Abstract
Both invasion-promoting MT1-MMP and its physiological inhibitorTIMP-2 play a significant role in tumorigenesis and are identified in the most aggressive cancers. Despite its antiproteolytic effects in vitro, clinical data suggest that TIMP-2 expression is positively associated with tumor recurrence, thus emphasizing the wide-ranging role of TIMP-2 in malignancies. To shed light on this role of TIMP-2, we report that low concentrations of TIMP-2, by interacting with MT1-MMP (a specific membrane receptor of TIMP-2), induce the MEK/ERK signaling cascade in fibrosarcoma HT1080 cells which express MT1-MMP naturally. TIMP-2 binding with cell surface-associated MT1-MMP stimulates phosphorylation of MEK1/2, which is upstream of ERK1/2, and the ERK1/2 substrate p90RSK. Consistent with volumes of literature, we confirmed that the activation of ERK stimulated cell migration. Both the transcriptional silencing of MT1-MMP and the inhibition of MEK1/2 reversed the signaling effects of TIMP-2/MT1-MMP while the active site-targeting MMP inhibitor GM6001 did not. Our data suggest that both the interactions of TIMP-2 with MT1-MMP, which activate the pro-migratory ERK signaling cascade,and the conventional inhibition of MT1-MMP’s catalytic activity by TIMP-2, play a role in the invasion-promoting function of MT1-MMP. The TIMP-2-induced stimulation of ERK signaling in cancer cells explains the direct, as opposed to the inverse, association of TIMP-2 expression with poor prognosis in cancer.
Keywords: MT1-MMP, TIMP-2, cell migration, ERK, MEK
The MMP family includes 24 individual proteinases.1 Among MMPs, MT1-MMP is the most relevant to cell locomotion, tumorigenesis and metastasis processes.2,3 An enhanced expression of MT1-MMP is directly associated with aggressive malignancies.4 In tumors, MT1-MMP acts both as a growth factor and as an oncogene and, as a result, usurps tumor growth control.5–7 MT1-MMP is directly involved in the pericellular proteolysis of the matrix, in the activation of soluble MMPs, and in the cleavage of the cell receptors.3 The presence of the transmembrane and the cytoplasmic domains distinguishes MT1-MMP from the soluble MMPs. The MT1-MMP zymogen also exhibits an N-terminal prodomain, a catalytic domain, a hinge and a C-terminal hemopexin (PEX) domain. To become active, the MT1-MMP zymogen requires proteolytic activation.8,9 The N-terminal prodomain is removed by furin during the secretion pathway of MT1-MMP and the catalytic site of the emerging MT1-MMP enzyme becomes liberated.
The catalytic domain of MT1-MMP binds TIMP-2 with high affinity.10 TIMP-2 is a member of a multigene family (TIMP-1 through −4) that binds noncovalently to active MMPs in a 1:1 molar ratio and inhibits their activity.11 In contrast with TIMP-2, TIMP-1 does not inhibit MT1-MMP.10 MMP/TIMP balance, including the balance of MT1-MMP with TIMP-2, is a major factor in the regulation of the proteolytic activity of MMPs.1
TIMP-2 contains two domains. The inhibitory N-terminal domain binds the active site of MMPs including MT1-MMP, blocking the access of substrates to the catalytic site.12 The C-terminal domain of TIMP-2 binds to the PEX domain of proMMP-2.13,14 This binding mode is essential for the cell surface activation of proMMP-2 by MT1-MMP.4,15 The peptide sequence of the PEX domain is conserved in the MMP family. This domain regulates the interactions of MT1-MMP with its cleavage targets including CD44 and the α integrins.16–18 The PEX is also responsible for the formation of the MT1-MMP homodimers, the presence of which is essential for the activation of MMP-2.19
Our earlier data suggest that MT1-MMP’s PEX binds the C-terminal domain of TIMP-2 in a manner similar to proMMP-2. It is likely that these interactions, rather than binding the N-terminal domain of TIMP-2 with the catalytic domain of MT1-MMP, are required for the induction of the Ras/Raf/ERK signaling cascade.20 The stimulation of this cascade appears to play a significant role in the functionality of MT1-MMP. Because in our earlier work we used transfected cells, which overexpressed MT1-MMP, we limited the significance of our findings. To extend our results, we now show that TIMP-2 binding to MT1-MMP, which is naturally expressed by cancer cells, activates the intracellular cascade that involves MEK1/2, ERK1/2 and the downstream ERK substrate p90RSK. We show that the MT1-MMP/TIMP-2 complex stimulates cell locomotion via a mechanism that is independent of and in addition to the proteolytic activity of MT1-MMP.21,22
Material and Methods
Reagents
Unless otherwise indicated, reagents were purchased from Sigma (St. Louis, MO). Human 18.5 kDa myelin basic protein (MBP) was from Biodesign International (Saco, ME). The individual catalytic domain of MT1-MMP (MT1-CAT) was purified from the E. coli inclusion bodies and refolded to restore its catalytic activity.23 Human TIMP-1, a rabbit anti-human antibody to the hinge (AB815) and a murine monoclonal antibody against the catalytic domain of MT1-MMP (3G4), a TMB/M substrate and a hydroxamate inhibitor (GM6001) were from Chemicon (Temecula, CA). The goat anti-human TIMP-2 antibody (AF971) was from R&D Systems (Minneapolis, MN). The rabbit antibodies to pERK1/2 Thr202/Tyr204 (D13.14.4E), pMEK1/2 Ser217/221 (41G9), p-p90RSK Ser-380 (9341), total ERK1/2 (137F7), total MEK1/2 (47E6), pSrc Tyr-416 (2101), total Src (2108) and a MEK inhibitor (U0126) were from Cell Signaling Technology (Danvers, MA). The secondary horseradish peroxidase (HRP)-conjugated antibodies were from GE Healthcare-Amersham (Piscataway, NJ). Rat tail Type I collagen was from Becton-Dickinson (San Diego, CA).
TIMP-2 purification
CHO cells deficient for dihydrofolate reductase (DHFR) were cultured in Minimum Essential Medium alpha (α-MEM) containing 10% FBS, 50 units/ml penicillin and 50 µg/ml streptomycin. Cells were electroporated with the human TIMP-2 cDNA in the bi-cistronic adenoviral vector that encoded methotrexate-resistant DHFR.24 Stable transfectants were selected using α-MEM supplemented with 500 nM methotrrexate. To amplify the DHFR-TIMP-2 gene number, the methotrexate levels in the medium were doubled each week to reach a 25 µM final concentration. TIMP-2 was purified from the 5-day culture medium (500 ml). The medium was concentrated 10-fold using a 400 ml ultrafiltration cell with a 10 kDa cut-off membrane (Amicon) and dialyzed against 20 mM Tris-HCl, pH 8.5, containing 1 mM CaCl2. The samples were chromatographed on a HiTrap Q Sepharose Fast Flow column (GE Healthcare-Amersham in 20 mM Tris-HCl, pH 8.5, containing 1 mM CaCl2. TIMP-2 was eluted with a 0–200 mM NaCl gradient. The purity of TIMP-2 was greater than 90%. The yield was 15% (5 mg TIMP-2 from 1 l medium).
Cells
Parental human fibrosarcoma HT1080 (HT cells) and breast carcinoma MCF-7 cells were from ATCC (Manassas, VA). HT1080 cells stably transfected with MT1-MMP in the pcDNA3-neo plasmid (HT-MT1 cells) and control cells transfected with the original pcDNA3-neo plasmid (HT-neo cells) were obtained and characterized earlier.25 Human glioma U251 cells (U-MT/PDX cells) cotransfected with both MT1-MMP and PDX were isolated and characterized ear-lier.26,27 Cells were cultured in DMEM/FBS and 10 µg/ml gentamicin.
siRNA constructs
Selection of the siRNA and siRNAscr constructs was based on our previous extensive investigations.25,27,28 We tested the transcription silencing efficiency of six MT1-MMP siRNA constructs (numbered 1 through 6) designed using the siRNA Designer software (www.promega.com/techserv/siRNADesigner). As controls, we evaluated several scrambled constructs (Supporting Information Table 1). The sequence of construct 1 was identical with that of MT1-MMP siRNA reported earlier.29 The efficiency of MT1-MMP silencing was determined using FACS, immunoblotting and gelatin zymography. The siRNA construct 3 was the most efficient in silencing MT1-MMP. HT1080 cells in which MT1-MMP transcription was silenced (MT1-MMP-siRNA cells) using the construct 3 and a scrambled siRNA-transfected cell control (siRNAscr cells) were isolated earlier.25 No off-target effects of the constructs were observed. To avoid clone-specific effects, transfected cell lines were generated as pools of 3–6 positive cell clones. Transfected cells were grown in the presence of 1 µg/ml puromycin.
