Abstract
The study of biomolecules in their native environments is a challenging task because of the vast complexity of cellular systems. Technologies developed in the last few years for the selective modification of biological species in living systems have yielded new insights into cellular processes. Key to these new techniques are bioorthogonal chemical reactions, whose components must react rapidly and selectively with each other under physiological conditions in the presence of the plethora of functionality necessary to sustain life. Herein we describe the bioorthogonal chemical reactions developed to date and how they can be used to study biomolecules.
Keywords: alkynes, azides, bioconjugation, bioorthogonal reactions, Staudinger ligation
1. Introduction
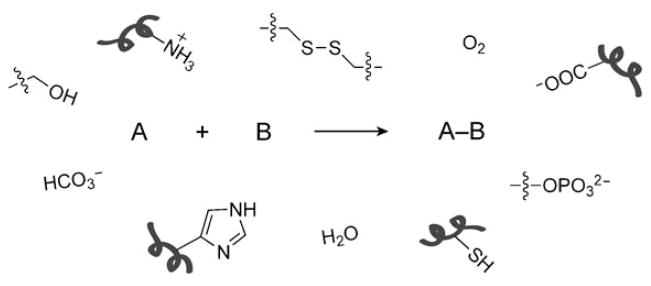
Chemists and biologists have begun to share a common interest in developing methods to study biomolecules in their native settings. Their combined efforts have brought forth innovations such as genetically encoded fluorescent proteins (for example, the green fluorescent protein, GFP), whose widespread use and impact was recognized with the 2008 Nobel Prize in Chemistry.[1,2] However, many biomolecules, such as nucleic acids, lipids, and glycans, as well as various posttranslational modifications, cannot be monitored with genetically encoded reporters. A growing area of chemical biology strives to probe these biomolecules in living systems by using bioorthogonal chemical reactions (that is, reactions that do not interfere with biological processes). Such reactions must have fast rates under physiological conditions and be inert to the myriad of functionalities found in vivo (Scheme 1).
Scheme 1.
A bioorthogonal chemical reaction. The reaction of compounds A and B bearing bioorthogonal functional groups proceeds in the presence of all the functionality found within living systems, some examples of which are indicated.
Here we provide a historical account of the development of bioorthogonal reactions, starting with their roots in protein bioconjugation. We discuss how unique sequences of natural amino acids have been designed to create orthogonal functionality for selective protein modification within complex samples. We then focus on the development of bioorthogonal transformations involving unnatural functional groups and methods to incorporate these unnatural groups into a variety of biomolecules. We conclude with a discussion of avenues toward new bioorthogonal chemical reactions and applications to unexplored biological processes.
2. Historical Perspective
During the past century, the chemical modification of biomolecules has evolved from a means of defining composition to a highly selective method for monitoring cellular events. Proteins, with their numerous side-chain functionalities, complex tertiary structures, and diverse biological functions, were early favorites for chemical modification, initially with the goal of defining their amino acid components.[3] The field of mechanistic enzymology benefited tremendously from the efforts of early protein chemists, as the methods they developed allowed for chemical alteration of side chains implicated in catalysis.[4] Protein modification methods have also been important in the biotechnology industry. Popular applications include PEGylation (modification of proteins with polyethylene glycol (PEG) groups) of therapeutic proteins to improve serum half-life[5,6] and the conjugation of cytotoxins or imaging agents to cancer-targeting elements, such as monoclonal antibodies.[7] The vast majority of examples involve classic residue-specific protein-modification methods, which continue to be vital tools used by chemical biologists.
