in This Study Stratified by Failure or Success of NVP-Containing ART after sdNVP Exposure
| ART failures (n = 7) | Controls (n = 19) | p Valuea | |
|---|---|---|---|
| CD4 count (cells/μl) at HAART initiation, median (IQR)b | 51 (46–84)c | 179 (156–231)d | 0.001 |
| Viral load (RNA copies/log10) at HAART initiation, median (IQR)b | 5.31 (4.91–5.88) | 4.90 (4.57–5.5) | 0.19 |
| Months after delivery before starting HAART, median (IQR) | 5.5 (3.75–8.63) | 9.5 (7–13.56) | 0.09 |
| Months to failure or duration of follow-up, median | 6 (5.5–6.6) | 31 (24.13–42.69) | — |
p-value by Wilcoxon rank sum test.
CD4 count and viral load are from samples immediately prior to antiretroviral therapy. IQR, interquartile range.
One value missing.
Two values missing.
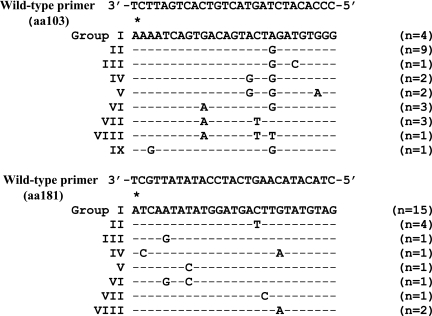
Cloning and sequencing of the 26 samples resulted in 530 sequences analyzed, with 18 to 24 clones available for each sample. All of the patients in this analysis had HIV-1 subtype C virus. Five of the 26 samples (19%) had detectable nevirapine resistance mutations (shown on Table 2) as determined by the IAS-USA list of HIV-1 drug resistance mutations; these included the 103N, 106A, 108I, 188H, and 190A mutations in reverse transcriptase. Only one sample had more than one resistance mutation detected (103N and 188H). Resistance was detected by TA cloning and sequencing in both those who failed therapy (three had resistance) and in individuals who did not fail therapy (two had resistance). Sequencing also revealed extensive nucleotide polymorphism in the binding site for the reverse primer in the allele-specific quantitative PCR assay. There were nine unique sequences identified in the K103 binding site (Groups I–IX) and eight unique sequences identified in the Y181 binding site (Groups I–VIII) as shown in Fig. 1.
Table 2.
Clinical and Drug Resistance Data from Women Exposed to sdNVP and Starting on NVP-Containing ARTa
| Patient | Time after sdNVP prior to HAART (months) | Time to failure/follow-up (months) | Viral load (log10) | CD4 count (cells/μl) | %103K (AAA) (AAG) | %103N (AAC) | %103N (AAT) | %181Y (TAT) | %181C (TGT) | Viroseq results at failure |
|---|---|---|---|---|---|---|---|---|---|---|
| 1b,c | 10 | 6 | 5.88 | 54 | 100 | — | — | 99.76 | 0.20 | 184V/190A |
| 2b,c | 5.25 | 6 | 4.45 | 46 | 97.99 | 1.95 | — | 99.88 | 0.10 | 103N |
| 3b | 9.5 | 5.5 | 5.88 | 48 | 100 | — | — | 98.1 | 0.35 | 181C/184V |
| 4b | 5.5 | 5.5 | 5.88 | 22 | 95.17 | 3.14 | 1.69 | 99.85 | 0.13 | 103N/181C/184V |
| 5b | 3.25 | 6.75 | 4.79 | n/a | 98.96 | 0.84 | 0.20 | 99.9 | — | — |
| 6b | 6 | 7.5 | 5.27 | 84 | 99.98 | — | — | 99.75 | 0.24 | 103N |
| 7b,c | 1 | 5 | 5.31 | 151 | 99.98 | — | — | 99.6 | 0.10 | 103N |
| 8c | 14.75 | 24 | 3.78 | 179 | 99.94 | — | — | 99.56 | 0.14 | |
| 9 | 12.25 | 25 | 5.88 | 292 | 98.65 | 1.34 | — | 99.87 | 0.12 | |
| 10 | 19 | 22.75 | 5.88 | n/a | 99.54 | 0.45 | — | 99.56 | 0.44 | |
| 11 | 13.75 | 25 | 4.80 | 171 | 100 | — | — | 99.62 | 0.35 | |
| 12 | 8.5 | 24.5 | 5.02 | 226 | 99.99 | — | — | 99.9 | — | |