Cell surface biotinylation
Cells were surface-biotinylated for 30 min on ice in PBS containing 0.1 mg/ml EZ-Link NHS-LC-biotin (Pierce, Rockford, IL). The residual biotin was quenched using 100 mM glycine. Cells were lysed in 50 mM N-octyl-β-d-glucopyranoside in TBS supplemented with 1 mM CaCl2, 1 mM MgCl2, and a protease inhibitor cocktail III (Calbiochem, San Diego, CA). Biotin-labeled proteins were precipitated using streptavidin-agarose beads. The precipitates were analyzed by immuno-blotting with the MT1-MMP antibody 3G4 or AB815 followed by the secondaryHRP-conjugated antibody and a TMB/M substrate.
Cell migration
The efficiency of cell migration was measured in wells of a 24-well, 8 µm pore size Transwell plate (Corning Costar, Lowell, MA). A 6.5 mm insert membrane was coated with 50 µl Type I collagen (100 µg/ml in DMEM) and then dried for 12 hr. The collagen coating was rehydrated in 0.5 ml DMEM for 30 min before the experiments. The inner chamber contained DMEM supplemented with 10% FBS as chemoattractant. Cells (1 × 104) were seeded in the outer chamber in serum-free DMEM. GM6001 (10 µM), U0126 (20 µM) and TIMP-2 (10–1000 ng/ml) or DMSO alone (0.01%) were added to both inner and outer chambers 15 min before plating the cells. Cells were allowed to migrate for 12 h. Cells were removed from the upper membrane surface with a cotton swab. Cells that migrate to the membrane’s undersurface were stained with 20% methanol-0.2 % Crystal Violet. The incorporated dye was extracted from cells using 0.25 ml SDS 1%. The A570 value of the extract was measured. Data are means ± SE from three individual experiments performed in triplicate. Statistical analysis was performed by the two tail unpaired t test. p values ≤0.05 were considered significant.
Gelatin zymography
Gelatin zymography of the serum-free medium samples was performed as described previously.28,30 Where indicated, TIMP-2 (10–1000 ng/ml) and GM6001 (10 µM) alone or in combination were added to the cells.
Immunoblotting
Cells were grown in DMEM/FBS for 18 h. The medium was replaced with DMEM containing TIMP-2 (10–1000 ng/ml), GM6001 (10 µM) and U0126 (10 µM) alone or in combination and incubation was continued for an additional 15 min or 24 h. Cells were washed with PBS containing 2 mM sodium orthovanadate and lysed in 50 mM N-octyl-β-d-glucopyranoside in TBS supplemented with 1 mM CaCl2, 1 mM MgCl2, 30 mM sodium fluoride, 10 mM sodium pyrophosphate, 2 mM sodium orthovanadate, 1 mM phenylmethylsulphonyl fluoride, and a protease inhibitor cocktail. Insoluble material was removed by centrifugation (10,000g; 30 min). The supernatant aliquots (20 µg total protein) were analyzed by immunoblotting with pERK1/2, pMEK1/2, p-p90RSK, total ERK1/2, total MEK1/2, pSrc, total Src and MT1-MMP antibodies followed by the secondary HRP-conjugated anti-body and a TMB/M substrate.25
MT1-MMP proteolysis of MBP
The cleavage reactions (20 µl) were performed in 50 mM HEPES, pH 6.8, containing 10 mM CaCl2, 0.5 mM MgCl2 and 50 µM ZnCl2. MBP (4 µg, 11 µM) was incubated for 1 h at 37°C with MT1-CAT (50 nM; an enzyme-substrate ratio 1:200). The increasing concentrations of TIMP-2 were added to MT1-CAT before the addition of MBP and the samples were preincubated for 10 min on ice. The reactions were stopped by a 5xSDS sample buffer and analyzed using SDS-PAGE.
Densitometry and statistical analysis
Quantitative analysis was obtained by scanning the images. The band density was digitized using the Multi Gauge V3.0 Image Analysis software (Fuji). The results are presented as a ratio between the readings of the sample and those of the corresponding loading control. Statistical analysis was performed by the Student’s t test. p values <0.05 were considered significant.
Results
Transcriptional silencing of MT1-MMP in HT1080 cells
The results we describe here represent the continuation of our published study in which we have used breast carcinoma MCF-7 cells which were stably transfected and, as a result, overexpressed MT1-MMP.20 We now specifically selected HT1080 cells because they express MT1-MMP naturally. To silence MT1-MMP, we stably transfected HT1080 cells with the siRNA construct and isolated MT1-MMP-siRNA cells. As a control, we isolated siRNAscr cells, which were transfected with the scrambled construct (Supporting Information Table 1).
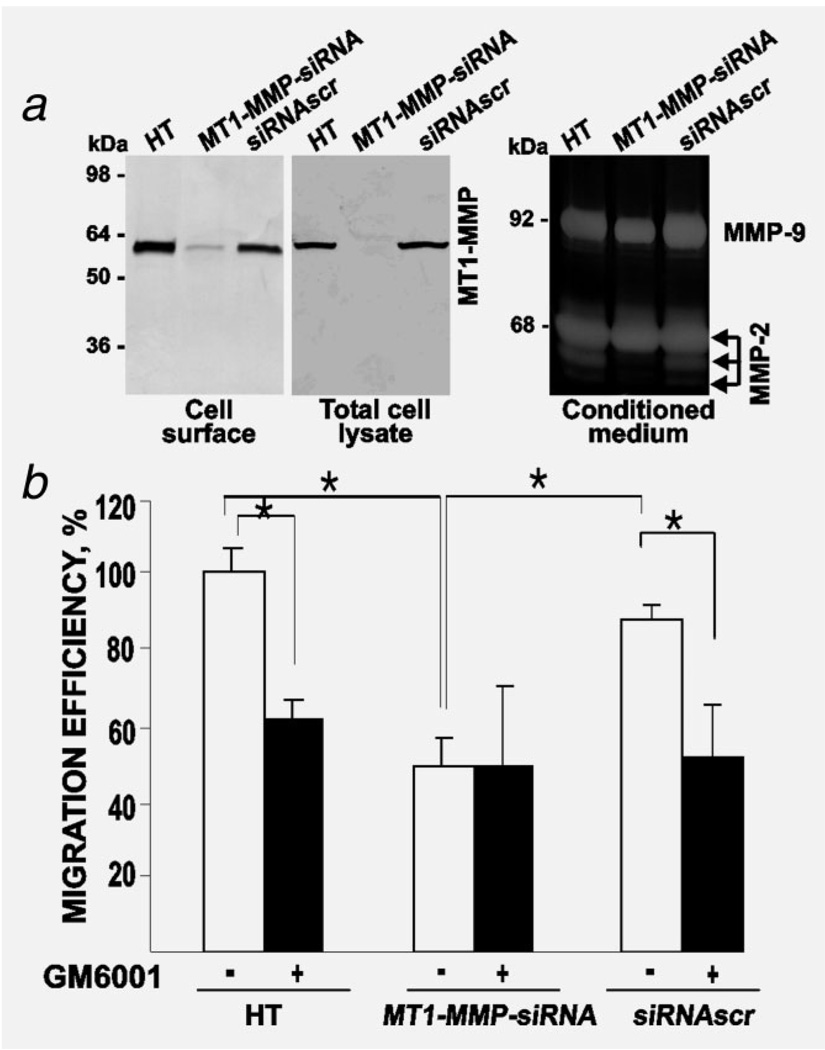
By using an immunoblotting analysis of the cell lysates we confirmed the near complete repression of MT1-MMP in MT1-MMP-siRNA cells. In turn, the scrambled siRNA construct did not exhibit any significant effect on MT1-MMP (Fig. 1). In both the original and the siRNAscr cells, MT1-MMP was represented only by the mature, 58 kDa, enzyme. Consistent with the presence of MT1-MMP on cell surfaces, only siRNAscr cells activated the secretory, naturally produced, 68 kDa proMMP-2. As a result, the 64 kDa intermediate and the 62 kDa enzyme of MMP-2 were generated and detected by gelatin zymography. Because of the silencing of MT1-MMP, MT1-MMP-siRNA cells did not activate MMP-2 efficiently. Consistent with the silencing of MT1-MMP, the migration efficiency of MT1-MMP-siRNA cells was reduced when compared with parental HT1080 cells and siRNAscr cells. Inhibition of MT1-MMP by GM6001 decreased the migration efficiency of the original HT1080 and siRNAscr cells to the levels we recorded with the untreated MT1-MMP-siRNA cells. In turn, GM6001 did not cause any additional suppression in MT1-MMP-siRNA cells.
Figure 1.
Silencing of MT1-MMP inhibits cell migration. (a) Silencing of MT1-MMP in HT1080 cells. Left – Immunoblotting of the cell lysate and biotin-labeled samples of HT, MT1-MMP-siRNA and siRNAscr cells. The MT1-MMP antibodies 3G4 and AB815 were used to analyze cell lysate and biotin-labeled samples, respectively. Right – gelatin zymography of the medium from HT, MT1-MMP-siRNA and siRNAscr cells. (b) Migration efficiency of HT, MT1-MMP-siRNA and siRNAscr cells. Cells (1 × 104) were allowed to migrate for 12 h. Where indicated, GM6001 (10 µM) was added. A 100% number denotes the migration efficiency of the untreated HT cells (5000 cells = 100%). The data are from three independent experiments performed in triplicate. *p < 0.05.