2.1. Classic Methods for Protein Modification
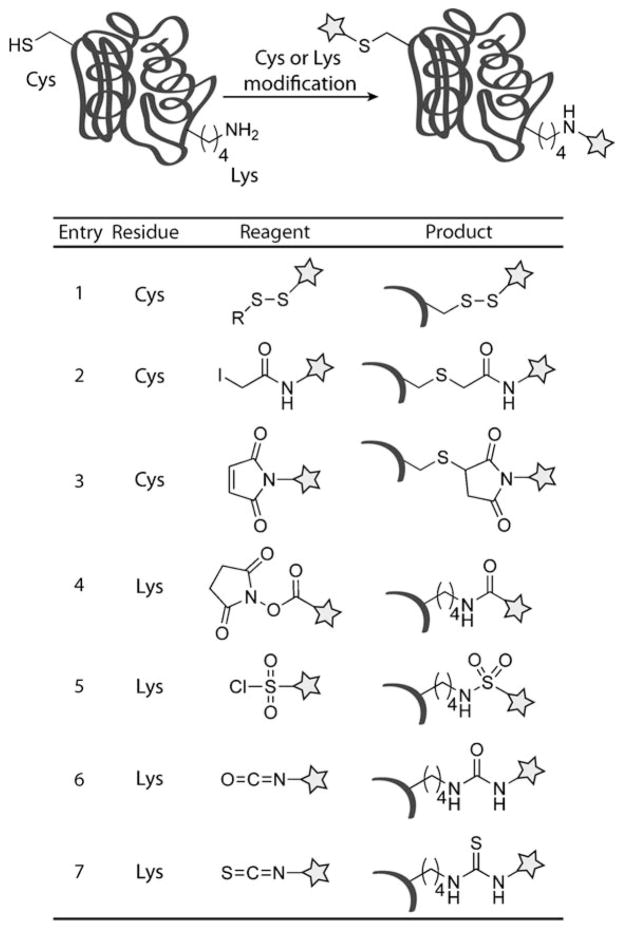
Classic protein bioconjugation primarily encompasses simple second-order reactions that selectively target the functionalities present in the side chains of the canonical, proteogenic amino acids.[3,8–10] Of those, cysteine and lysine are the most commonly modified residues. The thiol group of cysteine can undergo disulfide exchange to form mixed disulfides (Scheme 2, entry 1) as well as alkylation with alkyl halides or Michael addition with α,β-unsaturated carbonyl compounds to yield thioethers (Scheme 2, entries 2 and 3). Furthermore, as a relatively rare amino acid, cysteine can often be used for single-site modification. Although considerably more prevalent than cysteine, lysine residues are popular targets because of the abundance of methods to selectively modify primary amines. Lysine can react with activated esters, sulfonyl chlorides, isocyanates, or isothiocyanates to afford amides, sulfonamides, ureas, or thioureas, respectively (Scheme 2, entries 4–7). Lysine residues also undergo reductive amination reactions with aldehydes.[3,8–10] It is of note that these reagents can additionally modify the N termini of proteins.
Scheme 2.
Classic bioconjugation reactions for the modification of Cys and Lys residues. Cys residues can be modified through disulfide exchange, alkylation with iodoacetamide reagents, and Michael addition with maleimides (entries 1–3, respectively). Lys residues can be modified through amide, sulfonamide, urea, and thiourea formation with N-hydroxysuccinimide-activated esters, sulfonyl chlorides, isocyanates, and isothiocyanates (entries 4–7, respectively).
By comparison, the remaining 18 proteogenic amino acids have been minimally exploited for residue-selective modification. The phenol moiety of tyrosine has been modified through electrophilic aromatic substitution reactions with diazonium salts, iodine, or nitrous acid.[3,10] Glutamate and aspartate residues have been targeted for bioconjugation through coupling with amines via carbodiimides,[3,8–10] although the potential cross-linking of proteins limits the utility of this technique. Pyrocarbonates have also been used to successfully modify histidine residues.[3,8–10]
By using these classic methods, the conjugation of small-molecule probes, such as biotin and fluorophores, to proteins is quite routine. Similar methods are widely used to immobilize proteins on chromatography matrices, soluble polymers, plastic surfaces, and microarray chips.
2.2. Modern Methods for Protein Modification
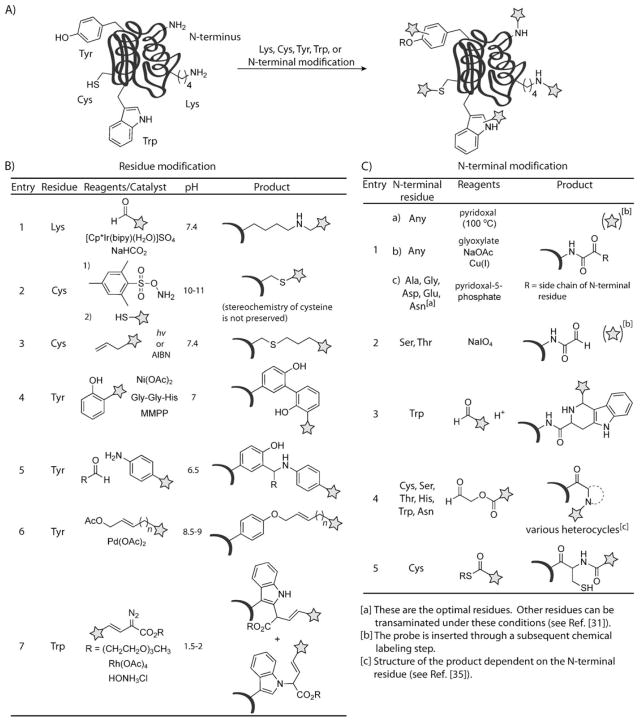
New methods have been developed for the modification of cysteine, lysine, tyrosine, and tryptophan (Scheme 3A). Many of these modern methods involve metal-mediated transformations.[11] Furthermore, the N terminus has emerged as a popular target for protein modification (Scheme 3A). For a more in-depth discussion of chemoselective protein modification methods, the reader is directed to the recent review by Hackenberger and Schwarzer.[12]
Scheme 3.
