Abstract
The X-ray structure at 2.0-Å resolution of the p90 ribosomal S6 kinase 2 C-terminal kinase domain revealed a C-terminal autoinhibitory αL-helix that was embedded in the kinase scaffold and determines the inactive kinase conformation. We suggest a mechanism of activation through displacement of the αL-helix and rearrangement of the conserved residue Glu500, as well as the reorganization of the T-loop into the active conformation.
The 90-kDa ribosomal S6 kinase 2 (RSK2) is broadly expressed in response to growth factors, peptide hormones, neurotransmitters, chemokines and other stimuli1–3. RSK2 is a serine/threonine kinase containing two distinct catalytically functional kinase domains connected by a linker region4,5. The C-terminal domain (CTD) phosphorylates the linker region6 and regulates the N-terminal domain, which phosphorylates various substrates3,4,7,8. No defined structure of RSK2 or of either kinase domain has been reported. However, sequence alignment with Ca2+/calmodulin-dependent kinase suggested the existence of an autoinhibitory helix outside the CTD RSK2 protein kinase domain, and its autoinhibitory role has been demonstrated in vivo9. Efficient activation of RSK2 requires interaction with extracellular signal–regulated protein kinases (ERKs) at a docking site in the RSK2 C terminus (residues 726–735)10,11 and subsequent phosphorylation of Thr577 in the CTD T-activation loop12.
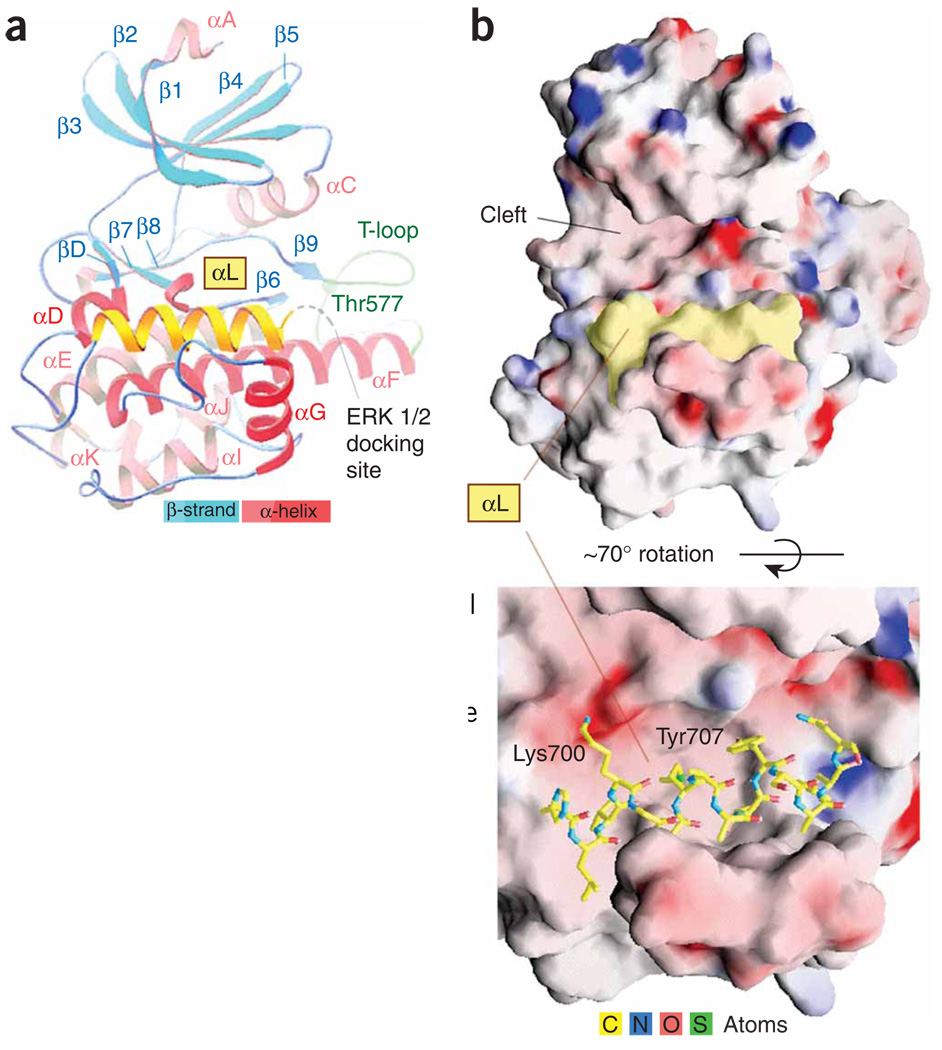
The CTD RSK2 adopted a classical bilobal kinase fold with an accessible catalytic cleft (Fig. 1 and Supplementary Fig. 1 online). The C-terminal segment (residues 696–710) formed another αL-helix, located underneath the catalytic cleft and embedded in the kinase