Abstract
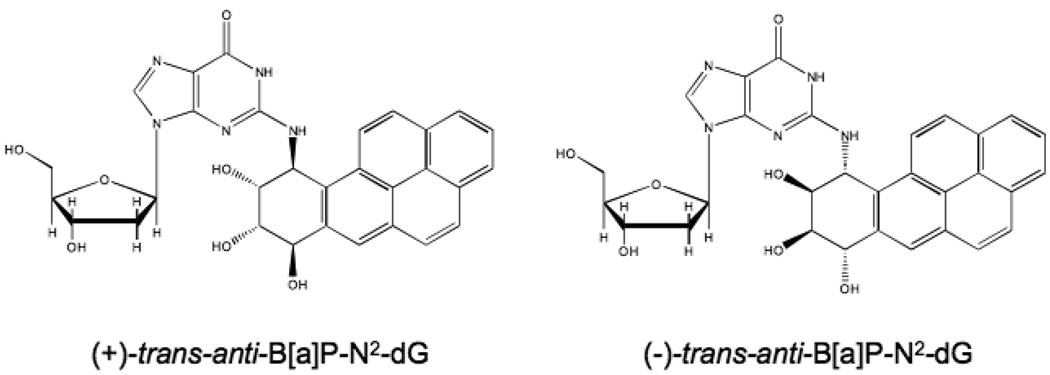
Benzo[a]pyrene (B[a]P) is a potent environmental carcinogen that is metabolized into diolepoxides that react with exocyclic amines in DNA. These DNA adducts have been shown to block DNA replication by high-fidelity polymerases and induce both base substitution and frame-shift mutations. To better understand the molecular mechanism of B[a]P-induced mutagenesis, a fluorescence resonance energy transfer (FRET) method was developed in which the (+) or (−)-trans-anti-B[a]P-N2-dG adducts, positioned in the active site of DNA polymerase I (Klenow fragment), serve as donor fluorophores to an acceptor molecule positioned on the DNA primer strand. FRET was measured for a primer that ended one nucleotide before the adduct position and one that ended across from the adduct and used to estimate the distances between the two fluorophores. These estimates are consistent with prior studies that suggest the adducts are positioned in the minor groove. A comparison of the FRET for the (+) and (−)-trans B [a]P adducts in the Klenow active site suggested that the (+)-trans adduct is positioned about 2 Å further from the acceptor, consistent with the structural differences observed in duplex DNA where it has been shown that the (+)-trans adduct is oriented towards the 5’-end of the template strand while the (−)-trans adduct lies towards the 3’end. Surprisingly, the adduct position did not change significantly when the primer was one nucleotide longer. The addition of either a correct (dCTP) or incorrect nucleotides showed only minor differences in FRET suggesting that the adduct did not undergo a large change in the position within the polymerase active site, as expected if the adduct inhibited the polymerase conformational change.
Benzo[a]pyrene (B[a]P1), one of the best studied chemical carcinogens, is known to form adducts to DNA following metabolic activation to highly reactive diolepoxide derivatives (1). These covalent DNA adducts have been shown to be strong blocks to replicative polymerases (2, 3) and presumably require bypass polymerases to overcome this inhibition (4). (±)-anti-B[a]P-diolepoxide, the most biologically relevant active metabolite of B[a]P (5), reacts predominantly with the N2 position of guanine to form four dieosteromeric adducts (6, 7) (the two trans isomers are shown in Fig. 1). The structures of these adducts in duplex DNA and at a primer-template junction have been studied using NMR and by other spectroscopic and computational methods (reviewed in (8)). These studies showed that in fully duplex DNA the trans isomers were found to reside predominantly in the minor groove of the helix allowing the retention of a properly aligned GC base pair. However, the orientation of these two adducts was quite different, with the (+)-trans adduct pointing towards the 5’-end of strand to which it is attached while the (−)-trans adduct is oriented towards the 3’-end (8). The structure of the (+)-trans adduct at the primer-template junction shows the B[a]P moiety no longer in the minor groove but instead stacked with the terminal base pair and rotated into a syn conformation (8).
Figure 1.
Structures of the (+)-trans-anti-B[a]P-N2-dG and the (−)-trans-anti-B[a]P-N2-dG adducts.
NMR structural studies on DNA containing a (+)-trans adduct at a primer-template junction have also shown that there is a substantial change in the position of the adduct when the primer is extended from a position one nucleotide before the adduct position to a position that terminates across from the adduct (9, 10). In the former case, the adduct is stacked with the last base-pair of the double-stranded segment of the primer-template, such that the modified guanine is displaced towards the major groove and adopts syn conformation. In contrast, when the primer is extended one nucleotide and contains a cytosine across from the modified guanine, the pyrenyl ring is positioned on the minor groove side of the primer-template pointing towards the 5’ end of the template strand similar to what is observed in fully duplex DNA.
A crystal structure of B. stearothermophilus polymerase I fragment (BF) has been reported where the polymerase is bound to a primer-template containing a (+)-trans-anti-B[a]P-N2-dG adduct in which a C in the primer is positioned across from the modified G (11). The adduct in this structure adopts a conformation that places the pyrene moiety into the minor groove but which causes substantial distortion of both strands of the DNA duplex. The normal interactions between the amino acid side chains in the active site and the terminal base pair are also disrupted, as well as the binding pocket for the incoming dNTP. A molecular dynamics simulation analysis has also been carried out using rat DNA polymerase β complexed with primer-template DNA containing the (+)-trans-anti-B[a]P-N2-dG adduct (12). The primer in this study was terminated before the adduct position and the pyrenyl ring was found to be stacked with the last base-pair of the double-stranded segment of the primer-template, resembling the above mentioned NMR structure except that the modified guanine retained the anti conformation. A second computational study suggested a different structure for the (+)-trans-B[a]P-N2-dG adduct positioned within the active site of the T7 DNA polymerase (13). In this study the primer was terminated before the adduct and the simulation was carried out in the presence of the most commonly incorporated nucleotide, dATP. The structure that was determined showed the modified G in a syn conformation and the B[a]P moiety positioned in the major groove, thus allowing the incoming dATP to be accommodated.
Prior studies with DNA polymerase I (Klenow fragment) (KF) have shown that the presence of the (+)-trans-anti-B[a]P-N2-dG adduct decreases the binding affinity to the primer template (14, 15). Moreover, the stability of the complex was further reduced in the presence of dNTPs suggesting that the conformational change to a closed ternary complex was inhibited by the presence of a B[a]P adduct (14). In addition, unlike what is observed on unmodified templates, the tryptic digestion pattern of KF bound to (+)-trans B[a]P-modified primer-templates was independent of the presence of any dNTP, further evidence that these adducts inhibited the formation of the closed form of the polymerase (14).