Modern bioconjugation reactions for protein modification. A) Selective modification of Lys, Cys, Tyr, Trp, and the N terminus. B) Modern methods to modify Lys, Cys, Tyr, and Trp. Lys is modified through a reductive amination using an Ir hydride as the reductant (entry 1; bipy = bipyridyl, Cp* = C5Me5). Cys is modified through a two-step labeling procedure which involves formation of dehydroalanine and subsequent Michael addition of a thiol (entry 2), or the photochemically promoted thiol-ene reaction (entry 3; AIBN = 2,2′-azobisisobutyronitrile). Tyr is modified by a nickel(II)-mediated radical coupling with magnesium monoperoxyphthalate (MMPP) as a stoichiometric oxidant (entry 4), a three-component Mannich reaction with aldehyde and aniline reagents (entry 5), or a palladium-catalyzed π-allylation (entry 6). Trp modification is performed using a rhodium carbenoid (entry 7). C) Methods for modification of the N terminus. Modification of the N terminus is achieved through transamination with aldehydes, oxidation with periodate, a Pictet–Spengler reaction between an N-terminal tryptophan and an aldehyde, formation of a bicyclic lactam with acyl-aldehyde reagents, or native chemical ligation with thioester reagents (entries 1–5, respectively).
2.2.1. Next-Generation Lysine and Cysteine Modification Reactions
While the classic bioconjuation techniques for lysine and cysteine have been widely used, methods for the selective modification of amines and thiols continue to be developed and optimized. McFarland and Francis reported a lysine-specific reductive alkylation reaction that proceeds through an iridium-catalyzed transfer hydrogenation. Unlike the classic reaction based on sodium cyanoborohydride, which requires acidic conditions, the iridium-mediated process proceeds in high yield at pH 7.4 (Scheme 3B, entry 1).[13]
Davis and co-workers have recently developed a two-step method for cysteine modification (Scheme 3B, entry 2).[14] The first step in this procedure is the transformation of cysteine into dehydroalanine by treatment with O-mesityl-enesulfonylhydroxylamine under basic conditions. The dehydroalanine residues then undergo a Michael addition with thiol reagents to yield a thioether linkage. The Michael addition is not stereospecific and, thus, a diastereomeric mixture of modified proteins is produced. Another emerging technique for the modification of cysteine that yields a thioether linkage is thiol-ene chemistry,[15] which involves the addition of a thiol across an alkene by a radical-based mechanism (Scheme 3B, entry 3).[16] The radical species can be generated by standard radical initiators or by irradiation with light,[15] with the latter method displaying greater functional-group tolerance and shorter reaction times.[17] Thiol-ene reactions have been performed on proteins functionalized with either thiols[18] or alkenes,[19] although their use in the direct modification of cysteine residues has yet to be reported, possibly because generation of a protein-associated thiyl radical could lead to unwanted side reactions.
2.2.2. Tyrosine and Tryptophan Modification
Much recent activity has focused on the modification of tyrosine and tryptophan residues, often by employing transition-metal-mediated processes that are compatible with aqueous conditions.[11] These residues are relatively rare on protein surfaces, and thus offer opportunities for controlled single-site modification.[20] The first example of a metal-mediated modification of tyrosine involved the oxidative coupling of two phenol groups. This method was first explored by Kodadek et al., who used a nickel(II) catalyst and a co-oxidant to cross-link two proteins (Scheme 3B, entry 4).[21] Finn and co-workers have since validated this as a bioconjugation method by coupling biotin and alkyne reagents to tyrosine residues on the capsid proteins of a virus particle.[22] Francis and co-workers have also explored the modification of tyrosine residues through a three-component Mannich reaction with aldehydes and anilines (Scheme 3B, entry 5),[20,23,24] as well as through palladium π-allyl chemistry (Scheme 3B, entry 6).[25]
Additionally, Antos and Francis have developed a bio-conjugation reaction for tryptophan, the rarest amino acid, in which they used a rhodium carbenoid that was generated in situ from [Rh2(OAc)4] and a diazo compound (Scheme 3B, entry 7).[26] However, this reaction requires acidic conditions (pH ≈ 2), which may affect the structure of some protein targets.