TIMP-2 induced ERK1/2 activation
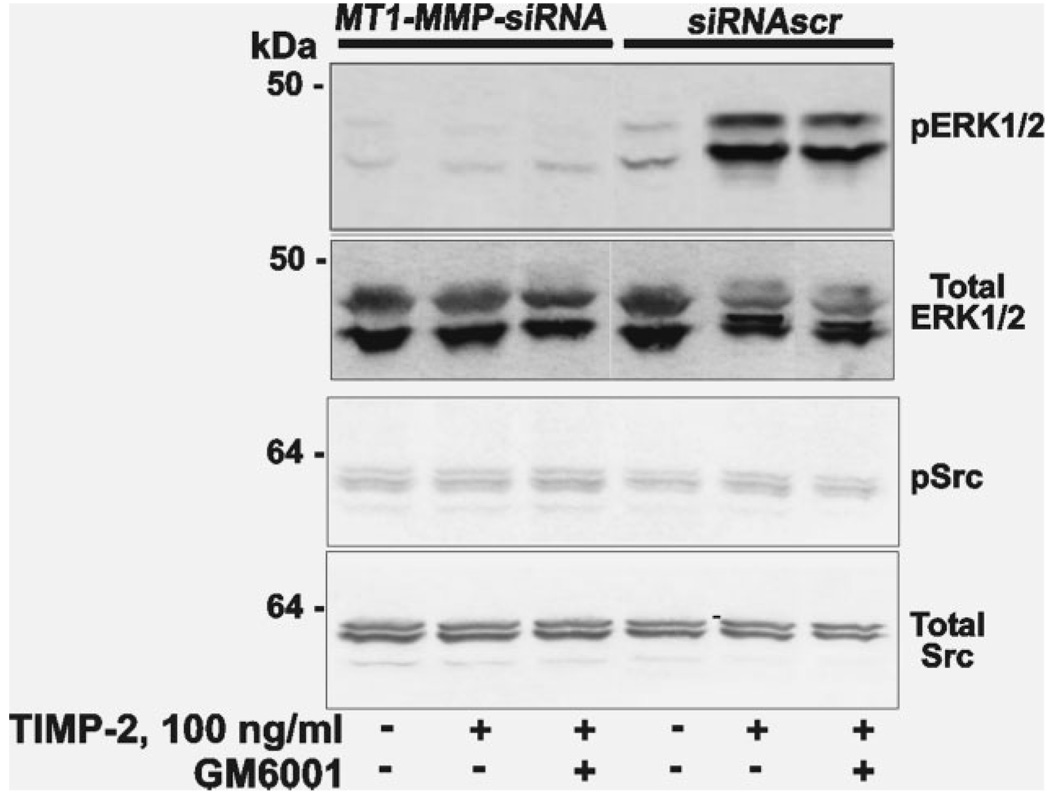
To identify the role of naturally expressed MT1-MMP in ERK1/2 activation, MT1-MMP-siRNA and siRNAscr cells were coincubated for 15 min with TIMP-2 (100 ng/ml) alone, and with TIMP-2 (100 ng/ml) together with GM6001. The cells were lysed. An immunoblotting analysis determined the levels of both ERK1/2 phosphorylation and total ERK1/2 in the lysates (Fig. 2). As a control, we determined the levels of pSrc and total Src. TIMP-2 induced a significant level of ERK1/2 phosphorylation in siRNAscr cells. GM6001 did not reverse the stimulatory effect of TIMP-2 on phosphorylation of ERK1/2 in siRNAscr cells. As we expected,20 TIMP-2 did not induce any significant activation of ERK1/2 in MT1-MMP-siRNA cells. In contrast with pERK1/2, TIMP-2 did not affect the levels of both pSrc and total Src in both MT1-MMP-siRNA and siRNAscr cells, thus supporting the specific activation of the ERK pathway.
Figure 2.
TIMP-2 binding with MT1-MMP activates ERK1/2. MT1-MMP-siRNA and siRNAscr cells were incubated for 15 min in DMEM. Where indicated, TIMP-2 (100 ng/ml) alone and in combination with GM6001 (10 µM) was added. Cell lysates were analyzed by immunoblotting for pERK1/2, total ERK1/2, pSrc and total Src.
TIMP-2 used in these tests was highly purified. Its purity was confirmed by both gel electrophoresis and immunoblotting. TIMP-2, at the molar ratio of MT1-CAT - TIMP-2 equal to 1:2, completely blocked the cleavage of MBP (a sensitive protein substrate of MT1-MMP) by MT1-CAT, thus indicating the high inhibitory potency of our TIMP-2. TIMP-2 (100 ng/ml) blocked the activation of MMP-2 in HT1080 cells (Supporting Information Fig. 1). These tests confirmed the inhibitory efficiency of TIMP-2 and suggested that TIMP-2 itself caused ERK1/2 activation in siRNAscr cells.
TIMP-2 stimulated cell migration
It has been demonstrated that excessively high, 2.5–10 µg/ml, nonphysiological concentrations of TIMP-2 repressed cell migration.6,7,31–33 To determine if low concentrations of TIMP-2 affect cell migration, MT1-MMP-siRNA and siR-NAscr cells were allowed to migrate for 12 h in the presence of increasing, 10–1,000 ng/ml, concentrations of TIMP-2 (Fig. 3). In parallel, we coincubated the cells for 24 h with the same concentrations of TIMP-2. The concentrations of TIMP-2 we used are comparable with those of endogenous TIMP-2 measured using ELISA in the medium conditioned for 24 h by HT1080 cells (1 × 106/ml).34
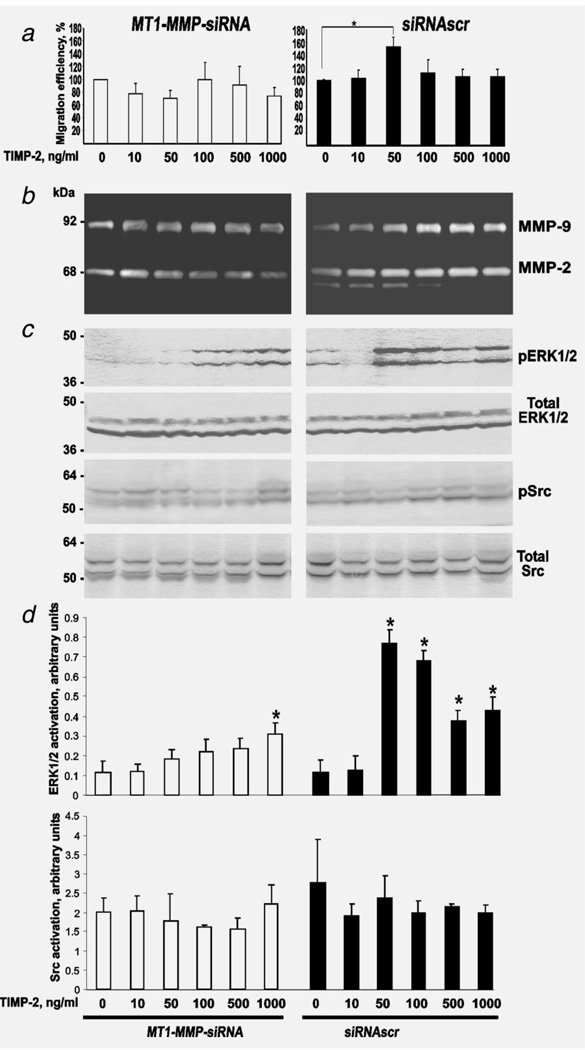
Figure 3.
TIMP-2 binding with MT1-MMP stimulates cell migration. (a) Migration efficiency of MT1-MMP-siRNA and siRNAscr cells. Cells (1 × 104) were allowed to migrate for 12 h in DMEM containing the indicated concentrations of TIMP-2. A 100% number denotes the migration efficiency of the untreated cells. The data are from three independent experiments performed in triplicate. *p < 0.05. (b) Gelatin zymography of the medium conditioned by MT1-MMP-siRNA and siRNAscr cells. Cells (1 × 105) were incubated for 24 h in 0.2 ml DMEM containing the indicated concentrations of TIMP-2. (c) Immunoblotting of cell samples. Cells (5 × 105) were incubated for 24 h in 1 ml DMEM containing the indicated concentrations of TIMP-2. Cell lysates were analyzed by immunoblotting for pERK1/2, total ERK1/2, pSrc and total Src. (c) and (d), Densitometric analysis. Mean ± S.E. of densitometric readings normalized to the corresponding loading control (pERK1/2 versus total ERK, and pSrc versus total Src). *p < 0.05. These experiments were repeated three times with comparable results.