| 13 | 0 | 31 | 2.94 | 261 | 99.84 | — | — | 99.91 | — | |
| 14 | 1 | 30 | 5.47 | 117 | 93.56 | 2.44 | 4.11 | 99.69 | 0.29 | |
| 15 | 9.5 | 17.5 | 4.51 | n/ad | 99.98 | — | — | 99.85 | 0.13 | |
| 16 | 1 | 24 | 4.75 | 156 | 100 | — | — | 99.79 | — | |
| 17 | 7.25 | 55.75 | 5.44 | 50 | 99.98 | — | — | 99.96 | — | |
| 18 | 13 | 47 | 4.74 | 206 | 99.98 | — | — | 99.96 | — | |
| 19 | 16.5 | 41.5 | 5.88 | 246 | 99.97 | — | — | 99.8 | 0.17 | |
| 20c | 38.5 | 17.5 | 4.90 | 132 | 96.89 | 3.11 | — | 99.96 | — | |
| 21 | 7 | 54 | 5.72 | 155 | 99.99 | — | — | 99.9 | — | |
| 22 | 12 | 41 | 3.73 | 294 | 99.99 | — | — | 99.98 | — | |
| 23 | 12 | 37 | 3.48 | 200 | 99.97 | — | — | 99.92 | — | |
| 24 | 9.5 | 41.75 | 5.02 | 174 | 96.5 | 3.5 | — | 99.95 | — | |
| 25 | 7 | 45 | 5.26 | 171 | 99.99 | — | — | 99.94 | — | |
| 26 | 1.5 | 43 | 4.86 | 179 | 99.88 | 0.13 | — | 99.95 | — |
Patients 1–7 were those who experienced virologic failure after starting NVP-containing ART after exposure to sdNVP. Cells with dash marks represent negative results (either no mutations are present when the cloning sequence was performed or the result is below the clinical cutoff on the ASPCR assay).
Virologic failure following treatment with NVP-containing ART.
Patient with detectable resistance by cloning and sequencing.
—, data not available.
FIG. 1.
Sequence variation in primer binding sites for ASPCR assay. Variations in the primer binding site sequences for the ASPCR assay necessitated creating nine different primer sets for the 103 position and eight different primer sets for the 181 position of reverse transcriptase with the wild-type primer binding site polymorphisms shown above. The asterisk (*) is over the site of interest where a nucleotide change confers drug resistance at both positions 103 and 181. The reverse wild-type primer as shown has an intentional mismatch at the (–2) position to enhance specificity. (n) represents the number of samples within this data set that matches each primer set.
Viroseq results on the seven failure samples showed that six of the seven patients had detectable NNRTI resistance at the time of failure (Table 2). Minor variant viruses with the K103N and/or Y181C nevirapine resistance mutations were detected by ASPCR in 17/26 (65%) of the samples tested: 7/7 (100 %) of the case (treatment failure) samples had resistance mutations detected and 10/19 (53%) of the control (treatment success) had resistance mutations detected. For all 17 samples in which resistance mutations were detected, the frequency of the resistance mutation(s) was well below the 20% threshold of the standard genotyping assay; the K103N mutations (AAC and AAT) were present at a median of 1.82% (range 0.13–4.11) and the Y181C mutation (TGT) was present with a median value of 0.17% (range 0.10–0.44). The K103N mutations were detected in 9/26 (35%) of the samples: 9/9 (100%) AAC, 3/9 (33%) AAC + AAT, 0/9 (0%) AAT alone. The Y181C mutation was detected in 13/26 (50%) of the samples, and mutations for both K103N and Y181C were detected in 5/26 (19%).