2.2.3. Chemical Modification of the N Terminus
The N termimus of a protein has unique pH-dependent reactivity and is thus an attractive target for single-site modification.[27] Its decreased pKa value relative to amino groups on lysine side chains renders selective acylation or alkylation possible, although difficult, in the presence of many competing lysine side chains. However, transamination reactions have been particularly successful for selective modification of the N terminus. Transamination of the N terminus dates back to 1956, when it was attempted by Bonetti and co-workers at 100°C, a temperature that often denatures proteins (Scheme 3C, entry 1a).[28] Almost a decade later, Dixon performed a transamination at room temperature by using glyoxylate, catalytic base, and copper(I), which facilitated imine formation between the N terminus and the glyoxylate group (Scheme 3C, entry 1b).[29]
Even with the improvements made by Dixon, the transamination reaction did not receive considerable attention until Francis and co-workers reported a biomimetic transamination that proceeds under physiological conditions without the need for metal or base additives (Scheme 3C, entry 1c).[30] Their method involves condensation of the N-terminal amine with pyridoxal-5-phosphate and subsequent hydrolysis to result in a pyruvamide. The protein can then be further modified through the ketone of the pyruvamide by reaction with hydrazide or aminooxy reagents (Section 4.1). Extensive characterization revealed that the transamination reaction proceeded best when Ala, Gly, Asp, Glu, or Asn occupied the N-terminal position. This reaction also occurred with many other N-terminal residues, but the yields were variable.[31]
Other chemical methods for N-terminal modification rely on a specific residue at the N terminus. For example, N-terminal serine or threonine residues undergo periodate oxidation to form glyoxylamides (Scheme 3C, entry 2).[32] The aldehyde moiety of the glyoxylamide can then be modified with hydrazide or aminooxy reagents (Section 4.1). A Pictet–Spengler reaction can selectively modify N-terminal tryptophan residues with aldehyde probes (Scheme 3C, entry 3).[33] There is also evidence that this reaction can be used to modify N-terminal histidine residues.[34] The advantage of the Pictet–Spengler reaction is that a carbon–carbon bond is obtained between the probe and protein in a one-step procedure, whereas hydrazide/aminooxy-based methods do not form an irreversible linkage between the probe and protein. Additionally, aldehyde probes can be selectively conjugated to proteins containing N-terminal cysteine residues (as well as Ser, Thr, Trp, His, and Asn) to give various heterocycles (Scheme 3C, entry 4).[35] N-Terminal cysteine residues have also been exploited in the highly successful method of protein modification known as native chemical ligation (Scheme 3C, entry 5).
2.2.4. Native Chemical Ligation
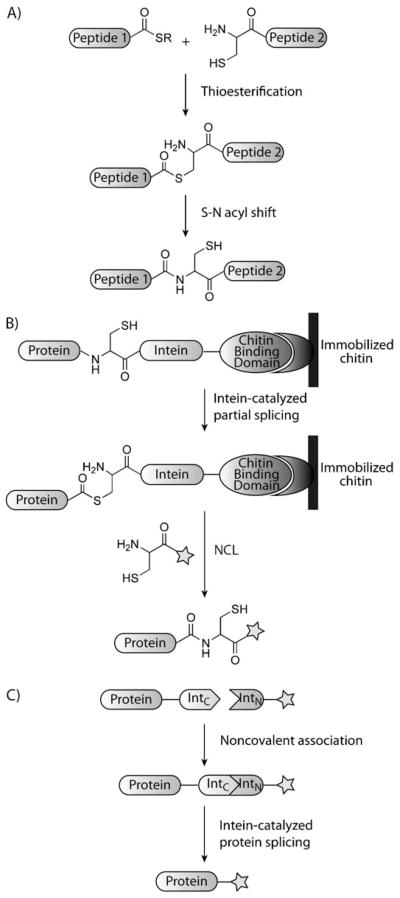
In 1994, Kent and co-workers reported the ligation of thioesters with N-terminal cysteine residues to give a “native” amide bond, a reaction now termed native chemical ligation (NCL, Scheme 4A).[36] Mechanistically, this transformation involves a rapid equilibration of thioesters that is interrupted by an irreversible intramolecular reaction with the N-terminal amine of the protein (an S-N acyl transfer), ultimately forming an amide bond. The S-N acyl transfer was first discovered by Wieland et al. in 1953, but was not applied as a protein modification technique until much later.[37] NCL can be used to selectively ligate two highly functionalized molecules under physiological conditions without the use of protecting groups. As such, NCL has become a powerful method for the modification, synthesis, and semisynthesis of proteins. Through NCL, proteins larger than the traditional limits of solid-phase peptide synthesis have been generated and studied.[38] Furthermore, NCL has allowed for portions of proteins to be isotopically labeled for structural biology studies[39,40] or for the selective addition of posttranslational modifications and chemical probes.[41]
Scheme 4.
Native chemical ligation and intein-based technologies. A) Native chemical ligation of two peptides: Peptide 1 contains a C-terminal thioester that undergoes thioesterification with the N-terminal cysteine of peptide 2. An S-N acyl transfer results in a native peptide bond. B) Expressed protein ligation: A protein is recombinantly expressed and fused to a mutated intein and a chitin-binding domain to facilitate purification. The intein is mutated so that it forms a thioester but does not undergo S-N acyl exchange, thereby allowing for the recombinant protein to be selectively cleaved from the immobilized chitin by a species containing an N-terminal cysteine. C) Protein-trans splicing: A protein is recombinantly expressed fused to a portion of a split intein (IntC). The complementary portion of the intein (IntN) is connected to an unnatural chemical species. When the two inteins associate noncovalently, splicing occurs to give a modified protein.
