Summary
Metals are essential elements of all living organisms. Among them, copper is required for a multiplicity of functions including mitochondrial oxidative phosphorylation and protection against oxidative stress. Here we will focus on describing the pathways involved in the delivery of copper to cytochrome c oxidase (COX), a mitochondrial metalloenzyme acting as the terminal enzyme of the mitochondrial respiratory chain. The catalytic core of COX is formed by three mitochondrially-encoded subunits and contains three copper atoms. Two copper atoms bound to subunit 2 constitute the CuA site, the primary acceptor of electrons from ferrocytochrome c. The third copper, CuB, is associated with the high-spin heme a3 group of subunit 1. Recent studies, mostly performed in the yeast Saccharomyces cerevisiae, have provided new clues about 1- the source of the copper used for COX metallation; 2- the roles of Sco1p and Cox11p, the proteins involved in the direct delivery of copper to the CuA and CuB sites, respectively; 3- the action mechanism of Cox17p, a copper chaperone that provides copper to Sco1p and Cox11p; 4- the existence of at least four Cox17p homologues carrying a similar twin CX9C domain suggestive of metal binding, Cox19p, Cox23p, Pet191p and Cmc1p, that could be part of the same pathway; and 5- the presence of a disulfide relay system in the intermembrane space of mitochondria that mediates import of proteins with conserved cysteines motifs such as the CX9C characteristic of Cox17p and its homologues. The different pathways are reviewed and discussed in the context of both mitochondrial COX assembly and copper homeostasis.
Keywords: Mitochondria, cytochrome oxidase, copper, metal homeostasis, superoxide dismutase
CYTOCHROME C OXIDASE: A MITOCHONDRIAL COPPER/HEME METALLOENZYME
Aerobic life depends on cellular copper homeostasis and distribution since this element is a critical component of enzymes involved in primary metabolism (1). Copper ions can undergo unique chemistry due to their ability to adopt distinct redox states, either oxidized [Cu(II)] or reduced [Cu(I)], and they serve as important catalytic cofactors in redox chemistry for proteins that carry out fundamental biological functions. A copper-containing metalloenzyme, mitochondrial cytochrome c oxidase (COX), is the final electron acceptor in the mitochondrial electron transport chain and is required for aerobic ATP production.
Over the last 15 years COX biogenesis has received significant attention because of its medical relevance. Defective COX biogenesis results in mitochondrial diseases frequently involving brain, skeletal muscle and heart (reviewed in (2-4)). In particular, mutations in genes required for COX copper metallation result in severe hepatopathies, encephalomyopathies, hypertrophic cardiomyopathy and muscular atrophy (5-9). Because most of the basic biological knowledge concerning mitochondrial copper homeostasis and insertion into COX derives from investigations in the yeast Saccharomyces cerevisiae, however, this review will primarily focus on these studies. Intracellular copper transport in mammals has been reviewed elsewhere (1, 10) but significant differences between yeast and human COX copper metallation will also be briefly discussed here.
Mitochondrial COX is a multi-subunit enzyme formed, depending on the eukaryotic organism, by 12 or more subunits (11) of dual genetic origin. The mitochondrial genome encodes the three catalytic core subunits which are also present in the bacterial enzyme (12), while the nucleus houses genes for nine other subunits that form a protective cage around the core. COX transverses the inner mitochondrial membrane with portions protruding into the intermembrane space and the matrix. The enzyme catalyzes electron transfer from cytochrome c to molecular oxygen. This reaction occurs concurrently with vectorial proton pumping from the matrix to the intermembrane space, thus contributing to the generation of a transmembrane proton gradient which is subsequently used by the mitochondrial ATP synthase to drive the synthesis of ATP.
COX is a heme/copper terminal oxidase which uses cytochrome c as an electron donor as mentioned above. The enzyme contains four redox-active metal centers which sequentially aid in reducing molecular oxygen to water: two iron centers, heme a and heme a3 (also referred to as cytochromes a and a3), and two copper centers, CuA and CuB. Electrons enter COX through a mixed valence dinuclear copper center, the CuA site, located in subunit 2. CuA transfers electrons to the low spin heme a located in subunit 1. From heme a, electrons are transferred intra-molecularly to the active site in subunit 1, composed of the high spin heme a3 and CuB, where oxygen (and other ligands such as CO, NO and CN−) binds. The mechanism of electron transfer through COX has been extensively studied and reviewed elsewhere (13-15). From an evolutionary point of view, the presence of the CuA center defines cytochrome c oxidase and distinguishes this enzyme from other terminal oxidases that use quinol instead of cytochrome c as the electron donor (13, 16, 17).
As a consequence of its central role in oxidative metabolism COX has been intensively studied by biochemical, genetic, spectroscopic, and crystallographic means (11, 18, 19). From these studies it is known that other metals such as zinc (in Cox4p) and magnesium (in the interface between Cox1p and Cox2p) are also bound to the enzyme, although the basis for this specific requirement is largely unknown. The zinc atom (Zn2+), located in a non-catalytic subunit, could play a role in structural stability of the complex (20). The magnesium/manganese site (Mg2+/Mn2+) is in close proximity to the H2O exit channel and is thought to aid in the stability and release of H2O produced by the reduction of O2 (21). Interestingly, it has been proposed that insertion of CuB is likely to facilitate the stable formation of the Mg2+/Mn2+ center (22), the two sites are approximately 13.5 Å apart, with imidazole side chains of ligands from each of these centers coming within 7 Å (11, 19).
Copper metallation of mitochondrial subunits 1 and 2 is essential for the maturation and stability of these subunits. In the absence of the copper centers (as well as in the absence of the heme prosthetic groups) the proteins are rapidly degraded and the COX holoenzyme fails to assemble (reviewed in (23, 24)). Thus, copper is not only required for COX catalytic function but also for its biogenesis, assembly and stability. The following sections will focus on briefly describing the pathways involved in cellular copper uptake, delivery to mitochondria and insertion into the COX catalytic core subunits.
CELLULAR COPPER HOMEOSTASIS AND DELIVERY TO MITOCHONDRIA
Copper is a transition metal ion essential for the function of key metabolic enzymes as explained above. However, given its high redox reactivity copper can be the source of reactive oxygen species and extremely toxic to the cell. Therefore, a network of transporters strictly controls the trafficking of copper in living systems. Once within the cell, copper is either sequestered by metallothioneins (such as Cup1p and Crs5p), directed to the vacuole which is important for proper copper detoxification, or delivered to the sites of its utilization by specialized proteins called copper chaperones. An abundant amount of literature concerning the structure and function of these proteins is now available (recently reviewed in (1, 25-27)).