In the present study, we have developed an intermolecular fluorescence resonance energy transfer (FRET) system where the intrinsic fluorescence of the pyrene moiety of B[a]P is employed as the donor fluorophore and 7-dimethylaminocoumarin-4-acetic acid (DMACA) used as the acceptor fluorophore. Fluorescence resonance energy transfer is a technique widely used for measuring distances between two positions on biological macromolecules on a scale of between 10 and 100 Å (16). The efficiency of energy transfer was determined when either the (+) or (−)-trans-anti-B[a]P-N2-dG adduct was present at the primer-template junction within the active site of KF. Quantum yields of both fluorophores were approximately equal to 0.04 and their excitation and emission spectra were virtually identical. Using a primer that ended one nucleotide before the modified guanine resulted in a more efficient energy transfer for the (−) trans adduct compared with the (+) isomer, suggesting that the (−) isomer was positioned closer to the acceptor fluorophore and thus pointing towards the 3’-end of the template strand. Extension of the primer by one nucleotide so that it ended opposite the modified guanine did not cause a large change in the FRET levels of the (+)-trans adduct, suggesting that no large movement in the adduct conformation had occurred. Finally, the addition of any dNTP did not result in significant changes the FRET efficiency for ether adduct. This is consistent with prior studies that showed that the B[a]P adducts inhibited the conformational change to a closed ternary complex (14).
MATERIALS AND METHODS
Materials
The Klenow fragment of E. coli DNA polymerase I (exonuclease deficient) was either purchased from USB (Cleveland, OH) or was over-expressed and purified from a strain provided by Dr. Catherine Joyce (Yale University) that carries a double mutation, D355A and E357A (17). T4 polynucleotide kinase and dNTP’s were purchased from USB (Cleveland, OH). Oligonucleotides were purchased from Operon (Huntsville, AL) or Midland Certified Inc. (Midland, TX). [γ-32P]ATP was from MP Biomedicals (Cleveland, OH). Racemic (±)-anti-BPDE was purchased from the National Cancer Institute Chemical Reference Standard Repository (Kansas City, MO). DMACA-SE was purchased from Invitrogen (Carlsbad, CA). Sub-micro quartz fluorescence cuvettes with a 15 µL capacity were purchased from Starna Cells (Atascadero, CA) Inc. All other chemicals and reagents were purchased from Fisher Scientific or VWR.
Synthesis and purification of DMACA-labeled and B[a]P-modified oligonucleotides
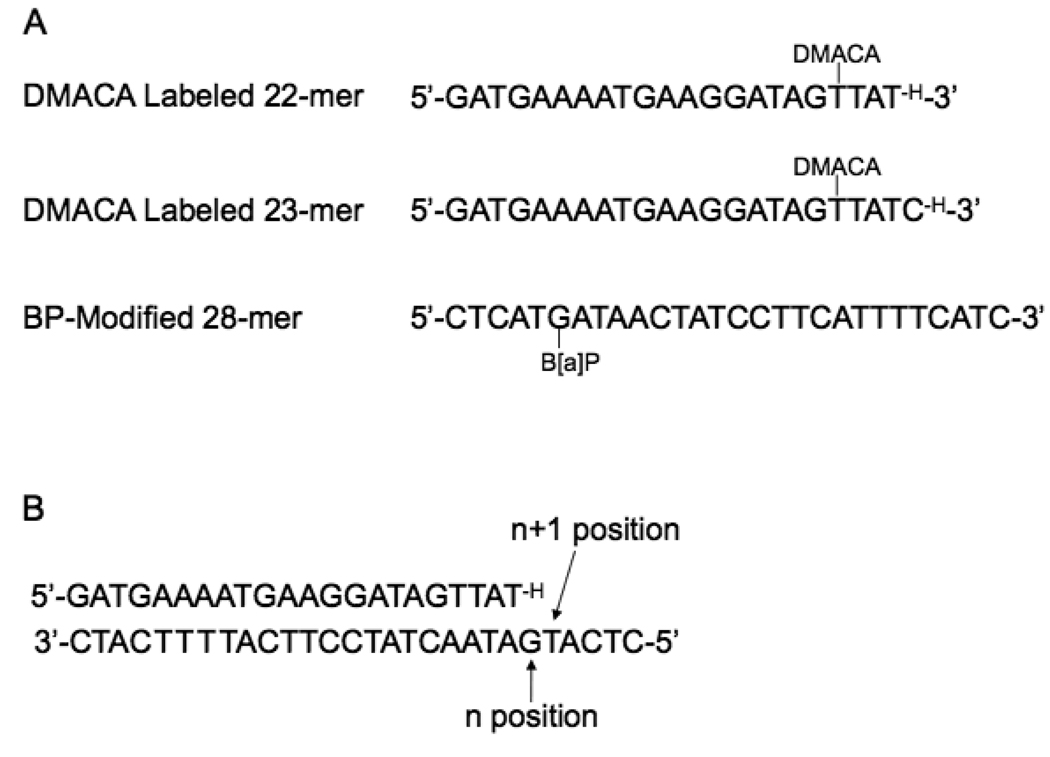
The sequences of the oligonucleotides used in the study are shown in Fig. 2. All oligonucleotides were purified by 20% denaturing polyacrylamide gel electrophoresis and desalted on centricon ultracel YM-3 microconcentrators (Amicon). The 22-mer and 23-mer chain terminated primers were labeled with DMACA at the thymidine four or five nucleotides, respectively, from the 3’ end using a C6 amine linker via succinylmidyl ester chemistry. Both the 22-mer and 23mer oligonucleotides (80 µL, 0.625 mM) were incubated in 20 fold excess of the NHS ester of DMACA in 20 µl of DMSO and 3 µL of diisopropylethylamine overnight at room temperature. Labeled oligonucleotides were separated from unlabeled by reverse-phase HPLC on a C18 column using a TEAA:acetonitrile gradient. The 28-mer oligonucleotides were modified by reaction with racemic (±)-anti-BPDE and the two major isomers were separated by reverse-phase HPLC and characterized as described in detail (18). After purification all samples were dried down completely, resuspended in ddH2O then dried to completion again. All samples were stored at −20 °C in Chelex-treated ddH2O.
Figure 2.