scaffold. It occupied a ‘cradle’ shaped by the αF and αG two-helix junction. The area of the αL-helix surface buried in the ‘cradle’ was ~800 Å2 (~50% of the total area) and was composed mostly (~80%) of nonpolar atoms. The position of the αL-helix in the ‘cradle’ was stabilized by one hydrogen bond (2.7 Å) between Tyr707 and Ser603 (αF-helix).
Figure 1.
Crystal structure of the CTD of RSK2. The autoinhibitory C-terminal αL-helix is shown in yellow. (a) The folding diagram. Disordered residues 715–740 are indicated in gray. (b) The ‘cradle’ position of the αL-helix on the potential surface (positive in blue, negative in red). See Supplementary Methods and Supplementary Table 1 online for details of the structure determination and crystallographic statistics, respectively. The sequence alignment is shown in Supplementary Figure 4 online.
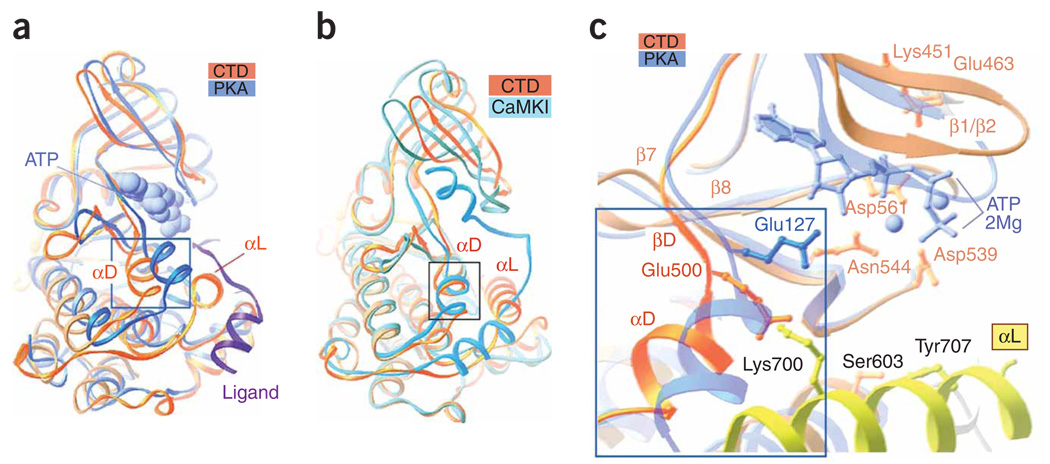
Superimposition of the structures of the RSK2 CTD and active cyclic AMP–dependent protein kinase (PKA) bound to an ATP analog and an inhibitory peptide (Fig. 2a and Supplementary Fig. 2a online) showed that the CTD had a normal arrangement of the N and C lobes with a slightly more open cleft conformation than found in PKA. Comparing the CTD and PKA invariant residues showed that Asp539 (RD-motif), Asn544 (catalytic loop) and Asp561 (DFG-motif), as well as the ionic pair between Glu463 (αC-helix) and Lys451 (β3-strand), essentially adopted the same conformation in both the CTD and PKA and fit the requirements for optimal phospho-transfer13. Despite extensive efforts to cocrystallize the protein with an unhydrolyzable ATP analog, we could not obtain ATP-bound crystals, suggesting that the CTD adopts an inactive conformation.