Briefly, yeast cells acquire copper, depending on extracellular copper concentrations, through the high affinity specific transporters Ctr1p (28) and Ctr3p (29) or through low affinity permeases such as the iron transporter Fet4p (30) and the metal transporter Smf1p (31). Extracellular copper mostly exists in the oxidized form Cu(II), but it is reduced to the Cu(I) form upon entering the cell by the membrane integral cupric reductases Fre1p and Fre2p (32). Once within the cell, copper is bound to cytosolic copper chaperones. Copper binds Atx1p for specific delivery to the secretory pathway. In yeast, Ccc2p, a P-type ATPase located in the trans-Golgi network, accepts copper from Atx1p (33-35) and transfers it into the lumen of the trans-Golgi network where it is incorporated into the multicopper ferroxidase Fet3p in a manner facilitated by the Gef1p chloride channel (36, 37). Active Fet3p forms a complex with the iron permease Ftr1p and both proteins translocate to the plasma membrane where they are responsible for high affinity iron uptake, thus connecting copper and iron metabolisms. In the cytosol, copper also binds Ccs1p which is required for the metallation and activation of the copper/zinc superoxide dismutase (Sod1p) (38). A small fraction of Sod1p and its metallochaperone, Ccs1p, localize to the intermembrane space of mitochondria (39). In both compartments the chaperone forms a Ccs1p–Sod1p heterodimer that facilitates copper transfer (40). Ccs1p-mediated copper insertion into Sod1p also catalyzes the formation of a critical disulfide bond in Sod1p which seems to be important for regulation of enzyme activity and for prevention of misfolding or aggregation (41). Alternative pathways for copper insertion into Sod1p have been proposed in mammals (42). Finally, copper also binds to Cox17p in the cytosol, a chaperone that has been proposed to have, similarly to Ccs1p, a dual location in the cytoplasm and in the mitochondrial intermembrane space. Mitochondrial Cox17p is necessary for copper delivery to cytochrome c oxidase (43) as discussed below.
Involvement of COX17 in copper homeostasis was originally proposed because the respiratory deficient phenotype of cox17 null mutant strains can be rescued by supplementation of copper to the media (43). It was proposed that COX17 could be shuttling copper from the cytoplasm into the mitochondrial intermembrane space (43, 44). This hypothesis has been challenged by the observation that while apo-Cox17p is predominantly a monomer with a simple hairpin structure, the protein-copper complex exists in a dimer/tetramer equilibrium (45) making its free passage though the outer membrane protein channel unlikely. In addition, it has been recently shown that the tethering of Cox17p to the inner mitochondrial membrane does not affect COX assembly (46). Two homologues of Cox17p, the small soluble proteins Cox19p and Cox23p, exhibit a cellular distribution similar to Cox17p when over-expressed and are required for COX assembly (47, 48). As stated above for Cox17p, it is unlikely that these proteins will act as copper shuttles from the cytosol to the mitochondrial compartments. It was recently shown that Cox19p and Cox23p are not required for the import or accumulation of copper in the mitochondrion (46). Mitochondria contain a matrix pool of copper bound by a low molecular weight non-proteinaceous ligand (46) which has been shown to be the copper source for metallation of COX and mitochondrial Sod1p (49). The ligand from the copper complex has been found in the cytoplasm and it has been suggested that it may recruit, in place of a copper chaperone, the copper that is translocated and stored within the mitochondrial matrix (49). Whatever the case, the identity of key proteins in the copper trafficking process to mitochondria remains elusive. Which is/are the predicted mitochondrial membrane copper permease/s and how is copper transported from the matrix pool to the copper chaperons located in the IMS are two obvious questions remaining.
COPPER DELIVERY TO COX
Mitochondrial copper homeostasis and delivery to cytochrome c oxidase appears to require the function of a growing number of gene products, some of them largely uncharacterized, listed in Table 1 (References (22, 43-45, 47, 48, 50-81)).
Table 1.
Genes encoding for mitochondrial proteins required for mitochondrial copper homeostasis and delivery to cytochrome c oxidase in the yeast Saccharomyces cerevisiae
| GENE | FUNCTION | REFERENCES* |
|---|---|---|
| Copper transfer to Cox1p and Cox2p | ||
|
| ||
| COX11 | Stable formation of the Cu(B) center in COX subunit 1 | (22, 50-54) |
| SCO1 | Transfer of copper to the Cu(A) center or reduction of cysteine residues in COX subunit 2. |
(55-62) |
|
| ||
| Copper homeostasis in the intermembrane space | ||
|
| ||
| COX17 | Transfer of Copper to Cox11p and Sco1p | (43-45, 63-69) |
| COX19 | Copper metabolism in the intermembrane space? | (47, 70) |
| COX23 | Copper metabolism in the intermembrane space? | (48) |
| CMC1 | Copper metabolism in the intermembrane space? | Horn and Barrientos, unpublished |
| PET191 | Uncharacterized | (71) |
|
| ||
| Import of COX copper chaperones | ||
|
| ||
| MIA40 | Import of mitochondrial copper chaperons containing twin CX9C motifs via a disulfide relay system |
(72-78) |
| ERV1 | Sulfhydryl oxidase involved in the reduction of Mia40p | (75, 77, 79-81) |
Due to space limitations, the list of references unfortunately does not include all the literature in the field but only some of the most relevant contributions
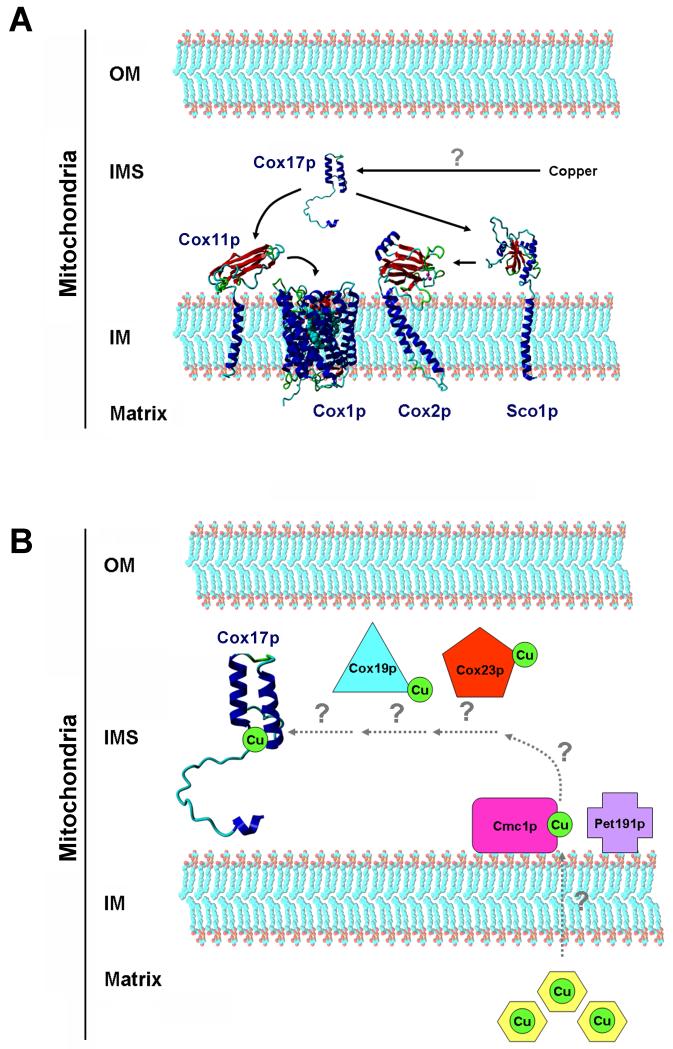