(A) Oligonucleotide sequences used in this study. The 22-mer and 23-mer were used as primers in the FRET analysis (DMACA denotes the position of the acceptor fluorophore) and the 28-mer containing the B[a]P adducts used as the template (B[a]P denotes the position of the donor adduct). (B) The complex of the 22-mer primer and the 28-mer template. The n position corresponds to the site across from the modified guanine. The n+1 position corresponds to the position one nucleotide past the modified guanine. The H at the end of the primer strand indicates that the primer was terminated with a dideoxy nucleotide.
Gel retardation assay
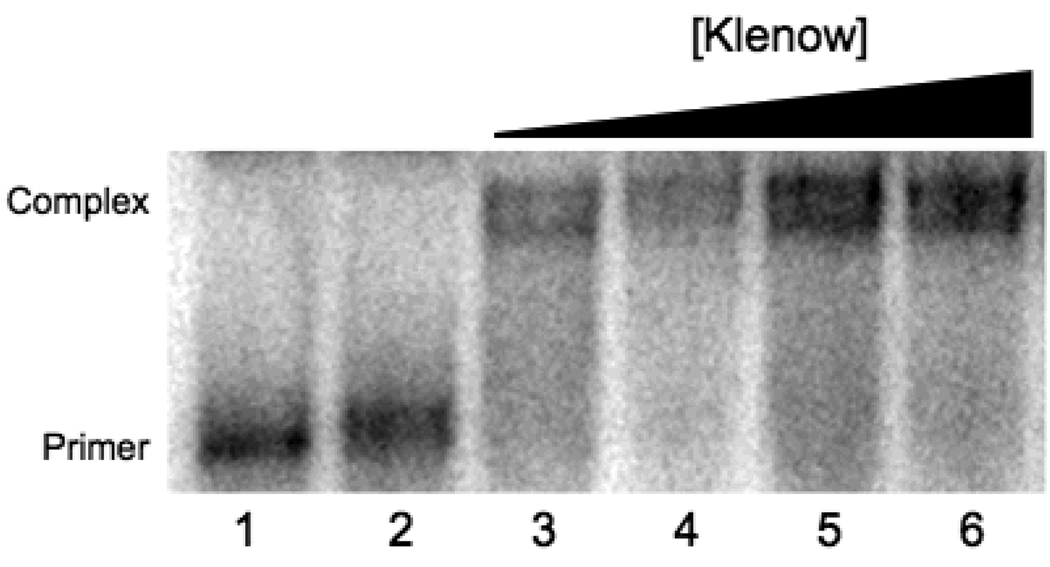
To determine the minimum amount of Klenow fragment necessary to bind virtually all of the primer-template, a gel retardation assay was performed similar to those described previously (19). The DMACA-linked 22-mer (Fig. 2A) was labeled with 32P using T4 polynucleotide kinase (USB, Cleveland, OH) as recommended by the supplier. This 22-mer was annealed with an equal molar amount of the B[a]P-linked template (Fig. 2) and the indicated amounts of Klenow fragment were incubated with this primer-template (50 nM) for 30 min at room temperature in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 0.05 mg/mL bovine serum albumin and 5% glycerol. The reaction mixtures were analyzed in a 5% non-denaturing polyacrylamide gel that was pre-equilibrated with 36 mM Tris borate buffer, pH 8.3 as described (19). The image was visualized using Molecular Dynamics PhosphorImager Storm and ImageQuant (Fig. 3).
Figure 3.
Klenow fragment binds efficiently to the modified primer-template. The DMACA-linked 22-mer primer shown in Figure 2A was labeled with 32P as described in “Materials and Methods”. This primer was annealed to the 28-mer containing the (−)-trans-anti-B[a]P-N2-dG adduct (Fig. 2) and 2.5 pmol of this primer-template was mixed with increasing concentrations of Klenow fragment in a final volume of 50 µL. The binding reactions were carried out as described in “Materials and Methods”. Lane 1, 22-mer primer; lane 2, annealed primer-template; lane 3, 1 µM KF; lane 4, 2 µM KF; lane 5, 3 µM KF; lane 6, 4 µM KF.
Quantum yield of B[a]P
The quantum yield was determined by relating the quantum yield of a quinine sulfate reference standard to that of B[a]P-labeled 28-mer/22-mer unlabeled primer duplex in the presence of a 20-fold excess of KF using the following equation: QB = QR [(IB/IR)(ODR/ODB)(nB2/nR2)] where QB and QR are the quantum yields, IB and IR are the intensities, ODB and ODR are the optical densities and nB and nR are the refractive indices of the solutions for the B[a]P-labeled 28-mer (B) and reference (R) fluorophores, respectively (16). The refractive indices determined for B and R were 1.3329 and 1.3340, respectively. For the (+)-trans adduct, the QD was 0.040 and for the (−)-trans adduct 0.041.
Overlap integral (J)
The J-overlap integral is the normalized spectral overlap of the donor emission spectrum and the excitation spectrum of the acceptor. The following equation was used to calculate J-overlap for the B[a]P-DMACA FRET pair used in this study: J(λ) = ∫ FD(λ) εA(λ)(λ)4dλ̣ where FD(λ) is the corrected fluorescence intensity of the donor in the wavelength range λ to λ+Δλ with the total intensity normalized to unity and εA is the extinction coefficient of the acceptor at λ in units of M−1 cm−1 (16). The J calculated for the B[a]P-DMACA FRET pair was 1.628×1014 M−1cm3(nm4).
Ensemble FRET assays
A FRET assay was developed similar to that described previously (20). Briefly, primer-template complexes (100–200 nM) were annealed in the presence of 50 mM Tris-HCl, pH 7.5, containing 10 mM MgCl2 and 1 mM dithiothreitol. Four separate annealing mixtures were necessary for each experiment, one containing both FRET donor (B[a]P) oligonucleotide and acceptor (DMACA) oligonucleotide, one duplex with the donor-labeled oligonucleotide, one duplex with the acceptor-labeled oligonucleotide, and one with the unmodified duplex, the last two needed for background subtraction (16). To each annealed primer-template excess KF was added to ensure complete binding to the primer template (final concentration, 4–8 µM, final volume 18 µL). Each sample was incubated for 30 min and then injected into a micro fluorescence cuvette and scanned on an Aviv 107F titrating spectrofluorometer. Each sample was excited at 350 nm (2 nm slit) using an excitation monochrometer and the emission spectrum was collected from 360–600 nm (4 nm slit) at 25 °C using an emission monochrometer. The spectra from the unlabeled duplex was subtracted from the spectra of the B[a]P-labeled duplex, the DMACA-labeled duplex, and the B[a]P-DMACA labeled duplex. Excitation using 310 nm, 320 nm, 330 nm, and 340 nm gave nearly identical emission spectra. The intensity of the donor-only fluorescence was then compared to that of the FRET complex and the difference used to calculate the efficiency of energy transfer. The distance between the FRET pair was determined using Förster’s equation; E = 1−(FDA/FD) = R06/(R06+r6) where E is the efficiency of energy transfer, FDA is the intensity of the donor in the presence of the acceptor, FD is the intensity of the donor in the absence of acceptor, and r is the actual distance between the two molecules (16). The Förster distance (R0) was determined as previously described (16).