Many of the applications of NCL have been enhanced by expressed protein ligation (EPL, Scheme 4B) and protein-trans splicing (PTS, Scheme 4C). EPL and PTS both rely on the biological relatives of NCL: self-splicing proteins.[42] Protein self-splicing is a natural phenomenon wherein a domain of a protein, referred to as an intein, is extruded in a posttranslational process that mechanistically mimics NCL. EPL is the ligation of a chemically prepared species (often containing an N-terminal cysteine) with a recombinantly expressed thioester-containing protein. PTS is similar to EPL in that a synthetically generated compound can be joined to a recombinately expressed protein through a native amide bond; however, in PTS the intein is split between the synthetic peptide and the expressed thioester-containing protein, and reassembly of the intein allows for splicing to occur.
EPL exploits inteins as a means to create a C-terminal thioester, which can then be modified through native chemical ligation (Scheme 4B). By using recombinant expression techniques, a desired polypeptide is fused with an intein that has been mutated so that it is unable to undergo S-N acyl transfer. Often a chitin-binding domain is added to the fusion protein on the C-terminal side of the intein to facilitate purification. The expressed fusion protein is isolated on a chitin affinity matrix, and, following the removal of all other proteins, the desired protein is cleaved from the chitin matrix by NCL with a synthetic peptide or other molecule containing an N-terminal cysteine residue.[43,44]
The PTS technique exploits the discovery that inteins can be separated into two polypeptides (IntC and IntN) that, upon noncovalent association, can produce an active intein capable of peptide splicing (Scheme 4C).[42] PTS has allowed for the extension of NCL into living systems, thus facilitating the study of protein–protein interactions,[45] the synthesis of cyclic peptides,[46,47] and the semisynthesis of proteins in vivo.[48] Yao and co-workers have also performed traditional NCL in E. coli to detect overexpressed N-terminal cysteine-containing proteins with thioester fluorophore conjugates.[49] In this study, only the proteins modified with the thioester fluorophore were detected, but there are many natural thioester-containing species that could also react with the proteins of interest, including coenzyme A (CoA) derivatives and polyketide and fatty acid synthases. These naturally occurring thioesters limit NCL primarily to in vitro techniques, such as the preparation of semisynthetic protein samples.
3. Achieving Bioorthogonality with Unique Amino Acid Sequences
Apart from the N terminus, single-site protein modification is difficult to achieve unless a single cysteine, tyrosine, or tryptophan residue can be engineered on the surface of the protein. Even in those cases, undesirable protein dimerization or solubility problems can result. However, several research groups have now demonstrated that combinations of natural amino acid side chains can create new functionalities that are both unique and armed for selective chemical or enzymatic modification. Such sequences can also be genetically encoded, thereby rendering their incorporation into a specific protein of interest straightforward.
3.1. Fluorogenic Biarsenical and Bisboronic Acid Reagents
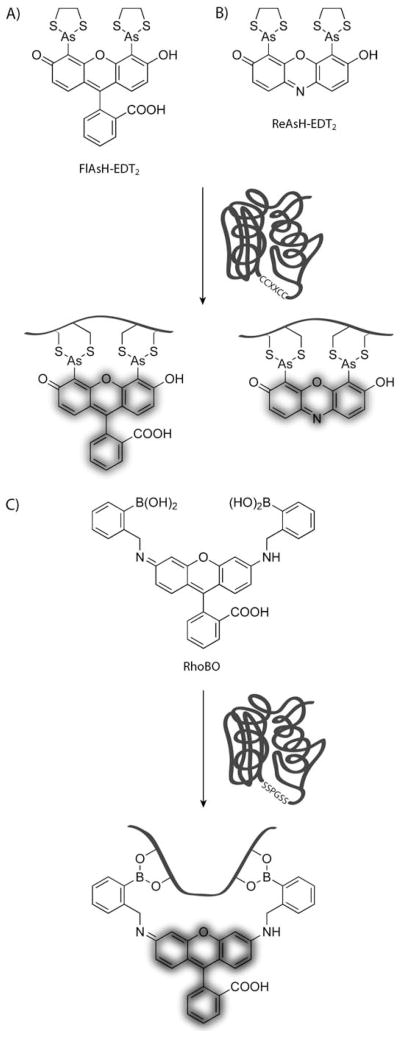
Fluorescent proteins, most notably GFP, have become essential tools for studying the localization, dynamics, and interactions of proteins within live cells and organisms.[50–53] Unfortunately, the large size of these fluorescent proteins (ca. 200 residues) can interfere with the functions of proteins fused to them.[54] This problem motivated Tsien and co-workers to develop an alternative method for labeling proteins with fluorophores that had a smaller genetically encoded tag. They discovered that the tetracysteine motif CCXXCC, when situated in a hairpin structure, reacts selectively with biarsenical-functionalized fluorescent dyes such as FlAsH and ReAsH (Scheme 5A and B, respectively; both shown as their bis(ethanedithiol) (EDT2) adducts).[55–57] Fortuitously, the biarsenical dyes undergo a dramatic enhancement of fluorescence upon binding to the protein. Thus, background fluorescence is low, excessive washing steps are not required, and real-time imaging can be performed.