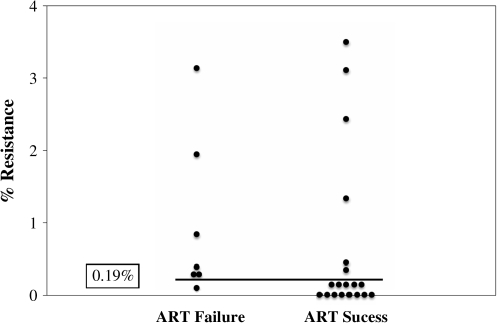
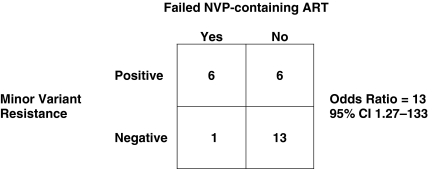
An ROC analysis of the resistance frequency data in the cases and controls was undertaken in an attempt to identify a clinically relevant threshold at which minor variants impact outcome. This analysis identified 0.19% as the threshold frequency at which the testing for minor variants in this assay maximized both sensitivity and specificity for predicting virologic failure. This exploratory threshold was then applied to the original set of 26 patients with a patient being designated to have resistance if the minor variant mutation was 0.19% or more of the viral population. Using this new threshold for minor variant resistance, 6/7 (86%) of the patients who failed therapy had NVP resistance mutations pre-ART vs. 6/19 (32%) of the patients who succeeded with therapy (Fig. 2). This represents an odds ratio of 13 (95% CI 1.27–133) for failing NVP-containing ART when a minor variant was detected in the plasma at the 0.19% threshold (Fig. 3).
FIG. 2.
Histogram showing number of cases of ART failure and success with clinical cutoff as determined by the ROC curve (0.19%). There are six of seven individuals in the failure group with minor variant resistance greater than 0.19% and six of 19 in the group that was successful with ART with resistance greater than 0.19%.
FIG. 3.
Odds ratio for ART failure with detectable minor variants in the pre-ART plasma sample. The odds of failing NVP-containing ART were 13 times greater if minor variants were present at a clinical threshold of 0.19% or more in the pre-ART plasma samples.
Discussion
sdNVP is frequently used in MTCT prevention programs as highly effective combination ART and even short-course antenatal zidovudine for PMTCT are not yet accessible to the majority of HIV-infected pregnant women in the developing world. Unfortunately, many studies have shown that at least transient drug resistance develops in a large fraction of women after sdNVP, and drug-resistant HIV variants persist for considerable periods in patient samples at frequencies below the limit of detection of the most commonly used commercial genotyping methods.15–18,26,27
The clinical importance of such drug-resistant “minor variants” following sdNVP remains unclear. Recent reports in settings other than sdNVP for PMTCT have suggested that minor variants may contribute to virologic failure.20–22,28 These studies were in both treatment-naive patients, in whom the minor variants may represent transmitted drug resistance, and in heavily treatment-experienced patients, in whom these minor variants likely represent archived, resistant virus that reemerges with the reintroduction of ART. Additionally, a new report demonstrated that minor variant resistance at the 103 position of reverse transcriptase after sdNVP was associated with early virologic failure with NNRTI-containing ART.23
The study described here was designed to determine if the presence of low-frequency drug variants, detected by highly sensitive methods, could predict virologic failure when NVP-based ART is initiated following sdNVP exposure. The ASPCR assay was performed at positions 103 and 181 of reverse transcriptase as these mutations were present most often after sdNVP in Botswana.29 The results of this study showed that patients with minor drug-resistant variants present in the plasma at a frequency greater than 0.19%, prior to ART initiation, had a higher odds of clinical failure (odds ratio = 13, 95% CI 1.27–133). Those that failed NVP-containing ART started medications closer to the time of nevirapine exposure compared to those who were successful with ART (5.5 months vs. 9.5 months, p = 0.09).