Fluorescence Anisotropy
Anisotropies of both the (+) and (−)-trans-B[a]P 28-mer templates annealed to the unlabeled 22-mer primer were determined using a Cary Eclipse Fluorescence Spectrophotometer (Varian, Palo Alto, CA) as described by Lakowicz (16). Briefly, solutions comprised of primer-templates (625 nM) prepared using the unmodified 22-mer primer and the 28-mer template modified with either the (+) or (−)-trans isomers in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol and 12.5 µM KF were incubated at room temperature for 30 min prior to conducting the experiments. The solution was then excited using 350 nm light polarized along the z-axis using a slit width of 2.5 nm. Emission intensities were then collected both parallel (I‖) and perpendicular (I⊥) to the polarized excitation beam at 400 nm with a slit width of 10 nm. Measured intensities were averaged over 1 s. The anisotropies (r) of the solution was calculated using the following equation: r = (I‖ − I⊥)/(I‖ + 2I⊥).
RESULTS
FRET system
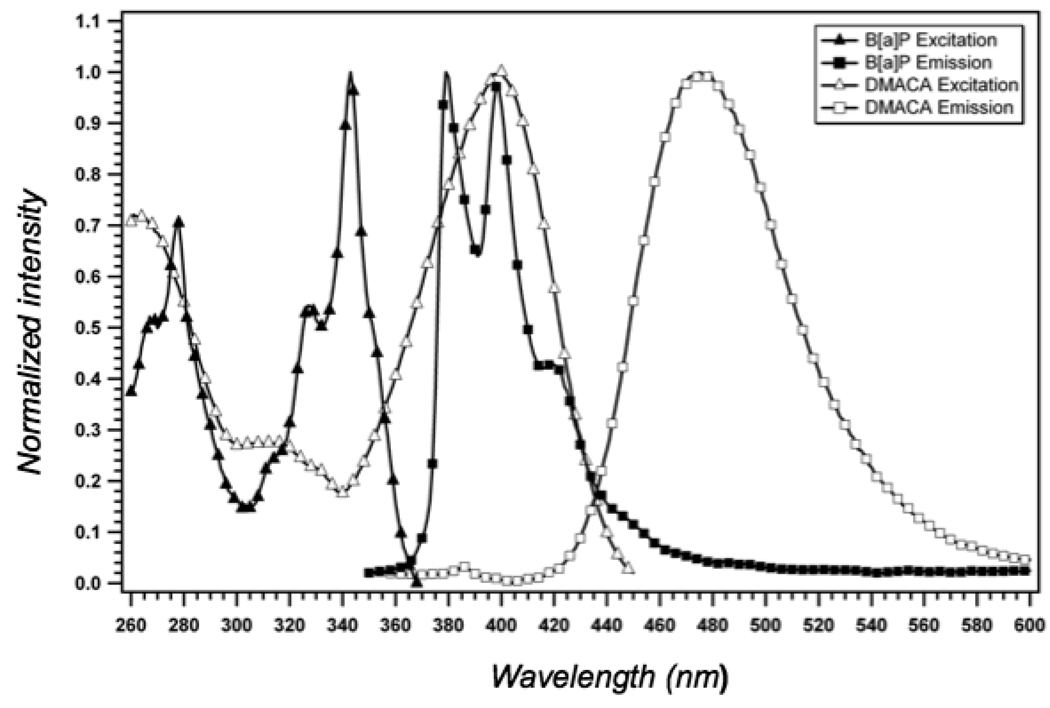
The development of a FRET system to measure the position of B[a]P adducts in the active site of a DNA polymerase required the placement of an acceptor fluorophore on the DNA primer-template whose excitation spectrum has good overlap (J-overlap integral) with the emission spectrum of B[a]P adducts in DNA. Shown in Fig. 4 are the absorption and emission spectra for the 22-mer DMACA-labeled primer annealed to the 28-mer template containing the (+)-trans B[a]P adduct (Fig. 1 and fig. 2) in the presence of KF. For these experiments sufficient polymerase was added to ensure that complex formation is the favored solution structure. Prior studies using primer-templates modified with B[a]P adducts indicated a KD of about 1 nM for the binding of KF to a B[a]P-modified primer-template (14). Knowing this value, we can calculate using Langmuir isotherm binding equation that at the concentrations of KF used for these studies (4 µM), the binary complex formation approaches 100%.
Figure 4.
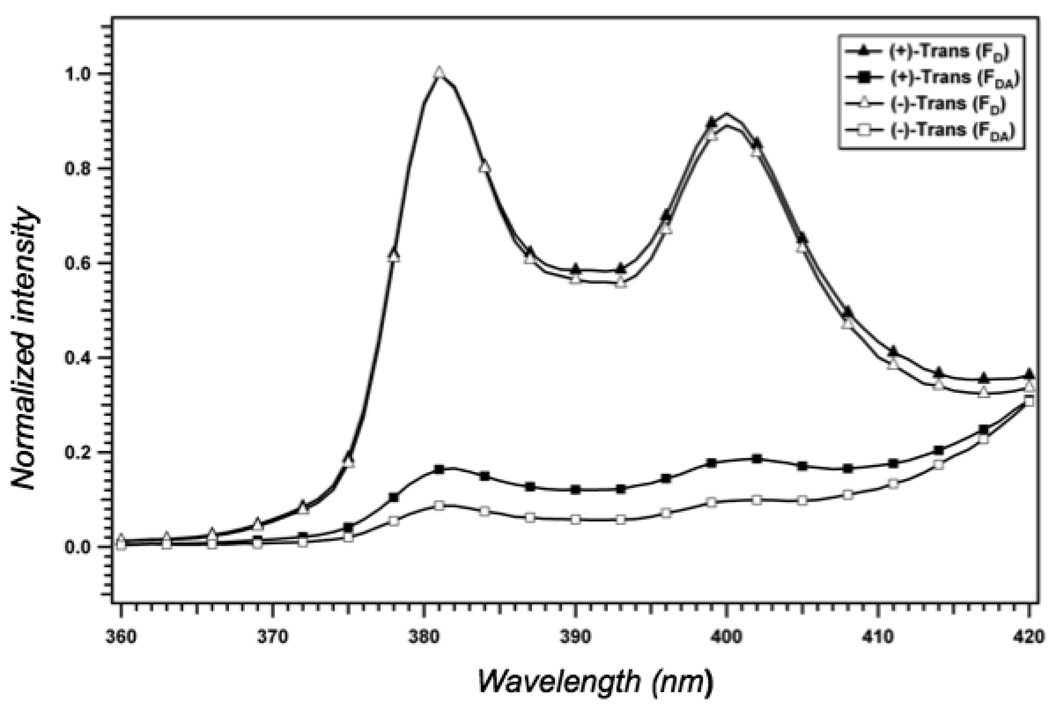
Normalized excitation and emission spectra for B[a]P (donor) and DMACA (acceptor). Each spectrum represents the excitation or emission spectrum of a primer-template (200 nM) in the presence of 4 µM KF. B[a]P excitation spectrum (▲); B[a]P emission spectrum (■); DMACA excitation spectrum (△); DMACA emission spectrum ( ).