COX copper metallation involves the copper chaperone Cox17p, essential for COX assembly (43), a small (~8 kDa) hydrophilic protein containing a CCXC metal binding motif which binds copper ions (44). Cox17p also contains a twin CX9C structural motif. While the first cysteine in the twin CX9C motif is part of the CCXC copper binding motif, the remaining three conserved cysteines are not important for Cox17p copper-binding function (67). Cox17p transfers copper ions to two additional chaperones (63) that facilitate copper insertion into the COX CuA and CuB active sites, respectively Sco1p (55) and Cox11p (22, 50) as depicted in Fig. 1A. These proteins are anchored to the mitochondrial inner membrane through a transmembrane α-helix and expose their copper binding sides in the IMS where copper transfer occurs (44, 51). In vitro experiments have shown that a soluble truncated form of both Cox11p and Sco1p are able to bind copper (51, 61). It is still not clear, however, how the transfer of copper occurs between Cox17p and these proteins because physical interactions among them have not been detected (63).
Figure 1.
Proteins proposed to be involved in mitochondrial copper metabolism and addition to COX subunits. (A) Copper insertion into the CuA and CuB sites in Cox2p and Cox1p, respectively. The soluble copper chaperone Cox17p transfers copper ions to two additional chaperones that facilitate copper insertion into the COX CuA and CuB active sites, respectively Sco1p and Cox11p. These proteins are anchored to the mitochondrial inner membrane through a transmembrane α-helix and expose their copper binding sides in the IMS, where copper transfer occurs. Solution structures of the apo- Cox17p, and of the globular intermembrane space domains of Sco1p and Cox11p and ligand-bound Cox1p and Cox2p structures were obtained from the protein data bank (PDB) website (http://www.rcsb.org/pdb/home/home.do) and modified to prepare the figure. The transmembrane domains of Cox11p and Sco1p were artificially generated and used to represent the tethering of these proteins to the inner membrane. The YASARA molecular-graphics, -modeling and -simulation program, developed by Elmer Krieger, was used to generate a cartoon model of the different proteins. (B) Hypothetical role of the several CX9C containing proteins in regulating copper transfer from a matrix copper pool, across the inner mitochondrial membrane through an uncharacterized transporter, towards Cox17p. The hypothetical model is explained in the text. A possible role of the CX9C containing proteins in redox homeostasis within the intermembrane space is not depicted here. OM, outer membrane, IMS, intermembrane space, IM inner membrane. Black arrows indicate experimentally proved copper transfer while grey arrows indicate strictly hypothetical copper transfer.
CuA site metallation
As stated above, the CuA site found in Cox2p is the ligand responsible for accepting electrons from cytochrome c. The two copper atoms in the CuA site are coordinated by a CX3C motif as well as two histidines, one methionine and the backbone of a glutamine. Mutagenesis of any of these residues results in protein instability and the inability of yeast to grow on non-fermentable carbon sources (82). The metallochaperone for the formation of the CuA center is the product of SCO1 (55) which was originally identified as essential for COX assembly in yeast (59) and subsequently as a multicopy suppressor of a cox17 null mutant (43). Sco1p transfers copper from Cox17p to Cox2p and has been shown to directly interact with Cox2p (60). The protein contains a single transmembrane domain with a large soluble copper binding domain facing the intermembrane space. Sco1p has a metal binding thioredoxin-like CX3C motif analogous to the copper binding motif of Cox2p and this motif is essential for its function as demonstrated by site-direct mutagenesis (62). NMR studies of the human Sco1p homologue both with Cu(I)-bound and Ni(II)-bound allowed Banci and coworkers to propose a model for the Sco1p-Cox2p copper transfer involving the formation of a transient Sco1p species carrying a disulfide moiety able to interact with a copper ion (83). Because the CuA center is formed by a Cu(I) ion and a Cu(II) ion, it remains to be elucidated if Sco1p mediates the transfer of both of the different valent ions or, alternatively, if two Cu(I) are inserted in Cox2p by Sco1p and the active site is successively oxidized. It was demonstrated that Sco1p is able to bind both Cu(I) and Cu(II) (61, 84, 85), although it is not clear if Sco1p receives both cations or only the monovalent copper Cu(I) from Cox17p (84). Although the experimental data could support a role for Sco1p in copper insertion, considering its structural similarity with the protein family of disulfide reductases it has been considered that it is more likely to be involved in the reduction of cysteines in the Cox2p copper binding site (58). This hypothesis is supported by data from the NMR models mentioned above where Sco1p could function as a thioredoxin to reduce the cysteine residues in the CuA site of Cox2p (83). This reduction is necessary for the co-factor incorporation (56, 86). Sco1p has the ability to form homodimeric complexes (60) which could facilitate the performance of both functions by the collaborative action of each monomer.
Interestingly, yeast SCO1 has a highly conserved homologue, SCO2 (87), with apparently non-overlapping functions. Deletion of SCO2 does not affect COX assembly (55). Although SCO2 over-expression does not suppress the COX assembly defect of a sco1 null mutant strain it is able to partially rescue a sco1 point mutant (55). Additionally, SCO2 over-expression also suppresses cox17 mutations, although less efficiently than SCO1, and only when the growth media is supplemented with copper (55). These data were interpreted to indicate that yeast Sco1p and Sco2p have overlapping but not identical functions (55).