Minor drug-resistant variants have been proposed as a possible cause of the higher than anticipated rates of ART failure in the Thailand PHPT-2 study,11 and our results support the hypothesis of a relationship between pre-ART initiation minor resistant variants and ART outcome. Our results also provide initial data for reexamining the clinically relevant threshold of drug-resistant viruses. Although resistance detected by standard genotyping in treatment-naive individuals is well documented to decrease the likelihood of successful ART and/or to delay the time to virologic suppression,30,31 these assays are limited to detecting drug resistance mutations representing approximately 20% or more of the virus population in a patient, and there is no logical reason to presuppose that the assay sensitivity threshold is also the threshold frequency of clinical relevance. As more sensitive methods such as ASPCR have been developed that allow detection and quantification of drug resistance mutations to frequencies as low as 0.10%, it has become possible to begin to reexamine the question of minimum drug resistance frequency that may impact clinical outcome.
A clinically relevant threshold of minor resistant variants after sdNVP that helps predict successful ART likely reflects the degree of viral decay of resistant virions that has occurred (with subsequent outgrowth of wild-type virus) to allow for clinical success with ART. This theory would explain the finding in Botswana that waiting at least until 6 months after NVP exposure significantly improved virologic success with ART, at levels matching those women who received zidovudine and placebo,12 but low levels of resistance can be overcome as seen in six patients in this study who were successful with ART despite the presence of minor variants above the level of 0.19%.
Viroseq testing at the time of failure in the six individuals who had baseline minor drug-resistant variants above 0.19% demonstrated NNRTI mutations in five of the six samples. Additionally, in three of the samples, the M184V mutation was detected, which confers resistance to lamivudine. It is likely that the baseline NNRTI resistance precluded full virologic suppression, allowing lamivudine resistance to emerge. This supports the importance of the baseline NNRTI resistance, even at these low levels, in some individuals.
This study analyzed patient samples immediately prior to starting ART and not at a specified time point (e.g., 3 or 6 months) after exposure to NVP. An examination of nevirapine resistance at a standardized time point in all patients who received sdNVP, determining resistance in both the plasma and proviral DNA reservoir, would allow correlation of both reservoirs with clinical outcome. Ideally, such a sample set would show clearly the viral kinetics of the resistant variants and determine if resistant virus decay is important for treatment success. It is unlikely to explain some of the variability in the results as some resistant variants are still detectable long after NVP exposure and not associated with treatment failure. This may be related to host factors or individual viral characteristics and cannot be resolved in this study.
Additionally, the threshold of clinical relevance that was selected was not differentiated between the two different mutations and there may be different thresholds of clinical relevance given the reported greater impact that the Y181C has on viral fitness compared to the K103N mutation.32 This possibly could be examined in a large sample set where stratification based on minor variant detection thresholds could be employed, but this may not be clinically significant given the recent report by Coovadia et al.23 In their study, when looking only at the K103N mutation, a threshold of 0.20% was found to be significant in predicting success of ART.
It is notable that the CD4 counts in the cases and controls were significantly different in this study (67.5 cells/μl vs. 188.8 cells/μl, p < 0.001). Persons with lower CD4 counts were more likely to fail therapy; this was also demonstrated in the Mashi study overall. However, there is no obvious biological explanation for why lower CD4 counts might contribute to minor variant drug resistance since the viral loads were not statistically different between the two groups. Indeed, in five of the six individuals who failed therapy with detectable low level drug resistance at treatment initiation, the genotype at the time of failure demonstrated NNRTI resistance. This certainly suggests that minor drug-resistant variants play a role in treatment failure, but these variants cannot be conclusively stated as the sole cause of failure, particularly as this study was unable to match patients by CD4 count. Future studies with a larger sample size should be stratified by CD4 count to clarify that minor variants contribute to clinical failure independent of CD4 count. It is unlikely that there were differences in nevirapine exposure to account for the different outcomes, as a subset of individuals examined in the Mashi study had detectable nevirapine levels in 95 of 96 (99%) women, suggesting very high adherence overall to the sdNVP intervention in the Mashi study.29
The ASPCR assay designed for this study, with its extensive primers and control plasmids, would be difficult to employ in many settings due to cost and the need for genotype information prior to primer selection. However, the data from this analysis do support the observation that waiting for a period of time after sdNVP exposure is likely to be beneficial, as these findings may represent the decay of resistant virions after sdNVP. The reality in resource-limited settings may be that waiting as long as possible after sdNVP exposure before starting ART, without jeopardizing patients' health, may better the odds of successful nevirapine-containing ART.