).
Each spectrum represents the excitation or emission spectrum of a primer-template complex bound to the active site of KF. As is evident from this analysis, excitation of the B[a]P adduct at 350 nm produces a fluorescence emission maximum at 380 nm, which matches very closely to the excitation maximum for DMACA. The Förster distance for the B[a]P–DMACA fluorophore pair was calculated using the equation Ro = (8.79 × 10−5 J QD n−4 κ2)1/6 where J is the J-overlap integral, QD is the quantum yield of the donor in the absence of acceptor, n is the refractive index of solution and κ2 is the geometric factor relating the orientation of the two dipoles (assumed to be 4/3 when, as in this case, the orientation of one of the fluorophore’s dipole is fixed along the axis of the DNA and the other is random). Using the calculated J-overlap of 1.628×1014 M−1cm3(nm4), B[a]P quantum yield of 0.04, the Ro for this FRET pair was determined to be 25.0 Å. The scalar distance between the fluorophores was then calculated as described in the Materials and Methods section using the equation E = (1−(FDA/FD)) = 1/(1+R6/Ro6) (16). Typical spectra showing the magnitude of the fluorescence energy transfer is shown in Fig. 5
Figure 5.
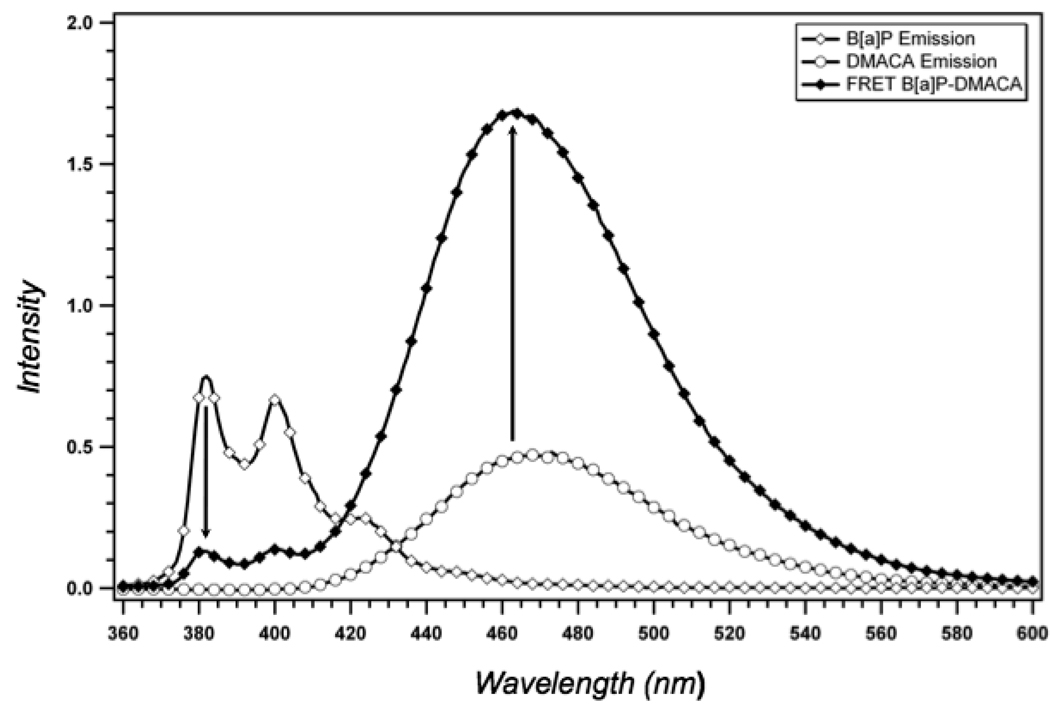
Spectra showing FRET between (+)-trans-anti-B[a]P-N2-dG and DMACA in the KF active site. Fluorescence emission from (+)-trans-anti-B[a]P-N2-dG is quenched when DMACA is present in the primer causing a concomitant increase in DMACA emission, indicative of resonance energy transfer. Emission spectrum of (+)-trans-anti B[a]P adduct in the pre-insertion position in the 28-mer template strand shown in Fig. 2A annealed to an unmodified 22-mer primer (□); emission spectrum of DMACA labeled 22-mer shown in Fig. 2A annealed to an unmodified 28-mer template (□); spectrum showing FRET between (+)-trans-anti-B[a]P-N2-dG and DMACA when both are present in the primer-template as shown in Figure 2 (□). The concentration of the primer-template and polymerase for each experiment was 200 nM and 4 µM, respectively.