We then determined the levels of pERK1/2 and total ERK1/2, and pSrc and total Src in the cells using immunoblotting analyses. We also examined the levels of active MT1-MMP by measuring the ability of the cells to activate MMP-2. No significant effects of TIMP-2 were observed in MT1-MMP-siRNA cells. In turn, we observed a significant increase of pERK1/2 at 50–1000 ng/ml TIMP-2 in MT1-MMP-siRNA cells. 50–100 ng/ml TIMP-2 induced the maximal level of ERK1/2 phosphorylation. The presence of the residual levels of MT1-MMP explains the minor increase in TIMP-2-induced ERK1/2 phosphorylation we observed in the incompletely silenced MT1-MMP-siRNA cells.
In contrast to MT1-MMP-siRNA cells, there was an obvious effect of TIMP-2 on the levels of MMP-2 activation, pERK1/2 and the migration efficiency in siRNAscr cells. The noticeable level of the activation of MMP-2 was observed at 10–50 ng/ml TIMP-2, while a further increase in TIMP-2 levels inhibited MMP-2 activation by siRNAscr cells. In turn, both the efficiency of migration and the phosphorylation of ERK1/2 were at their highest levels at 50 ng/ml TIMP-2. Over 100 ng/ml TIMP-2 repressed the levels of MMP-2 activation and the migration efficiency but maintained the pERK1/2 levels in siRNAscr cells.
In contrast to pERK1/2, TIMP-2 did not significantly affect total ERK1/2, pSrc and total Src in siRNAscr cells, although some minor variations in the low levels of pSrc and total Src were observed (Fig. 3). A quantitative densitometry analysis did not reveal any significant TIMP-2-induced changes in the pSrc-total Src ratio in MT1-MMP-siRNA and siRNAscr cells (Fig. 3). Incubation for 24 h with GM6001 alone or in combination with TIMP-2 had no effect on the levels of pSrc and total Src in MT1-MMP-siRNA and siR-NAscr cells (Supporting Information Fig. 2).
These results suggest that the migration efficiency of siR-NAscr cells is controlled by the proteolytic and the nonproteolytic mechanisms, both of which employ TIMP-2 as a mediator. In the nonproteolytic mechanism, stimulation of ERK1/2 phosphorylation requires lower concentrations of TIMP-2 compared to those which are required for the quantitative inhibition of the proteolytic activity of MT1-MMP.
TIMP-2 stimulated phosphorylation of MEK1/2
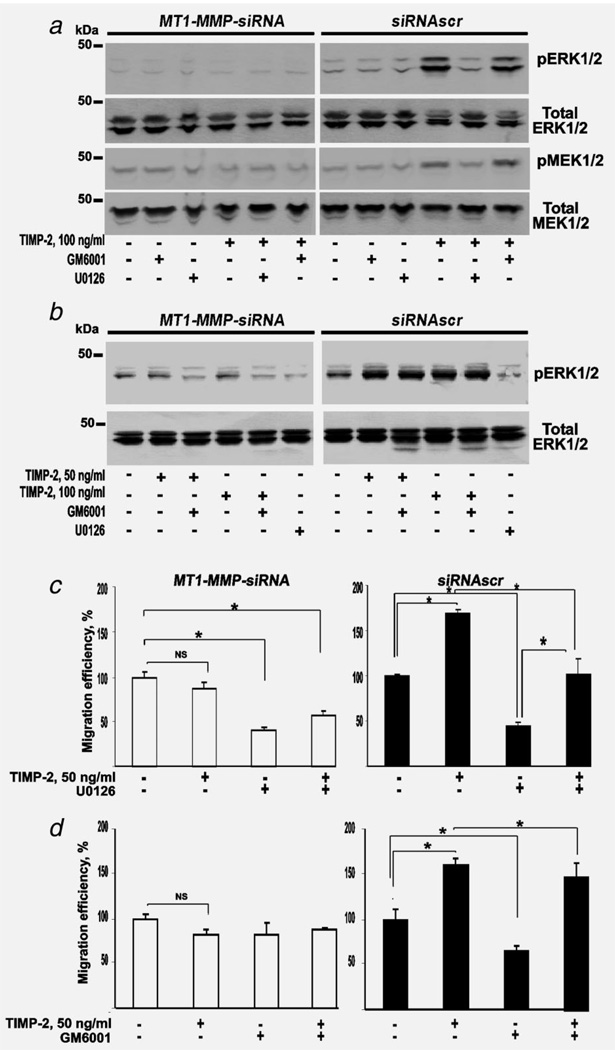
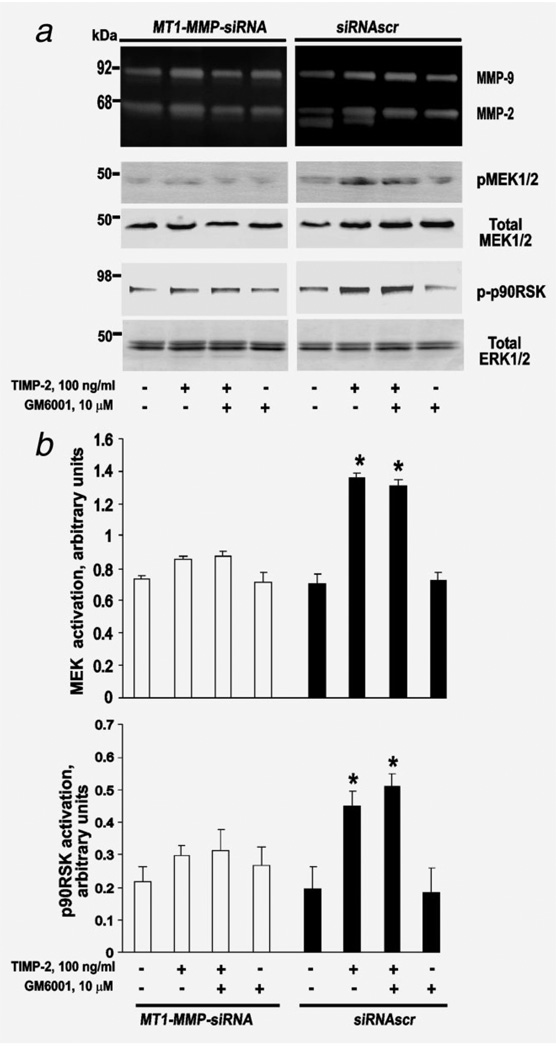
To corroborate our results, we tested if MEK1/2, which is directly upstream of ERK1/2, is activated by TIMP-2. For this purpose, we coincubated cells for 24 h with TIMP-2 (50 ng/ml and 100 ng/ml), GM6001 and U0126 alone and in combination (Fig. 4). We then used immunoblotting to measure the levels of MEK and ERK in cell lysates. No significant effect of TIMP-2, GM6001 and U0126 was observed in MT1-MMP-siRNA cells. In contrast, a significant effect was evident in siRNAscr cells. TIMP-2 strongly increased the levels of both pMEK1/2 and pERK1/2 in siRNAscr cells. U0126 reversed the stimulatory effect of TIMP-2 on pMEK1/2 and pERK1/2 while GM6001 alone or in combination with 100 ng/ml TIMP-2 had no significant effect on MEK and ERK (Fig. 4).
Figure 4.
TIMP-2 binding with MT1-MMP activates MEK1/2 upstream of ERK1/2. (a) MT1-MMP-siRNA and siRNAscr cells were incubated for 24 h in DMEM supplemented with TIMP-2 (100 ng/ml), GM6001 and U0126 (10 µM each) alone and in combination. Cell lysates were analyzed by immunoblotting for pERK1/2, total ERK1/2, pMEK1/2 and total MEK1/2. (b) Cells were incubated for 24 h in DMEM supplemented with TIMP-2 (50 ng/ml or 100 ng/ml) and GM6001 and U0126 (10 µM each) in combination or alone. Cell lysates were analyzed by immunoblotting for pERK1/2 and total ERK1/2. (c) Migration efficiency of MT1-MMP-siRNA and siRNAscr cells. Cells (1 × 104) were allowed to migrate for 12 h. Where indicated, TIMP-2 (50 ng/ml) and U0126 (20 µM) alone or in combination were added. (d) Cells where incubated with TIMP-2 (50 ng/ml) and GM6001 (10 µM) alone or in combination. A 100% number denotes the migration efficiency of the untreated cells. The data are from three independent experiments performed in triplicate. *p < 0.05. These experiments were repeated three times with comparable results.
Similar effects were observed with 50 ng/ml TIMP-2 (Fig. 4 and Supporting Information Fig. 3). These data demonstrated that the low, 50 ng/ml and 100 ng/ml, concentrations of TIMP-2 were comparably efficient in inducing the signaling in siRNAscr cells but not in MT1-MMP-siRNA cells. In agreement with these data, migration of siRNAscr cells was stimulated by TIMP-2 (50 ng/ml) and repressed by U0126. U0126 alone repressed the baseline migration rate of both MT1-MMP-siRNA and siRNAscr by 50%. TIMP-2 alone did not demonstrate any effect in MT1-MMP-siRNA cells.