The observation in this study that pre-ART minor drug-resistant variants in plasma samples after sdNVP were predictive of subsequent clinical failure suggests that detection of drug-resistant viruses with greater sensitivity than current standards has clinical value. More research should be performed to see that the clinical threshold with this ASPCR assay is maintained in larger data sets, to determine the relationship between CD4 count and minor variants, and also to explore the possibility that other viral reservoirs containing resistant viruses impact clinical success. The goal of PMTCT is for better, safer interventions designed to protect mother and infant. To do this, it is necessary to continue to try to understand the impact of sdNVP and how best to treat patients who develop nevirapine resistance as a consequence.
Acknowledgments
C.F.R. is supported by NIAID K08 AI067014 and Harvard University Centers for AIDS Research Grant P30 AI060354. M.E. and S.L. are supported by R01 HD037793 and R01 HD044391. C.L.B.'s present affiliation is the Partners AIDS Research Center at Massachusetts General Hospital; E.L. is currently enrolled at the Emory University School of Medicine.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.UNAIDS: 2008. Report on the Global HIV/AIDS Epidemic. Geneva: UNAIDS; 2008. [Google Scholar]
- 2.Guay LA. Musoke P. Fleming T. Bagenda D. Allen M. Nakabiito C. Sherman J. Bakaki P. Ducar C. Deseyve M. Emel L. Mirochnick M. Fowler MG. Mofenson L. Miotti P. Dransfield K. Bray D. Mmiro F. Jackson JB. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 3.Lallemant M. Jourdain G. Le Coeur S. Mary JY. Ngo-Giang-Huong N. Koetsawang S. Kanshana S. McIntosh K. Thaineua V. Single-dose perinatal nevirapine plus standard zidovudine to prevent mother-to-child transmission of HIV-1 in Thailand. N Engl J Med. 2004;351:217–228. doi: 10.1056/NEJMoa033500. [DOI] [PubMed] [Google Scholar]
- 4.Ayouba A. Tene G. Cunin P. Foupouapouognigni Y. Menu E. Kfutwah A. Thonnon J. Scarlatti G. Monny-Lobe M. Eteki N. Kouanfack C. Tardy M. Leke R. Nkam M. Nlend AE. Barre-Sinoussi F. Martin PM. Nerrienet E. Low rate of mother-to-child transmission of HIV-1 after nevirapine intervention in a pilot public health program in Yaounde, Cameroon. J Acquir Immune Defic Syndr. 2003;34:274–280. doi: 10.1097/00126334-200311010-00003. [DOI] [PubMed] [Google Scholar]
- 5.Stringer JS. Sinkala M. Chapman V. Acosta EP. Aldrovandi GM. Mudenda V. Stout JP. Goldenberg RL. Kumwenda R. Vermund SH. Timing of the maternal drug dose and risk of perinatal HIV transmission in the setting of intrapartum and neonatal single-dose nevirapine. AIDS. 2003;17:1659–1665. doi: 10.1097/01.aids.0000072647.21517.2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taha TE. Kumwenda NI. Hoover DR. Fiscus SA. Kafulafula G. Nkhoma C. Nour S. Chen S. Liomba G. Miotti PG. Broadhead RL. Nevirapine and zidovudine at birth to reduce perinatal transmission of HIV in an African setting: A randomized controlled trial. JAMA. 2004;292:202–209. doi: 10.1001/jama.292.2.202. [DOI] [PubMed] [Google Scholar]
- 7.Eshleman SH. Mracna M. Guay LA. Deseyve M. Cunningham S. Mirochnick M. Musoke P. Fleming T. Glenn Fowler M. Mofenson LM. Mmiro F. Jackson JB. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012) AIDS. 2001;15:1951–1957. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 8.Eshleman SH. Guay LA. Mwatha A. Cunningham SP. Brown ER. Musoke P. Mmiro F. Jackson JB. Comparison of nevirapine (NVP) resistance in Ugandan women 7 days vs. 6–8 weeks after single-dose nvp prophylaxis: HIVNET 012. AIDS Res Hum Retroviruses. 2004;20:595–599. doi: 10.1089/0889222041217518. [DOI] [PubMed] [Google Scholar]