FRET between (+) or (−)-trans-anti-B[a]P-N2-dG and DMACA in a primer-template bound to KF
To estimate and compare the distance between the (+) or (−)-trans B[a]P adducts and the acceptor fluorophore within the KF active site, a 22-mer primer containing a DMACA linked to the fourth nucleotide from the 3’-end was annealed to a 28-mer template containing a B[a]P adduct at the sixth position from the 5’end (Fig. 2A). This produced a primer-template construct in which the primer terminates one nucleotide before the adducted guanine (Fig. 2B). The efficiency of energy transfer from the B[a]P adduct to the DMACA acceptor was determined by measuring the intensity of the B[a]P (donor) emission at 380 nm in the absence of the acceptor FD and the intensity of the B[a]P emission at 380 nm in the presence of the acceptor FDA (Fig. 6). The (−)-trans adduct was more efficient at transferring energy to the acceptor as is evidenced by the greater reduction in FDA for the (−)-isomer compared with the (+) isomer. This strongly suggests that the (−)-trans adduct is closer to the DMACA acceptor when positioned in the KF active site, consistent with what would be expected if the (−)-trans adduct was pointing towards the 3’-end of the template strand and the (+)-trans adduct was pointing towards the 5’-end. These energy transfer differences correspond to an estimated distance of 16.9 Å for the (−)-trans adduct and 19.3 Å for the (+)-trans adduct (Table 1), resulting in a difference of approximately 2.4Å. These results are consistent with NMR structures of the (+) and (−)-trans adducts positioned in fully duplex DNA, which shows the same relative orientations of the adducts (8)
Figure 6.
Energy transfer is more efficient for the (−)-trans-anti-B[a]P-N2-dG adduct. The emission spectra was measured for the 28-mer template modified with either a (+) or (−)-transanti-B[a]P-N2-dG adduct (Fig. 2A) annealed to either an unmodified 22-mer or a DMACA-modified 22-mer (Fig. 2) in the presence of KF. Emission spectra were normalized to the peak excitation of the (+) or (−) adducts at 380 nm in the absence of the acceptor, DMACA. Emission spectra of the primer-template containing the (+)-trans adduct donor (▲); emission spectra of the primer-template containing the (−)-trans adduct donor (△); emission spectra of the primer-template containing the (+)-trans adduct donor and the DMACA acceptor (■); emission spectra of the primer-template containing the (−)-trans adduct donor and the DMACA acceptor ( ). The concentration of the primer-template and polymerase for each experiment was 200 nM and 4 µM, respectively.
). The concentration of the primer-template and polymerase for each experiment was 200 nM and 4 µM, respectively.
Table 1.
(−) or (+)-trans-anti-B[a]P-N2-dG Distances in the Presence of Klenow Fragment a
| Primer/Template | − | dCTP | dTTP | dATP | dGTP |
|---|---|---|---|---|---|
| Distance (Å) | |||||
| 22-mer/28-mer-(−)b | 16.9 ± 0.2 | 17.8 ± 0.2 | 17.8 ± 0.3 | 17.6 ± 0.4 | 18.8 ± 0.1 |
| 22-mer/28-mer-(+)c | 19.3 ± 0.4 | 18.9 ± 0.02 | 18.8 ± 0.2 | 19.0 ± 0.3 | 19.1 ± 0.1 |
| 23-mer/28-mer-(+) | 20.1 ± 0.3 | 19.6 ± 0.4 | 19.3 ± 0.2 | 19.0 ± 0.2 | 19.4 ± 0.03 |
Distances were determined using Förster resonance energy transfer equations as described in “Materials and Methods” using the primer-templates shown in Figure 2.
28-mer modified with(−)-trans-anti-B[a]P-N2-dG.
28-mer modified with (+)-trans-anti-B[a]P-N2-dG.
To ensure that the differences in FRET efficiencies observed for the (+) and (−)-trans adducts was not due to different environments in the polymerase active site, the anisotropy of each adduct was measured using an unlabeled 22-mer primer annealed to either the (+) or (−)-trans B[a]P modified 28-mer template bound to KF. The observed anisotropy for the two isomers was virtually identical (0.22 ± 0.03 for the (+)-trans isomer and 0.21 ± 0.04 for the (−)-trans isomer), indicating that the measured change in the FRET efficiencies observed for the two adducts is not due to differing active site environments.
FRET between (+)-trans-anti-B[a]P-N2-dG and DMACA using a primer that extends to a position across from the adduct
To determine if the relative position of the B[a]P adduct changed following nucleotide incorporation, a primer-template complex was constructed containing a (+)-trans B[a]P adduct and a 23-mer DMACA-linked primer providing a primer-template in which the primer is extended to a position across from the modified G (Fig. 2B). The distance between the (+)-trans adduct and the DMACA was calculated to be 20.1 Å, a value that appears to correspond to relatively little movement of the adduct compared with the position of the adduct observed using the 22-mer primer. This suggests that the position of the adduct is not significantly altered when the adducted base moves from the pre-insertion to the post-insertion site. However, this result does not completely exclude the less likely possibility that the distance remained the same even though a major conformational change had occurred.
The value calculated using the FRET intensities (20.1 Å) is in good agreement with a value we estimate (20.7 Å) based on a model of B-form DNA in solution represented by the following equation RDA = √((3.4 Δn + L)2 + d2 + a2 − 2da cos θ) (21) where RDA is the distance between the two dyes, Δn is the number of nucleotides between the dyes, L is the axial separation between the two dyes, d and a are the radial lengths of the donor and acceptor dyes (in this case we estimate d = 5 Å and a = 20 Å) and θ = Δn × 36° + θo. θ represents the angle between the two dyes and θo is the angle at Δn = 0. For dyes on the same strand θo = 0° and for dyes on opposite strands θo = 240°. For our calculation we assume that L = 0 based on the rigidity of B-form DNA.
Effect of the presence of dNTPs on FRET efficiencies
To determine whether or not a significant change in FRET efficiency occurs upon binding of an incoming nucleotide across from the adduct, FRET was measured using a dideoxy-terminated 22-mer primer that extended to the position one nucleotide before the B[a]P-modified guanine. These studies were also repeated with a dideoxy-terminated 23-mer primer that extended across from the modified guanine. With the exception of dGTP across from the (−)-trans adduct, none of the distances measured (Table 1) showed a large deviation from the distances determined in the absence of dNTP, consistent with prior studies that suggested that the (+)-trans B[a]P adduct inhibited the conformational change to a closed ternary complex (14), as well as the strong inhibition of this adduct on incorporation across from or past the adduct position by high-fidelity polymerases (11, 22). It is possible that the 1.9 Å change for the position of the (−)-trans adduct in the presence of dGTP is the result of a conformational change to a closed ternary complex.