It is noteworthy that humans also have two homologues of yeast Sco1p, named SCO1 and SCO2 (88), both of which are essential for COX assembly. Mutations in SCO1 (5) and SCO2 (6-8) actually result in severe mitochondrial disorders. Functional complementation studies have shown that expression of either human SCO1 or SCO2 does not complement a yeast strain carrying a null allele of sco1 (89). Interestingly, the expression of a chimera with the N terminus of yeast and the C terminus (that contains the CX3C copper binding domain) of human SCO1 is able to complement the yeast mutant, an ability that is lost when using the C terminus of SCO2 (89). The functional differences among the two yeast and human isoforms is explained by the fact that the two genes probably originated from a duplication that occurred separately in the two organisms (6).
In contrast with their yeast homologues, human SCO1 and SCO2 have been shown to perform independent, cooperative functions in the process of COX assembly (90). This is suggested by the fact that overexpression of each SCO protein in fibroblasts from patients with mutation in the other SCO protein results in a dominant negative phenotype (90). These results have suggested a model in which human COX17 delivers copper to SCO2 which in turn transfers it directly to the CuA site in COX subunit II in a reaction that is facilitated by SCO1 (90). Recently, SCO1 and SCO2 were shown to have additional regulatory roles in the maintenance of cellular copper homeostasis cooperating to regulate copper efflux under conditions of excessive cellular copper (91) and commented in (92). As stated above, the CuA site in COXII (as in yeast Cox2p) is of mixed valence. SCO2 could play a role in the formation of a heterodimer with SCO1 and this dimerization could be needed to create the Cu(II)-Cu(I) site (84).
CuB site metallation
The CuB site located in Cox1p is formed by one copper ion coordinated by three histidine ligands and present in close proximity to the heme a moiety. The metallochaperone for the formation of the CuB site of Cox1p is the product of COX11 (22, 50), as shown in the prokaryote Rhodobacter sphaeroides (22).
Similar to Sco1p, yeast Cox11p has one transmembrane domain and a soluble copper binding domain which faces the intermembrane space (51). Cox11p was shown to bind Cu(I) (50). The soluble C-terminal domain of Cox11p forms a dimer that coordinates one Cu(I) per monomer via three thiolate ligands. The two Cu(I) ions in the dimer exist in a binuclear cluster and appear to be ligated by three conserved cysteine residues (50). The mechanism of copper transfer to CuA remains to be elucidated.
Copper transfer from Cox11p to the Cox1p CuB site, deeply buried below the membrane surface, may occur co-translationally in nascent Cox1p polypeptides extruded across the inner membrane. This possibility is supported by the observation of a weak interaction of Cox11p with the mitochondrial ribosome (53). This interaction is probably mediated through a third partner because domain mapping of yeast Cox11p showed that the matrix domain of Cox11p is non-essential for its function (51).
Computer models suggest a direct interaction between Cox11p and Cox1p (93), an interaction that would serve not only for copper transfer to the CuB site but also to perform a second function by protecting an early Cox1p-hemeA maturation intermediate against oxidative damage (93).
COPPER TRAFFICKING IN THE INTERMEBRANE SPACE
How copper reaches Cox17p in the intermembrane space is still an open question. As mentioned above, the intermembrane space houses several other proteins, containing the twin CX9C motif present in Cox17p, which are required for COX assembly. These proteins include Cox19p and Cox23p. The recombinant form of Cox19p was reported to bind copper (70), but the COX assembly defect of a cox19 null mutant strain could not be rescued by copper supplementation to the media (47). Mutants of cox23 also fail to assemble COX but their respiratory deficient phenotype is complemented by exogenous copper supplementation, albeit only with concomitant overexpression of COX17 (48). Cox23p does not physically interact with Cox17p in a stable complex. Recent data showed that Cox23p is also required for mitochondrial copper homeostasis by functioning in a common pathway with Cox17p acting downstream of Cox23p (48). In this same line, Cox19p could participate in a different portion of the copper distribution pathway (48). At least two other uncharacterized small proteins residing in the intermembrane space also share some similarities with Cox17p and Cox19p. These proteins are Pet191p (71) and the product of the gene encoded in ORF YKL137W termed Cmc1p (CX9C containing Mitochondrial protein required for full expression of COX) by our group. Both Pet191p (94) and Cmc1p (Horn and Barrientos, submitted for publication) are located in the intermembrane space but bound to the inner membrane. Like Cox17p and Cox19p, Pet191p and Cmc1p contain the CX9C signature motif and their presence is required for full assembly of COX. Their copper binding ability is unknown at present, but it is conceivable that the CX9C domains could confer this property to Pet191p and Cmc1p as well as to Cox23p.
An important remaining question in the field concerns the role of this relatively large number of different CX9C-domain containing proteins. Their functions are certainly important in COX biogenesis and mitochondrial metabolism as suggested by the existence of putative homologues from yeast to human (2). In a hypothetical and simplistic way, depicted in Fig. 1B, we could envision copper transfer from the mitochondrial matrix pool across the inner membrane by a still uncharacterized transporter to one or both of the membrane bound small proteins. In a daisy chain transfer mechanism, copper would be subsequently transferred to the soluble Cox19p, Cox23p and ultimately to Cox17p for final delivery to Cox11p and Sco1p. Such a mechanism could appear slower and less efficient than a direct metallation of Cox17p from the copper pool. It could be useful, however, to facilitate and regulate the distribution of copper from the same matrix pool towards the two copper enzymes located in the intermembrane space, COX and Sod1p. Currently it is unknown how copper trafficking towards these enzymes is regulated. It is equally important to note that such a daisy chain transfer mechanism could be useful to accumulate a small bio-available copper pool in the intermembrane space ready to be directed towards Cox17p in a regulated fashion.
We speculate that at least two models could explain copper transfer through this pathway. A redox changing-based copper relay system for copper delivery from Cmc1p to the soluble copper chaperones Cox19p, Cox23p and Cox17p and subsequently to the COX copper site-specific Sco1p and Cox11p could exist. In support of this possibility, it has been shown Cox17p and Sco1p change redox states depending on copper binding (64, 66, 83, 95, 96). In the case of Cox17p, the yeast and mammalian homologues share six conserved cysteine residues which are involved in complex redox reactions as well as in metal binding and transfer (95). Cox17p exists in three oxidative states, each characterized by distinct metal-binding properties. Studies of mammalian COX17 have identified fully reduced COX17(0S-S) binding co-operatively to four Cu(I) atoms; COX17(2S-S), with two disulfide bridges, binding to one Cu(I) atom; and COX17(3S-S), with three disulfide bridges, which does not bind any metal ions (64). In the intermembrane space the protein is probably present mostly in the COX17(2S-S) form (66), a conformer that enables retention of the protein in the intermembrane space and is competent to transfer copper to SCO1 (97). Similar oxidative mechanisms of metal release by oxidation of cysteine residues is known to function in the metal release from zinc–thiolate clusters of metallothionein (98). Alternatively, or as a concurrent mechanism, one could speculate that a non-oxidative mechanism of copper transfer occurs, facilitated by the kinetic labiality of metal–thiolate clusters. In the case that Cox19p, Cox23p and Cox17p had different, perhaps sequentially higher, copper–binding affinities, the resulting scenario could create a better regulated mechanism or pathway for copper delivery to COX. In this line, it is known that Cox17p has a high affinity for copper with a Kd of 13fM (64) while Cox19p has a lower affinity for copper as indicated by the inconsistent measurement of copper from purified Cox19p fractions (70).