- 9.Eshleman SH. Hoover DR. Chen S. Hudelson SE. Guay LA. Mwatha A. Fiscus SA. Mmiro F. Musoke P. Jackson JB. Kumwenda N. Taha T. Resistance after single-dose nevirapine prophylaxis emerges in a high proportion of Malawian newborns. AIDS. 2005;19:2167–2169. doi: 10.1097/01.aids.0000194800.43799.94. [DOI] [PubMed] [Google Scholar]
- 10.Toni TD. Masquelier B. Lazaro E. Dore-Mbami M. Ba-Gomis FO. Tea-Diop Y. Kouakou K. Diby J. Sia E. Soppi S. Essien S. Schrive MH. Pinson P. Chenal H. Fleury HJ. Characterization of nevirapine (NVP) resistance mutations and HIV type 1 subtype in women from Abidjan (Cote d'Ivoire) after NVP single-dose prophylaxis of HIV type 1 mother-to-child transmission. AIDS Res Hum Retroviruses. 2005;21:1031–1034. doi: 10.1089/aid.2005.21.1031. [DOI] [PubMed] [Google Scholar]
- 11.Jourdain G. Ngo-Giang-Huong N. Le Coeur S. Bowonwatanuwong C. Kantipong P. Leechanachai P. Ariyadej S. Leenasirimakul P. Hammer S. Lallemant M. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N Engl J Med. 2004;351:229–240. doi: 10.1056/NEJMoa041305. [DOI] [PubMed] [Google Scholar]
- 12.Lockman S. Shapiro RL. Smeaton LM. Wester C. Thior I. Stevens L. Chand F. Makhema J. Moffat C. Asmelash A. Ndase P. Arimi P. van Widenfelt E. Mazhani L. Novitsky V. Lagakos S. Essex M. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356:135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 13.Deeks SG. Wrin T. Liegler T. Hoh R. Hayden M. Barbour JD. Hellmann NS. Petropoulos CJ. McCune JM. Hellerstein MK. Grant RM. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N Engl J Med. 2001;344:472–480. doi: 10.1056/NEJM200102153440702. [DOI] [PubMed] [Google Scholar]
- 14.Church JD. Jones D. Flys T. Hoover D. Marlowe N. Chen S. Shi C. Eshleman JR. Guay LA. Jackson JB. Kumwenda N. Taha TE. Eshleman SH. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagn. 2006;8:430–432. doi: 10.2353/jmoldx.2006.050148. ; quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flys T. Nissley DV. Claasen CW. Jones D. Shi C. Guay LA. Musoke P. Mmiro F. Strathern JN. Jackson JB. Eshleman JR. Eshleman SH. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis. 2005;192:24–29. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- 16.Lecossier D. Shulman NS. Morand-Joubert L. Shafer RW. Joly V. Zolopa AR. Clavel F. Hance AJ. Detection of minority populations of HIV-1 expressing the K103N resistance mutation in patients failing nevirapine. J Acquir Immune Defic Syndr. 2005;38:37–42. doi: 10.1097/00126334-200501010-00007. [DOI] [PubMed] [Google Scholar]
- 17.Johnson JA. Li JF. Morris L. Martinson N. Gray G. McIntyre J. Heneine W. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- 18.Loubser S. Balfe P. Sherman G. Hammer S. Kuhn L. Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS. 2006;20:995–1002. doi: 10.1097/01.aids.0000222071.60620.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer S. Boltz V. Martinson N. Maldarelli F. Gray G. McIntyre J. Mellors J. Morris L. Coffin J. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci USA. 2006;103:7094–7099. doi: 10.1073/pnas.0602033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kullsiek KH. Peng G. Simen B. Simons J. Egholm M. Novak R. MacArther R. Kozal M. Virologic success of different strategies for initial ART regimens is predicted by the type and detection level of minor drug-resistant variant detected by ultra deep sequencing: The CPCRA 058 FIRST study; 15th Conference on Retroviruses and Opportunistic Infections; Boston. Feb 3–6;2008 ; Abstract 878. [Google Scholar]