DISCUSSION
Bulky adducts in DNA, such as the lesions caused by benzo[a]pyrene, are known to be strong blocks to DNA synthesis by high-fidelity polymerases (2, 3, 11). Nucleotide incorporation across from B[a]P adducts by such polymerases occurs very slowly, with purines being the preferred base incorporated (3, 23, 24), while extension past such structures is even more strongly inhibited (2, 11, 22). Thus, it is clear that the position of these adducts in the polymerase active site disrupts the normal base paring that must occur with the incoming dNTP. Crystal structure (11) and molecular dynamic studies (13) have provided some details regarding the position of the (+)-trans-anti-B[a]P-N2-dG adduct in the active sites of two A-family polymerases. While the crystal structure of BF shows the B[a]P moiety is located in the minor groove with the base in the anti conformation when in the post-insertion complex, the B[a]P-dG is disordered in the pre-insertion complex and thus its position cannot be defined (11). The molecular dynamic studies of the T7 DNA polymerase show that the B[a]P adduct resides in the major groove with the B[a]P-dG in a syn conformation when in the pre-insertion complex in the presence of dATP, the nucleotide that is best incorporated in this system (13). It is possible that the transition of the adducted base from the anti to syn conformation must precede nucleotide incorporation.
The FRET analysis we have used in this study gives further insight regarding the position of B[a]P adducts in the active site of a high-fidelity polymerase. We have used for these studies the Klenow fragment of DNA polymerase I and two structurally related B[a]P adducts, (+) and (−)-trans-anti-B[a]P-dG, both of which reside in the minor groove in duplex DNA but point in opposite directions (8). The analysis we have performed is novel because we have used the adduct as the FRET pair donor, allowing us to directly compare the position of each adduct in the polymerase active site. We were also able to make these measurements when the adducted base is in the templating position or when there is a C in the primer positioned across from the modified G. Finally, the use of dideoxy-terminated primers allowed us to compare the adduct positions in the presence of each of the dNTPs in an effort to detect a conformational change to a closed ternary complex.
The distance between the (+)-trans-anti-B[a]P-dG and the DMACA acceptor was determined to be 19.3 Å when the primer extended to the position before the modified guanine and 20.1 Å for the complex with a primer one nucleotide longer. These values are in good agreement with the estimated distances based on a model of B-form DNA previously used to calculate distances in duplex DNA (21). The fact that the adduct position does not change significantly when it moves from the templating position to the post-insertion site of the polymerase suggests that the adduct may reside in a similar conformation in both structures, with the G in an anti conformation and the pyrenyl ring positioned in the minor groove (based on the post-insertion BF crystal structure (11)). The distance determined for the (−)-trans-anti-B[a]P-dG was about 2.4 Å less than what was found for the (+) isomer. These values are consistent with the structures of these adducts in duplex DNA (8), where the (+) isomer points towards the 5’-end of the template strand (i.e. away from the DMACA) while the (−) isomer points towards the 3’-end (i.e. closer to the DMACA) and supports the hypothesis that adduct structures observed in duplex DNA in solution are often maintained within the polymerase active site (25).
One potential drawback to measuring distances using FRET is that the orientation of the transition dipoles of the two fluorophores (i. e. the orientation factor, κ2) must be estimated to determine absolute distances. In the present study our assumption of 4/3 for this value assumes that one fluorophore (DMACA) is able to rotate freely in solution while the other is fixed along the axis of the DNA in the minor groove. It is possible that this orientation factor is incorrect and that the orientation of the fluorophores is either more random or more static. However, without additional structural information, these approximations are the best that can be applied to this system. Moreover, the focus of these experiments is not the determination of absolute distances but rather the change in the distances for each isomer and the effect of dNTPs on these measurements. Although the absolute distances would change with a more refined estimate of the orientation coefficient, the relative change between the efficiency of transfer between the different conditions should remain the same (26).
The presence of each of the dNTPs resulted in little to no movement of either adduct in the polymerase active site, the largest change being the (−)-trans isomer in the presence of dGTP, which moves about 1.9 Å. Based on the structures of BF polymerase (11) and molecular dynamics studies (13), one would anticipate a fairly large change in the position of the (+)-trans adduct when it moves from an anti to syn conformation that has been suggested to occur as the polymerase undergoes a conformational change to a closed ternary complex. Although some movement was observed when a dNTP was added in each case, it is not clear if the magnitude of the changes is consistent with a change in the structure of the polymerase-primer-template complex that has been proposed to occur upon nucleotide incorporation opposite a (+)-trans adduct (27). However, it is intriguing that that dGTP and dATP are the nucleotides best incorporated across from the (−)-trans adduct, unlike what is observed for the (+)-trans adduct where dATP alone is best incorporated (3). The lack of a measurable conformational change also agree with prior protease digestion studies that were unable to detect the formation of a closed ternary complex when either a (+)-trans- or (+)-cis-anti-B[a]P-N2-dG adduct was positioned in the KF active site (14). Finally, our studies do not rule out the possibility that a small level of this conformational change has occurred, but is not detected because the ensemble nature of this assay precludes measurement of a small structural subpopulation.
In summary, this study has demonstrated the utility of using FRET-based assays in determining structural characteristics of biologically relevant carcinogenic adducts in the context of a DNA polymerase active site. The results are consistent with structural and biochemical studies obtained using B[a]P adducts positioned in the active site of DNA polymerases and provides further support that the structures observed for the adducts in solution are maintained in the polymerase active sites. Because both fluorphores in this system are located on the primer-template, this analysis can easily be extended to other polymerases, including bypass polymerases that may be involved in synthesis past these adducts in vivo. Finally, it may be possible to use this FRET pair in single-molecule investigations that might be able measure DNA synthesis on primer-templates containing B[a]P adducts.
ACKNOWLEDGEMENTS
We thank Rachael Natoli for technical support.
This investigation was supported by U.S. Public Health Service Grant, CA40605 awarded by the Department of Health and Human Services.
Footnotes
Abbreviations: B[a]P, benzo[a]pyrene; BF, Bacillus fragment (large fragment of Bacillus stearothermophilus DNA polymerase I); (+)-trans-anti-B[a]P-N2-dG, 7(R) 8(S) 9(S) 10 (R) absolute configuration of the (+)-trans adduct attached to the N2 position of guanine; (−)-trans-anti-B[a]P-N2-dG, 7(S) 8(R) 9(R) 10 (S) absolute configuration of the (−)-trans adduct attached to the N2 position of guanine; KF, Klenow fragment (5’-3’ exonuclease deficient large fragment of Escherichia coli DNA polymerase I); FRET, fluorescence resonance energy transfer; DMACA, 7-dimethylaminocoumarin-4-acetic acid; Ro, Förster distance.