BIOGENESIS OF COX COPPER CHAPERONES
Proteins are imported into mitochondria through several mechanisms extensively reviewed in Neupert and Herrmann (99). The CX9C twin motifs present in Cox17p, Cox19p, Cox23p, Cmc1p and Pet191p are critical for the import of these proteins into the mitochondrial IMS through the recently described Mia40p pathway (73). After passage through the mitochondrial outer membrane TOM channel, these proteins are covalently trapped by Mia40p via disulfide bridges. Mia40p also contains cysteine residues which are oxidized by the sulfhydryl oxidase Erv1p, functioning as a disulfide relay system that catalyzes the import of proteins into the IMS by an oxidative folding mechanism (73). Once folded the protein is retained in the intermembrane space (76). The redox state of Cox17p, Cox19p and Cox23p is unknown but reduction of the cysteines could be required for metal binding as it has been recently proposed (64, 66) suggesting the existence of a mechanism of cysteine reduction in the IMS after the oxidative folding.
Concerning the disulfide system, it was initially unclear how Erv1p itself is oxidized to became competent for new rounds of Mia40p oxidation. Interestingly, it has been recently reported that a connection exists between the disulfide relay import system and the mitochondrial respiratory chain consisting of electron transfer from Erv1p to molecular oxygen via an interaction with cytochrome c (75, 80). Erv1p also utilizes molecular oxygen as an electron acceptor to generate hydrogen peroxide which is subsequently reduced to water by cytochrome c peroxidase (Ccp1). Oxidized Ccp1p is in turn reduced by the Erv1p-reduced cytochrome c (80). Cytochrome c efficiently oxidizes Mia40p in oxygen-limiting conditions (75). Although oxidized cytochrome c facilitates Mia40p oxidation, it was found to be non-essential in normoxic conditions suggesting the existence of additional electron acceptors for oxidizing reduced Erv1p (75).
CONCLUDING REMARKS AND OPEN QUESTIONS
The copper metallation of COX is an intricate process that requires the action of several metallochaperone complexes. The list of mitochondrial proteins that affect mitochondrial copper homeostasis and COX assembly is steadily growing. The continued search for protein candidates involved in these processes should allow for the identification of key proteins in the mitochondrial copper trafficking process, including putative copper permease/s located in the outer membrane, and a copper transporter located in the inner membrane to facilitate copper transport from the matrix pool to the intermembrane space. What is/are the role/s of the CX9C containing proteins in COX assembly, how is the traffic of copper towards COX and Sod1p regulated, and how mitochondrial copper metabolism affects cellular copper homeostasis are some of the remaining questions to be answered.
Understanding these and other fundamental aspects of mitochondrial copper metabolism and COX assembly will aid in our understanding of the intricate process of building a polypeptidic enzyme crucial for aerobic production of energy. The biochemical and biological answers obtained will also contribute to a better understanding of human mitochondrial diseases resulting from mutations in the genes and pathways involved in copper homeostasis.
ACKNOWLEDGEMENTS
We thank Dr. Flavia Fontanesi, Dr. Karine Gouget and Erin Jenewein for critically reading the manuscript. Our research is supported by National Institutes of Health Research Grant GM071775A (to A.B.) and a Research Grant from the Muscular Dystrophy Association (to A.B.).
REFERENCES
- 1.Pena MM, Lee J, Thiele DJ. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 1999;129:1251–60. doi: 10.1093/jn/129.7.1251. [DOI] [PubMed] [Google Scholar]
- 2.Solans A, Zambrano A, Barrientos A. Cytochrome c oxidase deficiency: from yeast to human. Preclinica. 2004;2:336–348. [Google Scholar]
- 3.Pecina P, Houstkova H, Hansikova H, Zeman J, Houstek J. Genetic defects of cytochrome c oxidase assembly. Physiol. Res. 2004;53:S213–23. [PubMed] [Google Scholar]
- 4.Shoubridge EA. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 2001;106:46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- 5.Valnot I, Osmond S, Gigarel N, Mehaye B, Amiel J, Cormier-Daire V, Munnich A, Bonnefont JP, Rustin P, Rotig A. Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet. 2000;67:1104–9. doi: 10.1016/s0002-9297(07)62940-1. Epub 2000 Sep 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papadopoulou LC, Sue CM, Davidson MM, Tanji K, Nishino I, Sadlock JE, Krishna S, Walker W, Selby J, Glerum DM, et al. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999;23:333–7. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 7.Jaksch M, Ogilvie I, Yao J, Kortenhaus G, Bresser HG, Gerbitz KD, Shoubridge EA. Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:795–801. doi: 10.1093/hmg/9.5.795. [DOI] [PubMed] [Google Scholar]
- 8.Sue CM, Karadimas C, Checcarelli N, Tanji K, Papadopoulou LC, Pallotti F, Guo FL, Shanske S, Hirano M, De Vivo DC, et al. Differential features of patients with mutations in two COX assembly genes, SURF-1 and SCO2. Ann. Neurol. 2000;47:589–95. [PubMed] [Google Scholar]
- 9.Salviati L, Sacconi S, Rasalan MM, Kronn DF, Braun A, Canoll P, Davidson M, Shanske S, Bonilla E, Hays AP, et al. Cytochrome c oxidase deficiency due to a novel SCO2 mutation mimics Werdnig-Hoffmann disease. Arch. Neurol. 2002;59:862–5. doi: 10.1001/archneur.59.5.862. [DOI] [PubMed] [Google Scholar]
- 10.Prohaska JR, Gybina AA. Intracellular copper transport in mammals. J. Nutr. 2004;134:1003–6. doi: 10.1093/jn/134.5.1003. [DOI] [PubMed] [Google Scholar]
- 11.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei MJ, Libeu CP, Mizushima T, et al. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–9. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 12.Saraste M. Structural features of cytochrome oxidase. Q. Rev. Biophys. 1990;23:331–66. doi: 10.1017/s0033583500005588. [DOI] [PubMed] [Google Scholar]