Figure 2.
Distinctive features of the RSK2 CTD. (a) The CTD superimposed on active PKA (chain A from PDB 1CDK) illustrates the different positions of the αD-helix. Boxed region corresponds to boxed region in c. The ligand (inhibitory peptide, purple ribbon) and an ATP analog (AMP-PNP; blue van der Waals surface representation) bound to PKA are included. (b) The CTD RSK2 superimposed on the autoinhibited Ca2+/calmodulin-dependent protein kinase I (PDB 1A06). (c) A comparison of the active sites of CTD and PKA. Selected RSK2 CTD residues are shown. Only residue Glu127 (analogous to Glu500 in the CTD structure), which differs in the two structures, is shown for PKA. The ATP analog and Mg2+ ions (balls) are shown in blue.
A notable difference between CTD RSK and active PKA entailed the position of the αD-helix, which was shifted by ~5 Å from the cleft by the αL-helix N terminus (Fig. 2a) and an outward extrusion of the T-loop (Fig. 1a and Supplementary Fig. 2a). A subsequent comparison of CTD RSK2 with inactive Ca2+/calmodulin-dependent protein kinases, characterized by a twisted position of the N-terminal lobe and possession of C-terminal autoinhibitory helices that interfere with the binding site for ATP and substrate, illustrated their differences (Fig. 2b and Supplementary Fig. 2b). The αL-helix of CTD RSK2 in the ‘cradle’ did not occupy the catalytic cleft, and its autoinhibitory function is not obvious. However, inactive CTD and Ca2+/calmodulin-dependent protein kinases showed a similar shifted position of the αD-helix compared with active PKA.
The noticeable shift of the CTD αD-helix was accompanied by spatial rearrangement of adjacent residues, including Glu500 (Fig. 2c), which was turned away from the ATP binding site. The analogous residue Glu127 of PKA participates in hydrogen bonding to the ribose hydroxyl groups of ATP. Similar interactions were observed for glutamic or conserved aspartic acid residues in other kinase structures (Glu166 of phosphoinositide-dependent protein kinase-1 (PDB 1H1W), Glu236 of protein kinase B (PDB 1O6L), Glu100 of death-associated protein kinase (PDB 1JKK), Asp100 of ERK2 (PDB 1GOL) and Asp86 of cyclin-dependent kinase-2 (PDB 1QMZ)). In addition, Glu127 of PKA also participates in the interaction with the positively charged arginine of the inhibitory peptide14. In the RSK2 CTD, Lys700 attracts and captures Glu500 (Fig. 2c), forming an ionic pair (3.4 Å, 4.0 Å distances), and inhibits Glu500 from binding to ATP. This defines an autoinhibitory role of the αL-helix and reveals the structural basis for RSK2 activation. The CTD crystal structure suggests that disrupting the Tyr707-Ser603 hydrogen bond promotes displacement of the αL-helix and disruption of the Lys700-Glu500 ionic interaction, resulting in CTD activation, as demonstrated in vivo by truncation eliminating the αL-helix and by single Y707A mutation of RSK2 (ref. 9). We demonstrated that Xenopus embryos injected with mRNAs of the Y707A mutant showed severe defects of the anterior-posterior axis compared with wild type (Supplementary Fig. 3).