Scheme 5.
A),B) Fluorogenic biarsenical reagents for site-specific labeling of recombinant proteins with tetracysteine motifs. A) Fluorescein arsenical hairpin binder (FlAsH). B) Resorufin arsenical hairpin binder (ReAsH). C) Fluorogenic bisboronic acid rhodamine reagent (RhoBo) for labeling proteins containing tetraserine motifs.
The biarsenical dyes are not without problems, however. They suffer from nonspecific hydrophobic interactions as well as reaction with other biological thiols, which renders the detection of low-abundance and disperse proteins difficult.[57,58] To overcome these problems, Tsien and co-workers have developed a labeling mixture comprising hydrophobic molecules to compete for nonspecific binding sites and excess ethanedithiol to compete with thiol-containing biomolecules.[59] Additionally, the peptide sequence has been optimized for an enhanced association constant, thus allowing more stringent washing conditions to be used.[60]
These improvements have led to the widespread use of the tetracysteine tag and biarsenical dyes. Applications include studies of mRNA translation,[61] G-protein-coupled receptor activation,[62] amyloid formation,[63] ATP-gated P2X receptors,[64] and transport of HIV-1 complexes.[65] In addition, Schepartz and co-workers used a modified system termed bipartite tetracysteine display to study protein–protein interactions and protein folding.[66] A similar split tetracysteine motif has been employed to study β-sheet formation.[67] Extensions of the method to imaging by electron microscopy,[68] pulse-chase experiments,[61] Western blotting,[69] and affinity chromatography[57] have been performed.
Recently, Schepartz and co-workers have reported that a bisboronic acid rhodamine-based dye (RhoBo, Scheme 5C) binds to tetraserine motifs with a nanomolar Kd value.[70] Similar to FlAsH and ReAsH, RhoBo is fluorogenic and cell-permeable, yet RhoBo does not utilize the cytotoxic element arsenic nor does it suffer from background fluorescence arising from thiol exchange. RhoBo was initially designed as a tool for monosaccharide detection,[71] but it binds monosaccharides with a significantly higher Kd value compared to peptides with SSPGSS motifs, thus allowing RhoBo to selectively label proteins in the presence of carbohydrates.[70] However, some endogenous proteins contain SSXXSS-like sequences that might lead to off-target labeling in cell-based systems.
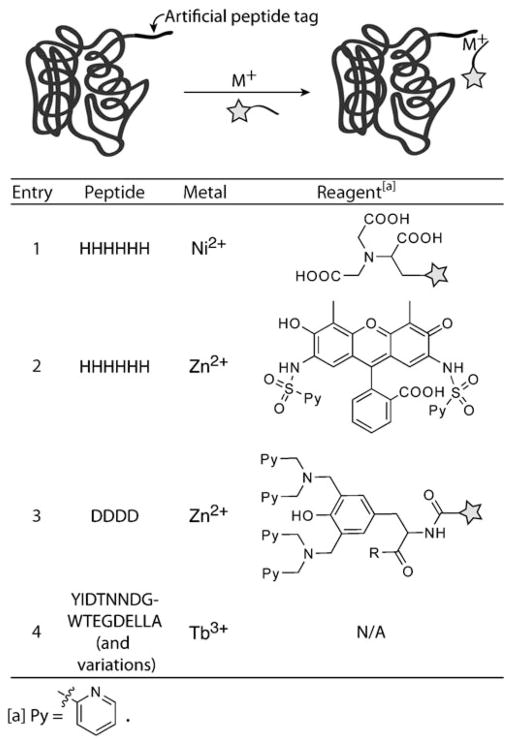
3.2. Peptide Tags Detected through Chelation of Transition Metals
The hexahistidine peptide, a popular purification tag, has been exploited for labeling proteins in cellular systems. The imidazole group chelates nickel nitrilotriacetate (Ni-NTA) with high affinity.[72] Rhodamine derivatives,[73] cyanine dyes,[74] and fluorescein[75] have been conjugated to Ni-NTA and used for imaging proteins containing His6 or His10 sequences (Scheme 6, entry 1). Two shortcomings of the technique derive from its reliance on nickel: the paramagnetic nature of nickel(II) often leads to fluorescence quenching,[76] and nickel(II) can be toxic.[77] Lippard and co-workers have reported a chlorinated fluorescein analogue whose fluorescence is not quenched by nickel(II) ions, thereby addressing the first limitation,[76] but the latter problem still remains. Hauser and Tsien were able to overcome the toxicity and quenching problems associated with polyhistidine chelators by switching from nickel(II) to zinc(II). The fluorescent tag termed HisZiFit (Scheme 6, entry 2) was designed to bind His6 in a ZnII-dependent manner, upon which its fluorescence is activated. This reagent was used to study the stromal interaction molecule STIM1.[78]
Scheme 6.