To investigate the effect of GM6001, we measured the efficiency of migration of cells in the presence of 50 ng/ml TIMP-2 and GM6001 alone and in combination (Fig. 4). In siRNAscr cells, TIMP-2 (50 ng/ml) and GM6001 increased and inhibited migration, respectively. TIMP-2 and GM6001 had no significant effect in MT1-MMP-siRNA cells. If the inhibitors were used jointly, GM6001 had only a minor effect on the TIMP-2-induced migration stimulation in siRNAscr cells.
TIMP-2 stimulated phosphorylation of p90RSK
To elucidate if TIMP-2 binding with MT1-MMP stimulated the terminal events of the Ras/Raf/MEK/ERK cascade such as phosphorylation of the downstream effector of ERK1/2 p90RSK, we coincubated cells for 24 h with TIMP-2 (100 ng/ ml) alone and in combination with GM6001. The levels of p-p90RSK were estimated using immunoblotting of the cell lysates. We also used gelatin zymography to determine the ability of cellular MT1-MMP to activate MMP-2. Our results showed that both TIMP-2 and GM6001 were without effect in MT1-MMP-siRNA cells (Fig. 5). As expected, GM6001 alone inhibited MT1-MMP and this event resulted in the detection of only proMMP-2 in the siRNAscr medium. GM6001 did not increase the level of pMEK1/2 in siRNAscr cells when compared with the untreated control. Compared to GM6001, TIMP-2 (100 ng/ml) was less efficient in inhibiting MMP-2 activation in siRNAscr cells. In contrast with GM6001, TIMP-2 up-regulated the levels of both pMEK1/2 and p-p90RSK in siRNAscr cells (Fig. 5).
Figure 5.
TIMP-2 binding with MT1-MMP activates ERK1/2 substrate p90RSK. (a) MT1-MMP-siRNA and siRNAscr cells were incubated for 24 h in DMEM supplemented with TIMP-2 (100 ng/ml) and GM6001 (10 µM) alone or in combination. Medium aliquots were analyzed by gelatin zymography (upper panel). Cell lysates were analyzed by immunoblotting (bottom panels) for pMEK1/2, total MEK1/2, p-p90RSK and total ERK1/2 (loading control). (b) Densitometric analysis. Mean ± S.E. of densitometric readings normalized to the corresponding loading control (pMEK versus total MEK, and p-p90RSK versus total p90RSK). *p < 0.05. These experiments were repeated three times with comparable results.
TIMP-2 binding with overexpressed MT1-MMP stimulated the MEK/ERK pathway and cell migration
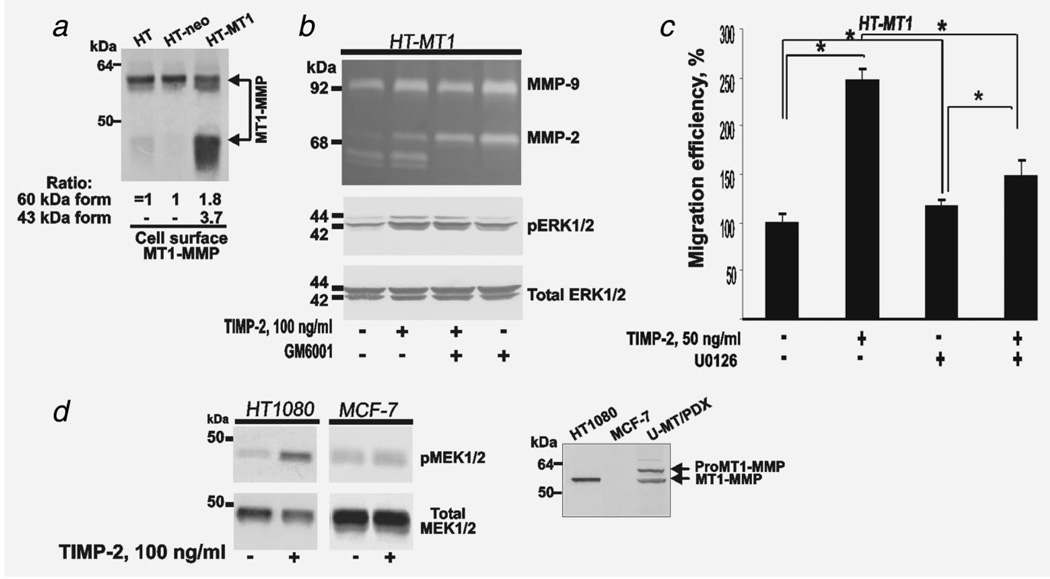
We used HT-MT1 cells, which overexpressed MT1-MMP, to recapitulate the effects we observed in siRNAscr cells, which produce MT1-MMP naturally. To assess cell surface-MT1-MMP, cells were surface-biotinylated. Biotin-labeled proteins were precipitated. The levels of MT1-MMP in the precipitates were determined by immunoblotting. Because of its self-degradation in HT-MT1 cells, MT1-MMP was predominantly represented by the 40–45 kDa inactive species but not by the full-length protease. Similar amounts of the full-length MT1-MMP were detected in the parental HT and HT-neo cells (Fig. 6). These data indicated that because of its complex with TIMP-2, MT1-MMP was largely inactivated and incapable of efficient self-proteolysis in HT1080 cells.35
Figure 6.
TIMP-2 binding with overexpressed MT1-MMP activates ERK1/2 and stimulates cell migration. (a) Cell surface expression of MT1-MMP. HT, HT-neo and HT-MT1 cells (2.5 × 106 cells each) were surface labeled with membrane-impermeable biotin and then lysed. The lysates (1 mg total protein each) were precipitated with streptavidin-agarose beads. The precipitates were analyzed by immunoblotting with the MT1-MMP antibody AB815. The MT1-MMP bands were scanned and the images were digitized. The ratio of the band density is shown at the bottom of the panel (HT cells = 100%). (b) TIMP-2 binding activates ERK1/2. HT-MT1 cells were incubated 24 h in DMEM supplemented with TIMP-2 (100 ng/ml) and GM6001 (10 µM) alone or in combination. Medium was analyzed by gelatin zymography (upper panel). Cell lysates were analyzed by immunoblotting for pERK1/2 and total ERK1/2 (bottom panels). (c) Migration efficiency of HT-MT1 cells. Cells (1 × 104) were allowed to migrate for 12 h. Where indicated, TIMP-2 (50 ng/ml) and U0126 (20 µM) alone or in combination were added to the cells. (d) TIMP-2 activates MEK1/2 in HT1080 cells but not in MT1-MMP-deficient breast carcinoma MCF-7 cells. Left - HT1080 and MCF-7 cells were incubated 24 h with TIMP-2 in DMEM. Cell lysates were analyzed by immunoblotting for pMEK1/2 and total MEK1/2. Right - HT1080, MCF7 and U-MT/PDX cell surface-biotinylated proteins were analyzed by immunoblotting with the MT1-MMP antibody 3G4.
MT1-MMP-overexpressing HT-MT1 cells activated proMMP-2 and generated active MMP-2 (Fig. 6). GM6001 fully inhibited MMP-2 activation by HT-MT1 cells while TIMP-2 (100 ng/ml) caused only a partial inhibition of MMP-2 activation. TIMP-2 (100 ng/ml) significantly up-regulated pERK1/2 in HT-MT1 cells. GM6001 did not reverse the stimulatory effects of TIMP-2 (Fig. 6). TIMP-2 (50 ng/ml) stimulated the migration efficiency of HT-MT1 cells (Fig. 6). U0126 alone did not affect the baseline migration efficiency in HT-MT1 cells. The pro-migratory effect of TIMP-2 was reversed by U0126, thus supporting the primary role of MEK/ERK in the pro-migratory TIMP-2-mediated pro-migratory pathway in HT1080 cells.36
TIMP-2 did not stimulate phosphorylation of MEK1/2 in breast carcinoma MCF-7 cells
We next determined if TIMP-2 was capable of activating MEK1/2 in MT1-MMP-deficient breast carcinoma MCF-7 cells. As controls we used parental HT1080 cells and glioma U251 cells (U-MT/PDX cells), which coexpressed MT1-MMP with PDX, an inhibitor of the MT1-MMP activator furin. Because of PDX, U-MT/PDX cells expressed both the proenzyme and the enzyme of MT1-MMP. Parental HT1080 cells exhibited only the MT1-MMP enzyme. No MT1-MMP was detected in MCF-7 cells (Fig. 6). These results were consistent with earlier observations.20,30,37,38 TIMP-2 induced MEK1/2 activation in HT1080 cells but not in MCF-7 cells (Fig. 6).