- 13.Hill BC. Modeling the sequence of electron transfer reactions in the single turnover of reduced, mammalian cytochrome c oxidase with oxygen. J. Biol. Chem. 1994;269:2419–25. [PubMed] [Google Scholar]
- 14.Brunori M, Giuffre A, Malatesta F, Sarti P. Investigating the mechanism of electron transfer to the binuclear center in Cu-heme oxidases. J. Bioenerg. Biomembr. 1998;30:41–5. doi: 10.1023/a:1020503410377. [DOI] [PubMed] [Google Scholar]
- 15.Brunori M, Giuffre A, Sarti P. Cytochrome c oxidase, ligands and electrons. J. Inorg. Biochem. 2005;99:324–36. doi: 10.1016/j.jinorgbio.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Babcock GT, Wikstrom M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992;356:301–9. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 17.Saraste M. Structure and evolution of cytochrome oxidase. Antonie Van Leeuwenhoek. 1994;65:285–7. doi: 10.1007/BF00872214. [DOI] [PubMed] [Google Scholar]
- 18.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–44. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 19.Ostermeier C, Harrenga A, Ermler U, Michel H. Structure at 2.7 A resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10547–53. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coyne HJ, 3rd, Ciofi-Baffoni S, Banci L, Bertini I, Zhang L, George GN, Winge DR. The characterization and role of zinc binding in yeast Cox4. J. Biol. Chem. 2007;282:8926–34. doi: 10.1074/jbc.M610303200. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt B, McCracken J, Ferguson-Miller S. A discrete water exit pathway in the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15539–42. doi: 10.1073/pnas.2633243100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiser L, Di Valentin M, Hamer AG, Hosler JP. Cox11p is required for stable formation of the Cu(B) and magnesium centers of cytochrome c oxidase. J. Biol. Chem. 2000;275:619–23. doi: 10.1074/jbc.275.1.619. [DOI] [PubMed] [Google Scholar]
- 23.Barrientos A, Barros MH, Valnot I, Rotig A, Rustin P, Tzagoloff A. Cytochrome oxidase in health and disease. Gene. 2002;286:53–63. doi: 10.1016/s0378-1119(01)00803-4. [DOI] [PubMed] [Google Scholar]
- 24.Stiburek L, Hansikova H, Tesarova M, Cerna L, Zeman J. Biogenesis of eukaryotic cytochrome c oxidase. Physiol. Res. 2006;55:S27–41. doi: 10.33549/physiolres.930000.55.S2.27. [DOI] [PubMed] [Google Scholar]
- 25.Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta. 2006;1763:759–72. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Bertini I, Cavallaro G. Metals in the “omics” world: copper homeostasis and cytochrome c oxidase assembly in a new light. J. Biol. Inorg. Chem. 2008;13:3–14. doi: 10.1007/s00775-007-0316-9. [DOI] [PubMed] [Google Scholar]
- 27.Huffman DL, O’Halloran TV. Function, structure, and mechanism of intracellular copper trafficking proteins. Annu. Rev. Biochem. 2001;70:677–701. doi: 10.1146/annurev.biochem.70.1.677. [DOI] [PubMed] [Google Scholar]
- 28.Dancis A, Haile D, Yuan DS, Klausner RD. The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 1994;269:25660–7. [PubMed] [Google Scholar]
- 29.Pena MM, Puig S, Thiele DJ. Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 2000;275:33244–51. doi: 10.1074/jbc.M005392200. [DOI] [PubMed] [Google Scholar]
- 30.Hassett R, Dix DR, Eide DJ, Kosman DJ. The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 2000;351:477–84. [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen A, Nelson H, Nelson N. The family of SMF metal ion transporters in yeast cells. J. Biol. Chem. 2000;275:33388–94. doi: 10.1074/jbc.M004611200. [DOI] [PubMed] [Google Scholar]
- 32.Georgatsou E, Mavrogiannis LA, Fragiadakis GS, Alexandraki D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 1997;272:13786–92. doi: 10.1074/jbc.272.21.13786. [DOI] [PubMed] [Google Scholar]
- 33.Pufahl RA, Singer CP, Peariso KL, Lin SJ, Schmidt PJ, Fahrni CJ, Culotta VC, Penner-Hahn JE, O’Halloran TV. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science. 1997;278:853–6. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- 34.Arnesano F, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Huffman DL, O’Halloran TV. Characterization of the binding interface between the copper chaperone Atx1 and the first cytosolic domain of Ccc2 ATPase. J. Biol. Chem. 2001;276:41365–76. doi: 10.1074/jbc.M104807200. [DOI] [PubMed] [Google Scholar]
- 35.Banci L, Bertini I, Cantini F, Felli IC, Gonnelli L, Hadjiliadis N, Pierattelli R, Rosato A, Voulgaris P. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat. Chem. Biol. 2006;2:367–8. doi: 10.1038/nchembio797. [DOI] [PubMed] [Google Scholar]
- 36.Davis-Kaplan SR, Askwith CC, Bengtzen AC, Radisky D, Kaplan J. Chloride is an allosteric effector of copper assembly for the yeast multicopper oxidase Fet3p: an unexpected role for intracellular chloride channels. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13641–5. doi: 10.1073/pnas.95.23.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan DS, Stearman R, Dancis A, Dunn T, Beeler T, Klausner RD. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc. Natl. Acad. Sci. U. S. A. 1995;92:2632–6. doi: 10.1073/pnas.92.7.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Culotta VC, Klomp LW, Strain J, Casareno RL, Krems B, Gitlin JD. The copper chaperone for superoxide dismutase. J. Biol. Chem. 1997;272:23469–72. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 39.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 2001;276:38084–9. doi: 10.1074/jbc.M105296200. Epub 2001 Aug 10. [DOI] [PubMed] [Google Scholar]
- 40.Lamb AL, Torres AS, O’Halloran TV, Rosenzweig AC. Heterodimer formation between superoxide dismutase and its copper chaperone. Biochemistry. 2000;39:14720–7. doi: 10.1021/bi002207a. [DOI] [PubMed] [Google Scholar]
- 41.Furukawa Y, Torres AS, O’Halloran TV. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 2004;23:2872–81. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll MC, Girouard JB, Ulloa JL, Subramaniam JR, Wong PC, Valentine JS, Culotta VC. Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5964–9. doi: 10.1073/pnas.0308298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glerum DM, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 1996;271:14504–9. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- 44.Beers J, Glerum DM, Tzagoloff A. Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 1997;272:33191–6. doi: 10.1074/jbc.272.52.33191. [DOI] [PubMed] [Google Scholar]