Site-specific protein modification through the detection of artificial peptide tags by metal-mediated chelation with chemical reagents. Polyhistidine peptides chelate NiII and are detected with Ni-NTA reagents (entry 1) or they chelate ZnII and are imaged with the fluorogenic dye HisZiFit (entry 2). Tetraaspartate peptides are detected with multinuclear zinc complexes (entry 3). Peptides that have been engineered to bind TbIII can be visualized following chelation through the luminescent properties of TbIII (entry 4).
Hamachi and co-workers have also exploited zinc(II) in the development of chelating probes that recognize tetraaspartate sequences (Scheme 6, entry 3).[79] Multinuclear zinc complexes were synthesized, conjugated to fluorescein and cyanine dyes, and used to image the muscarinic acetylcholine receptor in chinese hamster ovary (CHO) cells. Two strategies were explored to engineer fluorescence activation of these zinc complexes upon protein binding. The first involved the use of an Asp4GlyAsp4 tag and a pyrene chromophore. When both tetraaspartate motifs were chelated to the zinc complex, the pyrenes created an excimer complex with altered fluorescent properties.[80] The second method is based on a fluorophore that undergoes a spectroscopic change as a function of the pH value. The tetraaspartate motif created a local acidic environment that was reflected in the fluorescence of the bound dye.[81] More recently, a zinc reagent bearing a chloroacetamide group was designed to alkylate a cysteine residue positioned near the tetraaspartate motif.[82] This reagent extended the use of the dye to Western blotting and affinity-purification applications, which require more robust protein conjugation reactions.
Other metal-based peptide tags have been developed for imaging proteins in complex systems. From library screens, Imperiali and co-workers discovered peptides that form luminescent complexes with terbium(III) (Scheme 6, entry 4).[83,84] These lanthanide-binding sequences were used to detect tagged ubiquitin in cell lysates[85] and to study, through luminescence resonance energy transfer, peptide–protein interactions between phosphopeptides and Src homology 2 domains.[86] So far, the applications of lanthanide-binding tags have focused on in vitro studies because of the cell-impermeability of these metals and their potential toxicity if they are able to cross the cell membrane.[87] However, these issues might be overcome by using chelators that can deliver the metals to their in vivo targets.
3.3. Enzymatic Modification of Peptide Tags
Nature provides a suite of enzymes that covalently modify proteins with small-molecule cofactors, and many of these enzymes recognize short peptide sequences that can be transported into heterologous proteins.[88,89] In some cases, the enzymes will recognize unnatural modifications to their small-molecule substrates, thereby allowing introduction of bioorthogonal functional groups or novel moieties such as biophysical probes. These methods are growing in popularity as a means to selectively modify proteins in live cells.
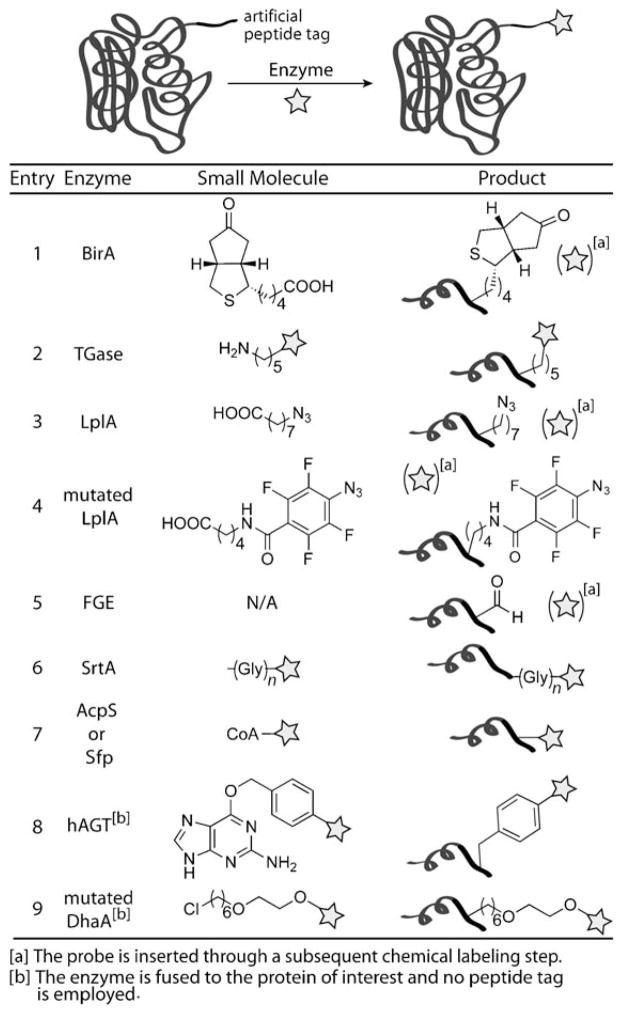
Biotin ligase has been artfully employed by Ting and co-workers for this purpose. The biotin ligase from E. coli, BirA, biotinylates a lysine residue within a 15-residue acceptor peptide (Scheme 7, entry 1) that is orthogonal to the peptide recognized by mammalian biotin ligases.[90] Consequently, mammalian proteins tagged with the BirA recognition motif can be selectively biotinylated and visualized with streptavidin-conjugated quantum dots.[91] Ting and co-workers also demonstrated that BirA can accept a ketone-containing analogue of biotin termed ketobiotin as a substrate.[90] After enzymatic transfer to the protein of interest, the ketobiotin can be covalently labeled with hydrazide or aminooxy compounds (Section 4.1). The tolerance of BirA for unnatural substrates was limited to conservatively modified biotin isosteres. However, ligases from P. horikoshii and yeast were able to catalyze the transfer of azido- and alkynylbiotin analogues to proteins, thus enabling detection by Staudinger ligation or CuAAC (Sections 4.2 and 4.3, respectively).[92]
Scheme 7.