Discussion
High levels of TIMP-2 positively correlate with an unfavorable prognosis in cancer patients.39–44 There are additional reports that both aberrant hypermethylation of the TIMP-2 promoter45,46 and TIMP-2 gene polymorphism47–49 affect cancer risk. Clearly, more studies are required to determine the multiple, concentrationdependent effects of TIMP-2 and other TIMP species including TIMP-1 in tumor progression and metastasis.50,51
In a manner counterintuitive to the conventional thinking, we recently suggested that in addition to extracellular proteolysis, the MT1-MMP/TIMP-2 interactions control cell functions through an additional, unconventional mechanism.20 TIMP-2 (but not structurally similar TIMP-1) binding to MT1-MMP induces, in a matter of minutes, an intracellular ERK1/2 signaling cascade. Our previous study, however, employed stably transfected, MT1-MMP-overexpressing cells. In turn, we now proved that the TIMP-2/MT1-MMP-mediated signaling mechanism also takes place in fibrosarcoma HT1080 cells which express MT1-MMP naturally.
The concentrations of TIMP-2 in tissues and biological fluids are in the 10–220 ng/ml range.52–56 The concentrations of endogenous TIMP-2 in the medium conditioned by HT1080 cells are also close to 100 ng/ml.34 Previous studies by others employed TIMP-2 concentrations which were 10-to 500-fold higher when compared with the physiological range.6,7,31–33 The physiological concentrations of TIMP-2 we used stimulated ERK signaling in HT1080 cells. We demonstrated that because of TIMP-2 binding with cell surface-associated naturally produced MT1-MMP and the subsequent stimulation of the intracellular signaling, the levels of pMEK1/2 that is directly upstream of ERK1/2, pERK1/2 itself and of the ERK1/2 substrate p-p90RSK increased in HT1080 cells. In turn, TIMP-2 binding with MT1-MMP did not affect pSrc. The TIMP-2/MT1-MMP-mediated signal generated a significant (2- to 3-fold) increase in the levels of pMEK/ pERK in the cells, thus suggesting the primary role of the TIMP-2/MT1-MMP interactions in the activation of ERK. TIMP-2 did not induce the ERK/MEK pathway in the cells which exhibited a low level of MT1-MMP (e.g., MT1-MMP-deficient breast carcinoma MCF-7 cells). The small-molecule, active site-targeting, inhibitor GM6001 did not reverse TIMP-2/MT1-MMP-mediated stimulation of the MEK-ERK-p90RSK cascade. We also confirmed that TIMP-2/MT1-MMP-dependent stimulation of intracellular signaling promotes cell migration.
These data agree well with the mechanism in which the C-terminal domain of TIMP-2 binds directly with the PEX domain of MT1-MMP.20 It is likely that this binding mode elicits intracellular signaling. Evidence suggests that this binding mode takes place in the formation of TIMP-2’s complex with proMMP-2.13,57 Because of the high homology between the PEX domains of MMP-2 and MT1-MMP, we believe that there is a structural rationale for a similar interaction of TIMP-2 with MT1-MMP.
It is highly likely that MT1-MMP itself cannot signal because its cytoplasmic tail is exceedingly short, because it does not exhibit any unusual sequences which are additional to the internalization signals and because the cytoplasmic tail-binding adaptor proteins have not been reliably identified.58 It may well be that an interaction of the MT1-MMP•TIMP-2 with a cellular signaling receptor is required to elicit the signaling through the latter and that either the low density lipoprotein-related receptor protein-1 (LRP1)59–63 or α3β1 in tegrin64 or both may play this signaling receptor role.
In sum, we suggest that both the interactions of TIMP-2 with MT1-MMP, which activate the signaling cascade, and the inhibition of the catalytic activity of MT1-MMP by TIMP-2 regulate the invasion-promoting function of MT1-MMP. The existence of this cascade in cancer cells supports a direct rather than an inverse association of TIMP-2 with a poor prognosis in certain cancer types. Non-MMP interactions may also affect the role TIMP-2 plays in malignancies. As a result, our hypothesis does not exclude a possibility that in some additional cancer types the interactions of the decreased levels of TIMP-2 with these non-MMP receptors counterbalance the pro-tumorigenic effects of the TIMP-2 binding with MT1-MMP.51 Collectively, we believe that the evidence we acquired validate the high importance we place on the TIMP-2/MT1-MMP-mediated signaling in tumor biology. Our follow-on studies focused on the identification of the cellular receptor, which selectively interacts with the MT1-MMP/TIMP-2 complex, are now in progress.
Acknowledgments
Grant sponsor: NIH; Grant numbers: CA83017, CA77470, RR020843; Grant sponsor: Susan G. Komen Breast Cancer Foundation; Grant number: BCTR123106; Grant sponsor: European Framework Program FP7 microenvimet
Abbreviations
- DMEM/FBS
Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum
- ERK1/2
extracellular signalregulated kinases 1/2
- HRP
horseradish peroxidase
- MBP
myelin basic protein
- MEK1/2
mitogen-activated protein kinases 1 and 2
- MMP-2
matrix metalloproteinase-2
- MT1-CAT
the individual catalytic domain of MT1-MMP
- MT1-MMP
membrane tType-1 matrix metalloproteinase
- p-p90RSK
phosphorylated p90RSK
- p90RSK
90 kDa ribosomal S6 kinase
- PDX
α1-antitrypsin Portland
- pERK1/2
phosphorylated ERK1/2
- PEX
hemopexin domain
- pMEK1/2
phosphorylated MEK1/2
- proMMP-2
the latent proenzyme of MMP-2
- TIMP-2
tissue inhibitor of metalloproteinases-2
Footnotes
Additional Supporting Information may be found in the online version of this article
References
- 1.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 2.Seiki M. Membrane-type 1 matrix metalloproteinase: a key enzyme for tumor invasion. Cancer Lett. 2003;194:1–11. doi: 10.1016/s0304-3835(02)00699-7. [DOI] [PubMed] [Google Scholar]
- 3.Strongin AY. Mislocalization and unconventional functions of cellular MMPs in cancer. Cancer Metastasis Rev. 2006;25:87–98. doi: 10.1007/s10555-006-7892-y. [DOI] [PubMed] [Google Scholar]
- 4.Zucker S, Pei D, Cao J, Lopez-Otin C. Membrane type-matrix metalloproteinases (MT-MMP) Curr Top Dev Biol. 2003;54:1–74. doi: 10.1016/s0070-2153(03)54004-2. [DOI] [PubMed] [Google Scholar]
- 5.Golubkov VS, Chekanov AV, Savinov AY, Rozanov DV, Golubkova NV, Strongin AY. Membrane type-1 matrix metalloproteinase confers aneuploidy and tumorigenicity on mammary epithelial cells. Cancer Res. 2006;66:10460–10465. doi: 10.1158/0008-5472.CAN-06-2997. [DOI] [PubMed] [Google Scholar]
- 6.Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 7.Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, Lopez-Otin C, Shapiro S, Inada M, Krane S, Allen E, Chung D, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golubkov VS, Chekanov AV, Shiryaev SA, Aleshin AE, Ratnikov BI, Gawlik K, Radichev I, Motamedchaboki K, Smith JW, Strongin AY. Proteolysis of the membrane type-1 matrix metalloproteinase prodomain: implications for a two-step proteolytic processing and activation. J Biol Chem. 2007;282:36283–36291. doi: 10.1074/jbc.M706290200. [DOI] [PubMed] [Google Scholar]
- 9.Pei D, Weiss SJ. Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature. 1995;375:244–247. doi: 10.1038/375244a0. [DOI] [PubMed] [Google Scholar]
- 10.Will H, Atkinson SJ, Butler GS, Smith B, Murphy G. The soluble catalytic domain of membrane type 1 matrix metalloproteinase cleaves the propeptide of progelatinase A and initiates autoproteolytic activation. Regulation by TIMP-2 and TIMP-3. J Biol Chem. 1996;271:17119–17123. doi: 10.1074/jbc.271.29.17119. [DOI] [PubMed] [Google Scholar]
- 11.Murphy G, Knauper V, Lee MH, Amour A, Worley JR, Hutton M, Atkinson S, Rapti M, Williamson R. Role of TIMPs (tissue inhibitors of metalloproteinases) in pericellular proteolysis: the specificity is in the detail. Biochem Soc Symp. 2003;70:65–80. doi: 10.1042/bss0700065. [DOI] [PubMed] [Google Scholar]