- 21.Johnson JA. Li JF. Wei X. Lipscomb J. Irlbeck D. Craig C. Smith A. Bennett DE. Monsour M. Sandstrom P. Lanier ER. Heneine W. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paredes R. Lalama C. Ribaudo H. Schackman B. Shikuma C. Meyer W., III Giguel F. Squires K. Gulick R. Kuritzkes D. Presence of minor populations of Y181C mutants detected by allele-specific PCR and risk of efavirenz failure in treatment-naive patients: Results of an ACTG 5095 case-cohort study; 15th Conference on Retroviruses and Opportu-nistic Infections; Boston. Feb 3–6;2008 ; Abstract 83. [Google Scholar]
- 23.Coovadia A. Hunt G. Abrams EJ. Sherman G. Meyers T. Barry G. Malan E. Marais B. Stehlau R. Ledwaba J. Hammer SM. Morris L. Kuhn L. Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clin Infect Dis. 2009;48:462–472. doi: 10.1086/596486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson V. Brun-Vezinet F. Clotet B. Gunthard H. Kuritzkes D. Pillay D. Schapiro J. Richman D. Update of the drug resistance mutations in HIV-1: Spring 2008. Top HIV Med. 2008;16:62–68. doi: 10.1007/s11750-007-0034-z. [DOI] [PubMed] [Google Scholar]
- 25.Rowley CF. Boutwell CL. Lockman S. Essex M. Improvement in allele-specific PCR assay with the use of polymorphism-specific primers for the analysis of minor variant drug resistance in HIV-1 subtype C. J Virol Methods. 2008;149:69–75. doi: 10.1016/j.jviromet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer S. Kearney M. Maldarelli F. Halvas EK. Bixby CJ. Bazmi H. Rock D. Falloon J. Davey RT., Jr. Dewar RL. Metcalf JA. Hammer S. Mellors JW. Coffin JM. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergroth T. Sonnerborg A. Yun Z. Discrimination of lamivudine resistant minor HIV-1 variants by selective real-time PCR. J Virol Methods. 2005;127:100–107. doi: 10.1016/j.jviromet.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Mellors J. Palmer S. Nissley D. Kearney M. Halvas E. Bixby C. Demeter L. Eshleman S. Bennett K. Hart S. Vaida F. Wantman M. Coffin J. Hammer SftASG. Low-frequency NNRTI-resistant variants contribute to failure of efavirenz-containing regimens; 11th Conference on Retroviruses and Opportunistic Infections; San Francisco. Feb 8–11;2004 ; Abstract A39. [Google Scholar]
- 29.Shapiro RL. Thior I. Gilbert PB. Lockman S. Wester C. Smeaton LM. Stevens L. Heymann SJ. Ndung'u T. Gaseitsiwe S. Novitsky V. Makhema J. Lagakos S. Essex M. Maternal single-dose nevirapine versus placebo as part of an antiretroviral strategy to prevent mother-to-child HIV trans-mission in Botswana. AIDS. 2006;20:1281–1288. doi: 10.1097/01.aids.0000232236.26630.35. [DOI] [PubMed] [Google Scholar]
- 30.Grant RM. Hecht FM. Warmerdam M. Liu L. Liegler T. Petropoulos CJ. Hellmann NS. Chesney M. Busch MP. Kahn JO. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288:181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 31.Little SJ. Holte S. Routy JP. Daar ES. Markowitz M. Collier AC. Koup RA. Mellors JW. Connick E. Conway B. Kilby M. Wang L. Whitcomb JM. Hellmann NS. Richman DD. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 32.Collins JA. Thompson MG. Paintsil E. Ricketts M. Gedzior J. Alexander L. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J Virol. 2004;78:603–611. doi: 10.1128/JVI.78.2.603-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]