- 45.Heaton DN, George GN, Garrison G, Winge DR. The mitochondrial copper metallochaperone Cox17 exists as an oligomeric, polycopper complex. Biochemistry. 2001;40:743–51. doi: 10.1021/bi002315x. [DOI] [PubMed] [Google Scholar]
- 46.Cobine PA, Ojeda LD, Rigby KM, Winge DR. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem. 2004;279:14447–55. doi: 10.1074/jbc.M312693200. [DOI] [PubMed] [Google Scholar]
- 47.Nobrega MP, Bandeira SC, Beers J, Tzagoloff A. Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J. Biol. Chem. 2002;277:40206–11. doi: 10.1074/jbc.M207348200. [DOI] [PubMed] [Google Scholar]
- 48.Barros MH, Johnson A, Tzagoloff A. COX23, a homologue of COX17, is required for cytochrome oxidase assembly. J. Biol. Chem. 2004;279:31943–7. doi: 10.1074/jbc.M405014200. Epub 2004 May 15. [DOI] [PubMed] [Google Scholar]
- 49.Cobine PA, Pierrel F, Bestwick ML, Winge DR. Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J. Biol. Chem. 2006;281:36552–9. doi: 10.1074/jbc.M606839200. [DOI] [PubMed] [Google Scholar]
- 50.Carr HS, George GN, Winge DR. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 2002;277:31237–42. doi: 10.1074/jbc.M204854200. Epub 2002 Jun 12. [DOI] [PubMed] [Google Scholar]
- 51.Carr HS, Maxfield AB, Horng YC, Winge DR. Functional analysis of the domains in Cox11. J. Biol. Chem. 2005;280:22664–9. doi: 10.1074/jbc.M414077200. Epub 2005 Apr 19. [DOI] [PubMed] [Google Scholar]
- 52.Tzagoloff A, Capitanio N, Nobrega MP, Gatti D. Cytochrome oxidase assembly in yeast requires the product of COX11, a homolog of the P. denitrificans protein encoded by ORF3. EMBO J. 1990;9:2759–64. doi: 10.1002/j.1460-2075.1990.tb07463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khalimonchuk O, Ostermann K, Rodel G. Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the Cu(B) site formation of cytochrome c oxidase. Curr. Genet. 2005;47:223–33. doi: 10.1007/s00294-005-0569-1. Epub 2005 Mar 18. [DOI] [PubMed] [Google Scholar]
- 54.Banting GS, Glerum DM. Mutational analysis of the Saccharomyces cerevisiae cytochrome c oxidase assembly protein Cox11p. Eukaryot. Cell. 2006;5:568–78. doi: 10.1128/EC.5.3.568-578.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glerum DM, Shtanko A, Tzagoloff A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:20531–5. doi: 10.1074/jbc.271.34.20531. [DOI] [PubMed] [Google Scholar]
- 56.Abajian C, Rosenzweig AC. Crystal structure of yeast Sco1. J. Biol. Inorg. Chem. 2006;11:459–46. doi: 10.1007/s00775-006-0096-7. [DOI] [PubMed] [Google Scholar]
- 57.Beers J, Glerum DM, Tzagoloff A. Purification and characterization of yeast Sco1p, a mitochondrial copper protein. J. Biol. Chem. 2002;277:22185–90. doi: 10.1074/jbc.M202545200. Epub 2002 Apr 10. [DOI] [PubMed] [Google Scholar]
- 58.Chinenov YV. Cytochrome c oxidase assembly factors with a thioredoxin fold are conserved among prokaryotes and eukaryotes. J. Mol. Med. 2000;78:239–42. doi: 10.1007/s001090000110. [DOI] [PubMed] [Google Scholar]
- 59.Krummeck G, Rodel G. Yeast SCO1 protein is required for a post-translational step in the accumulation of mitochondrial cytochrome c oxidase subunits I and II. Curr. Genet. 1990;18:13–5. doi: 10.1007/BF00321109. [DOI] [PubMed] [Google Scholar]
- 60.Lode A, Kuschel M, Paret C, Rodel G. Mitochondrial copper metabolism in yeast: interaction between Sco1p and Cox2p. FEBS Lett. 2000;485:19–24. doi: 10.1016/s0014-5793(00)02176-1. [DOI] [PubMed] [Google Scholar]
- 61.Nittis T, George GN, Winge DR. Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 2001;276:42520–6. doi: 10.1074/jbc.M107077200. Epub 2001 Sep 6. [DOI] [PubMed] [Google Scholar]
- 62.Rentzsch A, Krummeck-Weiss G, Hofer A, Bartuschka A, Ostermann K, Rodel G. Mitochondrial copper metabolism in yeast: mutational analysis of Sco1p involved in the biogenesis of cytochrome c oxidase. Curr. Genet. 1999;35:103–8. doi: 10.1007/s002940050438. [DOI] [PubMed] [Google Scholar]
- 63.Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J. Biol. Chem. 2004;279:35334–40. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- 64.Palumaa P, Kangur L, Voronova A, Sillard R. Metal-binding mechanism of Cox17, a copper chaperone for cytochrome c oxidase. Biochem J. 2004;382:307–14. doi: 10.1042/BJ20040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Punter FA, Glerum DM. Mutagenesis reveals a specific role for Cox17p in copper transport to cytochrome oxidase. J. Biol. Chem. 2003;278:30875–80. doi: 10.1074/jbc.M302358200. Epub 2003 Jun 4. [DOI] [PubMed] [Google Scholar]
- 66.Voronova A, Meyer-Klaucke W, Meyer T, Rompel A, Krebs B, Kazantseva J, Sillard R, Palumaa P. Oxidative switches in functioning of mammalian copper chaperone Cox17. Biochem. J. 2007;408:139–48. doi: 10.1042/BJ20070804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heaton D, Nittis T, Srinivasan C, Winge DR. Mutational analysis of the mitochondrial copper metallochaperone Cox17. J. Biol. Chem. 2000;275:37582–7. doi: 10.1074/jbc.M006639200. [DOI] [PubMed] [Google Scholar]
- 68.Abajian C, Yatsunyk LA, Ramirez BE, Rosenzweig AC. Yeast cox17 solution structure and Copper(I) binding. J. Biol. Chem. 2004;279:53584–92. doi: 10.1074/jbc.M408099200. Epub 2004 Oct 1. [DOI] [PubMed] [Google Scholar]
- 69.Amaravadi R, Glerum DM, Tzagoloff A. Isolation of a cDNA encoding the human homolog of COX17, a yeast gene essential for mitochondrial copper recruitment. Hum. Genet. 1997;99:329–33. doi: 10.1007/s004390050367. [DOI] [PubMed] [Google Scholar]
- 70.Rigby K, Zhang L, Cobine PA, George GN, Winge DR. Characterization of the cytochrome c oxidase assembly factor Cox19 of Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:10233–42. doi: 10.1074/jbc.M610082200. [DOI] [PubMed] [Google Scholar]
- 71.McEwen JE, Hong KH, Park S, Preciado GT. Sequence and chromosomal localization of two PET genes required for cytochrome c oxidase assembly in Saccharomyces cerevisiae. Curr. Genet. 1993;23:9–14. doi: 10.1007/BF00336742. [DOI] [PubMed] [Google Scholar]