- 12.Worley JR, Thompkins PB, Lee MH, Hutton M, Soloway P, Edwards DR, Murphy G, Knauper V. Sequence motifs of tissue inhibitor of metalloproteinases 2 (TIMP-2) determining progelatinase A (proMMP-2) binding and activation by membrane-type metalloproteinase 1 (MT1-MMP) Biochem J. 2003;372:799–809. doi: 10.1042/BJ20021573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72- kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 14.Willenbrock F, Crabbe T, Slocombe PM, Sutton CW, Docherty AJ, Cockett MI, O’shea M, Brocklehurst K, Phillips IR, Murphy G. The activity of the tissue inhibitors of metalloproteinases is regulated by C-terminal domain interactions: a kinetic analysis of the inhibition of gelatinase A. Biochemistry. 1993;32:4330–4337. doi: 10.1021/bi00067a023. [DOI] [PubMed] [Google Scholar]
- 15.Rapti M, Knauper V, Murphy G, Williamson RA. Characterization of the AB loop region of TIMP-2. Involvement in pro-MMP-2 activation. J Biol Chem. 2006;281:23386–23394. doi: 10.1074/jbc.M604423200. [DOI] [PubMed] [Google Scholar]
- 16.Belkin AM, Akimov SS, Zaritskaya LS, Ratnikov BI, Deryugina EI, Strongin AY. Matrix-dependent proteolysis of surface transglutaminase by membrane-type metalloproteinase regulates cancer cell adhesion and locomotion. J Biol Chem. 2001;276:18415–18422. doi: 10.1074/jbc.M010135200. [DOI] [PubMed] [Google Scholar]
- 17.Mori H, Tomari T, Koshikawa N, Kajita M, Itoh Y, Sato H, Tojo H, Yana I, Seiki M. CD44 directs membrane-type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. EMBO J. 2002;21:3949–3959. doi: 10.1093/emboj/cdf411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratnikov BI, Rozanov DV, Postnova TI, Baciu PG, Zhang H, Discipio RG, Chestukhina GG, Smith JW, Deryugina EI, Strongin AY. An alternative processing of integrin alpha(v) subunit in tumor cells by membrane type-1 matrix metalloproteinase. J Biol Chem. 2002;277:7377–7385. doi: 10.1074/jbc.M109580200. [DOI] [PubMed] [Google Scholar]
- 19.Itoh Y, Takamura A, Ito N, Maru Y, Sato H, Suenaga N, Aoki T, Seiki M. Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J. 2001;20:4782–4793. doi: 10.1093/emboj/20.17.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’alessio S, Ferrari G, Cinnante K, Scheerer W, Galloway AC, Roses DF, Rozanov DV, Remacle AG, Oh ES, Shiryaev SA, Strongin AY, Pintucci G, et al. Tissue inhibitor of metalloproteinases-2 binding to membrane-type 1 matrix metalloproteinase induces MAPK activation and cell growth by a non-proteolytic mechanism. J Biol Chem. 2008;283:87–99. doi: 10.1074/jbc.M705492200. [DOI] [PubMed] [Google Scholar]
- 21.Dufour A, Sampson NS, Zucker S, Cao J. Role of the hemopexin domain of matrix metalloproteinases in cell migration. J Cell Physiol. 2008;217:643–651. doi: 10.1002/jcp.21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishida Y, Miyamori H, Thompson EW, Takino T, Endo Y, Sato H. Activation of matrix metalloproteinase-2 (MMP-2) by membrane type 1 matrix metalloproteinase through an artificial receptor for proMMP-2 generates active MMP-2. Cancer Res. 2008;68:9096–9104. doi: 10.1158/0008-5472.CAN-08-2522. [DOI] [PubMed] [Google Scholar]
- 23.Ratnikov B, Deryugina E, Leng J, Marchenko G, Dembrow D, Strongin A. Determination of matrix metalloproteinase activity using biotinylated gelatin. Anal Biochem. 2000;286:149–155. doi: 10.1006/abio.2000.4798. [DOI] [PubMed] [Google Scholar]
- 24.Sounni NE, Devy L, Hajitou A, Frankenne F, Munaut C, Gilles C, Deroanne C, Thompson EW, Foidart JM, Noel A. MT1-MMP expression promotes tumor growth and angiogenesis through an up-regulation of vascular endothelial growth factor expression. FASEB J. 2002;16:555–564. doi: 10.1096/fj.01-0790com. [DOI] [PubMed] [Google Scholar]
- 25.Rozanov DV, Savinov AY, Williams R, Liu K, Golubkov VS, Krajewski S, Strongin AY. Molecular signature of MT1-MMP: transactivation of the downstream universal gene network in cancer. Cancer Res. 2008;68:4086–4096. doi: 10.1158/0008-5472.CAN-07-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deryugina EI, Bourdon MA, Luo GX, Reisfeld RA, Strongin A. Matrix metalloproteinase-2 activation modulates glioma cell migration. J Cell Sci. 1997;110:2473–2482. doi: 10.1242/jcs.110.19.2473. [DOI] [PubMed] [Google Scholar]
- 27.Golubkov VS, Boyd S, Savinov AY, Chekanov AV, Osterman AL, Remacle A, Rozanov DV, Doxsey SJ, Strongin AY. Membranetype-1 matrix metalloproteinase (MT1-MMP) exhibits an important intracellular cleavage function and causes chromosome instability. J Biol Chem. 2005;280:25079–25086. doi: 10.1074/jbc.M502779200. [DOI] [PubMed] [Google Scholar]
- 28.Golubkov VS, Chekanov AV, Doxsey SJ, Strongin AY. Centrosomal pericentrin is a direct cleavage target of membrane type-1 matrix metalloproteinase in humans but not in mice: potential implications for tumorigenesis. J Biol Chem. 2005;280:42237–42241. doi: 10.1074/jbc.M510139200. [DOI] [PubMed] [Google Scholar]
- 29.Ueda J, Kajita M, Suenaga N, Fujii K, Seiki M. Sequencespecific silencing of MT1-MMP expression suppresses tumor cell migration and invasion: importance of MT1-MMP as a therapeutic target for invasive tumors. Oncogene. 2003;22:8716–8722. doi: 10.1038/sj.onc.1206962. [DOI] [PubMed] [Google Scholar]
- 30.Rozanov DV, Deryugina EI, Ratnikov BI, Monosov EZ, Marchenko GN, Quigley JP, Strongin AY. Mutation analysis of membrane type-1 matrix metalloproteinase (MT1-MMP). The role of the cytoplasmic tail Cys(574), the active site Glu(240), and furin cleavage motifs in oligomerization,processing, and self-proteolysis of MT1- MMP expressed in breast carcinoma cells. J Biol Chem. 2001;276:25705–25714. doi: 10.1074/jbc.M007921200. [DOI] [PubMed] [Google Scholar]
- 31.Albini A, Melchiori A, Santi L, Liotta LA, Brown PD, Stetler-Stevenson WG. Tumor cell invasion inhibited by TIMP-2. J Natl Cancer Inst. 1991;83:775–779. doi: 10.1093/jnci/83.11.775. [DOI] [PubMed] [Google Scholar]
- 32.Filippov S, Koenig GC, Chun TH, Hotary KB, Ota I, Bugge TH, Roberts JD, Fay WP, Birkedal-Hansen H, Holmbeck K, Sabeh F, Allen ED, et al. MT1-matrix metalloproteinase directs arterial wall invasion and neointima formation by vascular smooth muscle cells. J Exp Med. 2005;202:663–671. doi: 10.1084/jem.20050607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehti K, Allen E, Birkedal-Hansen H, Holmbeck K, Miyake Y, Chun TH, Weiss SJ. An MT1-MMP-PDGF receptor-beta axis regulates mural cell investment of the microvasculature. Genes Dev. 2005;19:979–991. doi: 10.1101/gad.1294605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maquoi E, Frankenne F, Noel A, Krell HW, Grams F, Foidart JM. Type IV collagen induces matrix metalloproteinase 2 activation in HT1080 fibrosarcoma cells. Exp Cell Res. 2000;261:348–359. doi: 10.1006/excr.2000.5063. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez-Barrantes S, Toth M, Bernardo MM, Yurkova M, Gervasi DC, Raz Y, Sang QA, Fridman R. Binding of active (57 kDa) membrane type 1-matrix metalloproteinase (MT1-MMP) to tissue inhibitor of metalloproteinase (TIMP)-2 regulates MT1-MMP processing and pro-MMP-2 activation. J Biol Chem. 2000;275:12080–12089. doi: 10.1074/jbc.275.16.12080. [DOI] [PubMed] [Google Scholar]
- 36.Friday BB, Adjei AA. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin Cancer Res. 2008;14:342–346. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]
- 37.Deryugina EI, Bourdon MA, Jungwirth K, Smith JW, Strongin AY. Functional activation of integrin alpha V beta 3 in tumor cells expressing membrane-type 1 matrix metalloproteinase. Int J Cancer. 2000;86:15–23. doi: 10.1002/(sici)1097-0215(20000401)86:1<15::aid-ijc3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 38.Remacle AG, Chekanov AV, Golubkov VS, Savinov AY, Rozanov DV, Strongin AY. O-glycosylation regulates autolysis of cellular membrane type-1 matrix metalloproteinase (MT1-MMP) J Biol Chem. 2006;281:16897–16905. doi: 10.1074/jbc.M600295200. [DOI] [PubMed] [Google Scholar]