REFERNCES
- 1.Harvey RG. Polycyclic Aromatic Hydrocarbons: Chemistry and Carcinogenicity. Cambridge: University Press; 1991. [Google Scholar]
- 2.Alekseyev YO, Romano LJ. In vitro replication of primer-templates containing benzo[a]pyrene adducts by exonuclease-deficient Escherichia coli DNA polymerase I (Klenow fragment): effect of sequence context on lesion bypass. Biochemistry. 2000;39:10431–10438. doi: 10.1021/bi000552u. [DOI] [PubMed] [Google Scholar]
- 3.Shibutani S, Margulis LA, Geacintov NE, Grollman AP. Translesional synthesis on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 1993;32:7531–7541. doi: 10.1021/bi00080a027. [DOI] [PubMed] [Google Scholar]
- 4.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Newbold RF, Brooks P. Exceptional mutagenicity of a benzo[a]pyrene diol epoxide in cultured mammalian cells. Nature. 1976;261:52–54. doi: 10.1038/261052a0. [DOI] [PubMed] [Google Scholar]
- 6.Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem Res Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 7.Jerina DM, Chadha A, Cheh AM, Schurdak ME, Wood AW, Sayer JM. Covalent bonding of bay-region diol epoxides to nucleic acids. Adv Exp Med Biol. 1991;283:533–553. doi: 10.1007/978-1-4684-5877-0_70. [DOI] [PubMed] [Google Scholar]
- 8.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 9.Cosman M, Hingerty BE, Geacintov NE, Broyde S, Patel DJ. Structural alignments of (+)- and (−)-trans-anti-benzo[a]pyrene-dG adducts positioned at a DNA template-primer junction. Biochemistry. 1995;34:15334–15350. doi: 10.1021/bi00046a043. [DOI] [PubMed] [Google Scholar]
- 10.Feng B, Gorin A, Hingerty BE, Geacintov NE, Broyde S, Patel DJ. Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction. Biochemistry. 1997;36:13769–13779. doi: 10.1021/bi970069s. [DOI] [PubMed] [Google Scholar]
- 11.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J Biol Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 12.Singh SB, Beard WA, Hingerty BE, Wilson SH, Broyde S. Interactions between DNA polymerase beta and the major covalent adduct of the carcinogen (+)-anti-benzo[a]pyrene diol epoxide with DNA at a primer-template junction. Biochemistry. 1998;37:878–884. doi: 10.1021/bi9720639. [DOI] [PubMed] [Google Scholar]
- 13.Perlow RA, Broyde S. Evading the proofreading machinery of a replicative DNA polymerase: induction of a mutation by an environmental carcinogen. J Mol Biol. 2001;309:519–536. doi: 10.1006/jmbi.2001.4674. [DOI] [PubMed] [Google Scholar]
- 14.Alekseyev YO, Dzantiev L, Romano LJ. Effects of benzo[a]pyrene DNA adducts on Escherichia coli DNA polymerase I (Klenow fragment) primer-template interactions: evidence for inhibition of the catalytically active ternary complex formation. Biochemistry. 2001;40:2282–2290. doi: 10.1021/bi002245u. [DOI] [PubMed] [Google Scholar]
- 15.Rechkoblit O, Amin S, Geacintov NE. Primer length dependence of binding of DNA polymerase I Klenow fragment to template-primer complexes containing site-specific bulky lesions. Biochemistry. 1999;38:11834–11843. doi: 10.1021/bi990614k. [DOI] [PubMed] [Google Scholar]
- 16.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2nd ed. Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- 17.Joyce CM, Derbyshire V. Purification of Escherichia coli DNA polymerase I and Klenow fragment. Methods Enzymol. 1995;262:3–13. doi: 10.1016/0076-6879(95)62003-6. [DOI] [PubMed] [Google Scholar]
- 18.Arghavani MB, SantaLucia J, Jr, Romano LJ. Effect of mismatched complementary strands and 5'-change in sequence context on the thermodynamics and structure of benzo[a]pyrene-modified oligonucleotides. Biochemistry. 1998;37:8575–8583. doi: 10.1021/bi980237w. [DOI] [PubMed] [Google Scholar]
- 19.Dzantiev L, Romano LJ. Interaction of Escherichia coli DNA polymerase I (Klenow fragment) with primer-templates containing N-acetyl-2-aminofluorene or N-2-aminofluorene adducts in the active site. J Biol Chem. 1999;274:3279–3284. doi: 10.1074/jbc.274.6.3279. [DOI] [PubMed] [Google Scholar]
- 20.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 21.Sabanayagam CR, Eid JS, Meller A. Using fluorescence resonance energy transfer to measure distances along individual DNA molecules: corrections due to nonideal transfer. J Chem Phys. 2005;122:061103. doi: 10.1063/1.1854120. [DOI] [PubMed] [Google Scholar]
- 22.Alekseyev YO, Romano LJ. Effects of benzo[a]pyrene adduct stereochemistry on downstream DNA replication in vitro: evidence for different adduct conformations within the active site of DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:4467–4479. doi: 10.1021/bi015850l. [DOI] [PubMed] [Google Scholar]
- 23.Hanrahan CJ, Bacolod MD, Vyas RR, Liu T, Geacintov NE, Loechler EL, Basu AK. Sequence specific mutagenesis of the major (+)-anti-benzo[a]pyrene diol epoxide-DNA adduct at a mutational hot spot in vitro and in Escherichia coli cells. Chem Res Toxicol. 1997;10:369–377. doi: 10.1021/tx9601925. [DOI] [PubMed] [Google Scholar]
- 24.Lipinski LJ, Ross HL, Zajc B, Sayer JM, Jerina DM, Dipple A. Effect of single benzo[a]pyrene diol epoxide-deoxyguanosine adducts on the action of DNA polymerases in vitro. Int J Oncol. 1998;13:269–273. doi: 10.3892/ijo.13.2.269. [DOI] [PubMed] [Google Scholar]
- 25.Broyde S, Wang L, Zhang L, Rechkoblit O, Geacintov NE, Patel DJ. DNA adduct structure-function relationships: comparing solution with polymerase structures. Chem Res Toxicol. 2008;21:45–52. doi: 10.1021/tx700193x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clegg RM, Murchie AI, Zechel A, Lilley DM. Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 1993;90:2994–2998. doi: 10.1073/pnas.90.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu P, Oum L, Beese LS, Geacintov NE, Broyde S. Following an environmental carcinogen N2-dG adduct through replication: elucidating blockage and bypass in a high-fidelity DNA polymerase. Nucleic Acids Res. 2007;35:4275–4288. doi: 10.1093/nar/gkm416. [DOI] [PMC free article] [PubMed] [Google Scholar]