- 72.Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuan Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–46. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–69. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 74.Gabriel K, Milenkovic D, Chacinska A, Muller J, Guiard B, Pfanner N, Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J. Mol. Biol. 2007;365:612–20. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 75.Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 2007;179:389–95. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muller JM, Milenkovic D, Guiard B, Pfanner N, Chacinska A. Precursor Oxidation by Mia40 and Erv1 Promotes Vectorial Transport of Proteins into the Mitochondrial Intermembrane Space. Mol. Biol. Cell. 2007 Oct 31; doi: 10.1091/mbc.E07-08-0814. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rissler M, Wiedemann N, Pfannschmidt S, Gabriel K, Guiard B, Pfanner N, Chacinska A. The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J. Mol. Biol. 2005;353:485–92. doi: 10.1016/j.jmb.2005.08.051. [DOI] [PubMed] [Google Scholar]
- 78.Terziyska N, Lutz T, Kozany C, Mokranjac D, Mesecke N, Neupert W, Herrmann JM, Hell K. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 2005;579:179–84. doi: 10.1016/j.febslet.2004.11.072. [DOI] [PubMed] [Google Scholar]
- 79.Allen S, Balabanidou V, Sideris DP, Lisowsky T, Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 2005;353:937–44. doi: 10.1016/j.jmb.2005.08.049. Epub 2005 Sep 15. [DOI] [PubMed] [Google Scholar]
- 80.Dabir DV, Leverich EP, Kim SK, Tsai FD, Hirasawa M, Knaff DB, Koehler CM. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–11. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee J, Hofhaus G, Lisowsky T. Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett. 2000;477:62–6. doi: 10.1016/s0014-5793(00)01767-1. [DOI] [PubMed] [Google Scholar]
- 82.Speno H, Taheri MR, Sieburth D, Martin CT. Identification of essential amino acids within the proposed CuA binding site in subunit II of Cytochrome c oxidase. J. Biol. Chem. 1995;270:25363–9. doi: 10.1074/jbc.270.43.25363. [DOI] [PubMed] [Google Scholar]
- 83.Banci L, Bertini I, Calderone V, Ciofi-Baffoni S, Mangani S, Martinelli M, Palumaa P, Wang S. A hint for the function of human Sco1 from different structures. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8595–600. doi: 10.1073/pnas.0601375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horng YC, Leary SC, Cobine PA, Young FB, George GN, Shoubridge EA, Winge DR. Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 2005;280:34113–22. doi: 10.1074/jbc.M506801200. [DOI] [PubMed] [Google Scholar]
- 85.Balatri E, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Solution structure of Sco1: a thioredoxin-like protein Involved in cytochrome c oxidase assembly. Structure. 2003;11:1431–43. doi: 10.1016/j.str.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 86.Ye Q, Imriskova-Sosova I, Hill BC, Jia Z. Identification of a disulfide switch in BsSco, a member of the Sco family of cytochrome c oxidase assembly proteins. Biochemistry. 2005;44:2934–42. doi: 10.1021/bi0480537. [DOI] [PubMed] [Google Scholar]
- 87.Smits PH, De Haan M, Maat C, Grivell LA. The complete sequence of a 33 kb fragment on the right arm of chromosome II from Saccharomyces cerevisiae reveals 16 open reading frames, including ten new open reading frames, five previously identified genes and a homologue of the SCO1 gene. Yeast. 1994;10:S75–80. doi: 10.1002/yea.320100010. [DOI] [PubMed] [Google Scholar]
- 88.Petruzzella V, Tiranti V, Fernandez P, Ianna P, Carrozzo R, Zeviani M. Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics. 1998;54:494–504. doi: 10.1006/geno.1998.5580. [DOI] [PubMed] [Google Scholar]
- 89.Paret C, Ostermann K, Krause-Buchholz U, Rentzsch A, Rodel G. Human members of the SCO1 gene family: complementation analysis in yeast and intracellular localization. FEBS Lett. 1999;447:65–70. doi: 10.1016/s0014-5793(99)00266-5. [DOI] [PubMed] [Google Scholar]
- 90.Leary SC, Kaufman BA, Pellecchia G, Guercin GH, Mattman A, Jaksch M, Shoubridge EA. Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 2004;13:1839–48. doi: 10.1093/hmg/ddh197. Epub 2004 Jun 30. [DOI] [PubMed] [Google Scholar]
- 91.Leary SC, Cobine PA, Kaufman BA, Guercin GH, Mattman A, Palaty J, Lockitch G, Winge DR, Rustin P, Horvath R, et al. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. 2007;5:9–20. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 92.Briere JJ, Tzagoloff A. The scoop on Sco. Mol. Cell. 2007;25:176–8. doi: 10.1016/j.molcel.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 93.Khalimonchuk O, Bird A, Winge DR. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- 94.Khalimonchuk O, Winge DR. Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamcr.2007.10.016. doi:10.1016/j.bbamcr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arnesano F, Balatri E, Banci L, Bertini I, Winge DR. Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding. Structure. 2005;13:713–22. doi: 10.1016/j.str.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 96.Williams JC, Sue C, Banting GS, Yang H, Glerum DM, Hendrickson WA, Schon EA. Crystal structure of human SCO1: implications for redox signaling by a mitochondrial cytochrome c oxidase “assembly” protein. J. Biol. Chem. 2005;280:15202–11. doi: 10.1074/jbc.M410705200. Epub 2005 Jan 19. [DOI] [PubMed] [Google Scholar]
- 97.Banci L, Bertini I, Ciofi-Baffoni S, Leontari I, Martinelli M, Palumaa P, Sillard R, Wang S. Human Sco1 functional studies and pathological implications of the P174L mutant. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15–20. doi: 10.1073/pnas.0606189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maret W. Cellular zinc and redox states converge in the metallothionein/thionein pair. J. Nutr. 2003;133:1460S–2S. doi: 10.1093/jn/133.5.1460S. [DOI] [PubMed] [Google Scholar]
- 99.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–49. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]