- 39.Roca F, Mauro LV, Morandi A, Bonadeo F, Vaccaro C, Quintana GO, Specterman S, De Kier Joffe EB, Pallotta MG, Puricelli LI, Lastiri J. Prognostic value of E-cadherin, beta-catenin, MMPs (7 and 9), and TIMPs (1 and 2) in patients with colorectal carcinoma. JSurg Oncol. 2006;93:151–160. doi: 10.1002/jso.20413. [DOI] [PubMed] [Google Scholar]
- 40.Ruokolainen H, Paakko P, Turpeenniemi-Hujanen T. Tissue and circulating immunoreactive protein for MMP-2 and TIMP-2 in head and neck squamous cell carcinoma–tissue immunoreactivity predicts aggressive clinical course. Mod Pathol. 2006;19:208–217. doi: 10.1038/modpathol.3800506. [DOI] [PubMed] [Google Scholar]
- 41.Swellam M, Arab LR, Bushnak HA. Clinical implications of HER-2/neu overexpression and proteolytic activity imbalance in breast cancer. IUBMB Life. 2007;59:394–401. doi: 10.1080/15216540701395074. [DOI] [PubMed] [Google Scholar]
- 42.Remacle A, Mccarthy K, Noel A, Maguire T, Mcdermott E, O’higgins N, Foidart JM, Duffy MJ. High levels of TIMP-2 correlate with adverse prognosis in breast cancer. Int J Cancer. 2000;89:118–121. doi: 10.1002/(sici)1097-0215(20000320)89:2<118::aid-ijc3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 43.Grignon DJ, Sakr W, Toth M, Ravery V, Angulo J, Shamsa F, Pontes JE, Crissman JC, Fridman R. High levels of tissue inhibitor of metalloproteinase-2 (TIMP-2) expression are associated with poor outcome in invasive bladder cancer. Cancer Res. 1996;56:1654–1659. [PubMed] [Google Scholar]
- 44.Visscher DW, Hoyhtya M, Ottosen SK, Liang CM, Sarkar FH, Crissman JD, Fridman R. Enhanced expression of tissue inhibitor of metalloproteinase-2 (TIMP-2) in the stroma of breast carcinomas correlates with tumor recurrence. Int J Cancer. 1994;59:339–344. doi: 10.1002/ijc.2910590308. [DOI] [PubMed] [Google Scholar]
- 45.Galm O, Suzuki H, Akiyama Y, Esteller M, Brock MV, Osieka R, Baylin SB, Herman JG. Inactivation of the tissue inhibitor of metalloproteinases-2 gene by promoter hypermethylation in lymphoid malignancies. Oncogene. 2005;24:4799–4805. doi: 10.1038/sj.onc.1208599. [DOI] [PubMed] [Google Scholar]
- 46.Ivanova T, Vinokurova S, Petrenko A, Eshilev E, Solovyova N, Kisseljov F, Kisseljova N. Frequent hypermethylation of 5′ flanking region of TIMP-2 gene in cervical cancer. Int J Cancer. 2004;108:882–886. doi: 10.1002/ijc.11652. [DOI] [PubMed] [Google Scholar]
- 47.Kubben FJ, Sier CF, Meijer MJ, Van Den Berg M, Van Der Reijden JJ, Griffioen G, Van De Velde CJ, Lamers CB, Verspaget HW. Clinical impact of MMP and TIMP gene polymorphisms in gastric cancer. Br J Cancer. 2006;95:744–751. doi: 10.1038/sj.bjc.6603307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pulukuri SM, Patibandla S, Patel J, Estes N, Rao JS. Epigenetic inactivation of the tissue inhibitor of metalloproteinase-2 (TIMP-2) gene in human prostate tumors. Oncogene. 2007;26:5229–5237. doi: 10.1038/sj.onc.1210329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vairaktaris E, Yapijakis C, Yiannopoulos A, Vassiliou S, Serefoglou Z, Vylliotis A, Nkenke E, Derka S, Critselis E, Avgoustidis D, Neukam FW, Patsouris E. Strong association of the tissue inhibitor of metalloproteinase-2 polymorphism with an increased risk of oral squamous cell carcinoma in Europeans. Oncol Rep. 2007;17:963–968. [PubMed] [Google Scholar]
- 50.Jung KK, Liu XW, Chirco R, Fridman R, Kim HR. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 2006;25:3934–3942. doi: 10.1038/sj.emboj.7601281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stetler-Stevenson WG. Tissue inhibitors of metalloproteinases in cell signaling: metalloproteinase-independent biological activities. Sci Signal. 2008;1 doi: 10.1126/scisignal.127re6. re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker EA, Leaper DJ, Hayter JP, Dickenson AJ. The matrix metalloproteinase system in oral squamous cell carcinoma. Br J Oral Maxillofac Surg. 2006;44:482–486. doi: 10.1016/j.bjoms.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 53.Murawaki Y, Ikuta Y, Kawasaki H. Clinical usefulness of serum tissue inhibitor of metalloproteinases (TIMP)-2 assay in patients with chronic liver disease in comparison with serum TIMP-1. Clin Chim Acta. 1999;281:109–120. doi: 10.1016/s0009-8981(98)00215-0. [DOI] [PubMed] [Google Scholar]
- 54.Pasieka Z, Stepien H, Czyz W, Pomorski L, Kuzdak K. Concentration of metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 in the serum of patients with benign and malignant thyroid tumours treated surgically. Endocr Regul. 2004;38:57–63. [PubMed] [Google Scholar]
- 55.Ulrich D, Lichtenegger F, Eblenkamp M, Repper D, Pallua N. Matrix metalloproteinases, tissue inhibitors of metalloproteinases, aminoterminal propeptide of procollagen type III, and hyaluronan in sera and tissue of patients with capsular contracture after augmentation with Trilucent breast implants. Plast Reconstr Surg. 2004;114:229–236. doi: 10.1097/01.prs.0000129079.19089.6c. [DOI] [PubMed] [Google Scholar]
- 56.Larsen MB, Stephens RW, Brunner N, Nielsen HJ, Engelholm LH, Christensen IJ, Stetler-Stevenson WG, Hoyer-Hansen G. Quantification of tissue inhibitor of metalloproteinases 2 in plasma from healthy donors and cancer patients. Scand J Immunol. 2005;61:449–460. doi: 10.1111/j.1365-3083.2005.01585.x. [DOI] [PubMed] [Google Scholar]
- 57.Murphy G, Stanton H, Cowell S, Butler G, Knauper V, Atkinson S, Gavrilovic J. Mechanisms for pro matrix metalloproteinase activation. APMIS. 1999;107:38–44. doi: 10.1111/j.1699-0463.1999.tb01524.x. [DOI] [PubMed] [Google Scholar]
- 58.Uekita T, Itoh Y, Yana I, Ohno H, Seiki M. Cytoplasmic tail-dependent internalization of membrane-type 1 matrix metalloproteinase is important for its invasion-promoting activity. J Cell Biol. 2001;155:1345–1356. doi: 10.1083/jcb.200108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hackeng CM, Relou IA, Pladet MW, Gorter G, Van Rijn HJ, Akkerman JW. Early platelet activation by low density lipoprotein via p38MAP kinase. Thromb Haemost. 1999;82:1749–1756. [PubMed] [Google Scholar]
- 60.Hu K, Yang J, Tanaka S, Gonias SL, Mars WM, Liu Y. Tissue-type plasminogen activator acts as a cytokine that triggers intracellular signal transduction and induces matrix metalloproteinase-9 gene expression. J Biol Chem. 2006;281:2120–2127. doi: 10.1074/jbc.M504988200. [DOI] [PubMed] [Google Scholar]
- 61.Ma Z, Thomas KS, Webb DJ, Moravec R, Salicioni AM, Mars WM, Gonias SL. Regulation of Rac1 activation by the low density lipoprotein receptor-related protein. J Cell Biol. 2002;159:1061–1070. doi: 10.1083/jcb.200207070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mantuano E, Inoue G, Li X, Takahashi K, Gaultier A, Gonias SL, Campana WM. The hemopexin domain of matrix metalloproteinase-9 activates cell signaling and promotes migration of schwann cells by binding to low-density lipoprotein receptor-related protein. J Neurosci. 2008;28:11571–11582. doi: 10.1523/JNEUROSCI.3053-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webb DJ, Nguyen DH, Gonias SL. Extracellular signal-regulated kinase functions in the urokinase receptor-dependent pathway by which neutralization of low density lipoprotein receptor-related protein promotes fibrosarcoma cell migration and matrigel invasion. J Cell Sci. 2000;113:123–134. doi: 10.1242/jcs.113.1.123. [DOI] [PubMed] [Google Scholar]
- 64.Seo DW, Li H, Guedez L, Wingfield PT, Diaz T, Salloum R, Wei BY. Stetler-Stevenson WG. TIMP-2 mediated inhibition of angiogenesis: an MMP-independent mechanism. Cell. 2003;114:171–180. doi: 10.1016/s0092-8674(03)00551-8. [DOI] [PubMed] [Google Scholar]