The ‘cradle’ location of the αL-helix has two key consequences leading to the inactive kinase conformation: the displacement of the αD-helix, accompanied by misalignment of the active site residue Glu500 (Fig. 2a,c), and the extrusion of the T-loop to an inactive conformation (Fig. 1a and Supplementary Fig. 2a). Unlike that of other kinase domains, the scaffold of the RSK2 CTD is stabilized by the αL-helix, which occupies the ‘cradle’, rather than by the T-loop. On the basis of this structure of the CTD of RSK2, ERKs are likely to be involved in abolishing the autoinhibitory function of the CTD. The ERKs binding site (residues 726–735) is located at the RSK2 C terminus close to the αL-helix (residues 696–710). ERK docking to the C terminus should disrupt the Tyr707-Ser603 hydrogen bond and displace the αL-helix from its inhibitory position in the ‘cradle’. The αL-helix displacement will release Glu500 from its ionic interaction with Lys700 and allow readjustment of the αD-helix position and proper alignment of Glu500 for ATP binding. The αL-helix displacement might also be associated with rearrangement of the phosphorylated T-loop and repositioning of it to the front of the catalytic cleft. Predictive conformational changes upon RSK2 CTD activation are similar to the autoinhibitory C-terminal αK-helix realignment in the homologous MAPK-activated protein kinase 2, as shown by X-ray crystallography of the constitutively active (PDB 1NXK) protein (Supplementary Fig. 2c–e).
Supplementary Material
ACKNOWLEDGMENTS
Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Basic Energy Sciences, under contract DE-AC02-06CH11357. Part of this work was conducted at the Northeastern Collaborative Access Team, Sector 24-ID, supported by award RR-15301 from the US National Center for Research Resources at the National Institutes of Health. Use of the General Medicine and Cancer Institutes Collaborative Access Team Sector 23-ID was funded with federal funds from the US National Cancer Institute (Y1-CO-1020) and National Institute of General Medical Science (Y1-GM-1104). Other funding was provided by The Hormel Foundation and US National Institutes of Health grants CA111356, CA111536, CA077646, CA081064 and CA120388.
Footnotes
Accession codes. Protein Data Bank: Coordinates and structure factors have been deposited with accession codes 2QR8 (native) and 2QR7 (selenomethionine).
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
M.M. conducted cloning, protein purification and crystallization. V.T. performed data collection and X-ray structure determination. V.T. and M.M. performed structural analysis. S.-Y.L. conducted experiments with frog embryos. K.Y. and Y.-Y.C. assisted in the experiments. V.T. and M.M. wrote the manuscript with contributions from A.B. Z.D. supervised and ensured implementation of the project.
Reprints and permissions information is available online at http://npg.nature.com/ reprints and permissions
References
- 1.Erikson E, Maller JL. Proc. Natl. Acad. Sci. USA. 1985;82:742–746. doi: 10.1073/pnas.82.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sturgill TW, Ray LB, Erikson E, Maller JL. Nature. 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 3.Xing J, Ginty DD, Greenberg ME. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 4.Jones SW, Erikson E, Blenis J, Maller JL, Erikson RL. Proc. Natl. Acad. Sci. USA. 1988;85:3377–3381. doi: 10.1073/pnas.85.10.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher TL, Blenis J. Mol. Cell. Biol. 1996;16:1212–1219. doi: 10.1128/mcb.16.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chrestensen CA, Sturgill TW. J. Biol. Chem. 2002;277:27733–27741. doi: 10.1074/jbc.M202663200. [DOI] [PubMed] [Google Scholar]
- 7.Swanson KD, Taylor LK, Haung L, Burlingame AL, Landreth GE. J. Biol. Chem. 1999;274:3385–3395. doi: 10.1074/jbc.274.6.3385. [DOI] [PubMed] [Google Scholar]
- 8.Sassone-Corsi P, et al. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 9.Poteet-Smith CE, Smith J, Lannigan DA, Freed TA, Sturgill TW. J. Biol. Chem. 1999;274:22135–22138. doi: 10.1074/jbc.274.32.22135. [DOI] [PubMed] [Google Scholar]
- 10.Smith JA, Poteet-Smith CE, Malarkey K, Sturgill TW. J. Biol. Chem. 1999;274:2893–2898. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 11.Roux PP, Richards SA, Blenis J. Mol. Cell. Biol. 2003;23:4796–4804. doi: 10.1128/MCB.23.14.4796-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalby KN, Morrice N, Caudwell FB, Avruch J, Cohen P. J. Biol. Chem. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- 13.Huse M, Kuriyan J. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 14.Knighton DR, et al. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.