Protein modification through the enzymatic elaboration of peptide tags. Biotin ligase (BirA) catalyzes the attachment of biotin (not shown) or ketobiotin to proteins containing the appropriate peptide substrate (entry 1). Transglutaminase (TGase) catalyzes the attachment of primary amine-containing probes to proteins tagged with polyglutamine sequences (entry 2). Lipoic acid ligase (LplA) or a mutated LplA catalyzes the attachment of an alkyl (entry 3) or aryl azido-lipoic acid derivative (entry 4) to proteins containing the appropriate peptide substrate. The formylglycine-generating enzyme (FGE) catalyzes the transformation of a Cys to a formylglycine in proteins that contain the motif CXPXR (entry 5). Sortase A catalyzes the attachment of a polyglycine-containing probe to proteins that contain an LPXTG motif near the C terminus (entry 6). Phosphopantetheinyl transferases (AcpS or Sfp) catalyze the attachment of a coenzyme A (CoA) probe to proteins containing the appropriate protein/peptide substrate (entry 7). A protein is recombinantly expressed as a fusion with the human repair protein O6-alkylguanine-DNA alkyltransferase (hAGT). The enzyme hAGT is alkylated through removal of the benzyl group and probe from O6-benzylguanosine derivatives (entry 8). A protein is recombinantly expressed as a fusion with an engineered haloalkane dehalogenase (DhaA). The mutated DhaA enzyme is covalently modified by alkyl chloride probes (entry 9).
The success of the biotin ligase method prompted Ting and co-workers to consider other enzyme-mediated strategies to tag proteins. They first focused their attention on transglutaminase (TGase; Scheme 7, entry 2),[93] which had been employed previously for the in vitro modification of glutamine-tagged proteins with amine-conjugated probes.[94,95] Lin and Ting extended its applications to protein labeling on live cells.[93] More recently, Ting and co-workers used lipoic acid ligase (LplA) to site-specifically modify proteins with short-chain azido fatty acids (Scheme 7, entry 3).[96] Additionally, a mutant LplA transferred aryl azides onto proteins for photo-cross-linkng applications (Scheme 7, entry 4).[97] Lipoic acid ligase and biotin ligase have also been used to orthogonally label two independent proteins in the same cell.[96]
Some posttranslational modifications have intrisic orthogonal reactivity that allows for direct labeling of the protein at the modification site. This is the case for the aldehyde-containing formylglycine (FGly) residue formed by the action of the formylglycine-generating enzyme (FGE).[98,99] FGE recognizes a six-residue motif, in which a cysteine residue is oxidized to FGly. Normally found in type I sulfatases, the motif can be transported into heterologous proteins where it is nonetheless recognized by FGE. We have exploited the FGE consensus as a genetically encoded aldehyde tag for site-specific protein modification (Scheme 7, entry 5).[100–102] Coexpression of the tagged protein alongside FGE directly produces the aldehyde-functionalized protein. The aldehyde can be modified by using a variety of methods, such as condensations with aminooxy or hydrazide probes (Section 4.1).[100] While most organisms have endogenous FGE activity, Carrico et al. verified that conversion of cysteine into FGly is enhanced if FGE is overexpressed to result in more enzyme available to oxidize cysteine residues to FGly. The aldehyde tag has been employed to modify proteins expressed in E. coli as well as in mammalian cells, including secreted, cytosolic, and membrane-associated proteins.[103]
Site-specific protein modification has also been accomplished by use of bacterial sortases (Scheme 7, entry 6). Sortase A (SrtA), the most commonly used enzyme, naturally catalyzes the conjugation of proteins to the bacterial cell wall.[104] The enzyme recognizes a peptide sequence (LPXTG) near the C terminus of its target site, cleaves the Thr–Gly bond, and forms an amide bond between the new C-terminal threonine residue and the N-terminal glycine of a polyglycine species.[105] SrtA requires the polyglycine sequence but will tolerate heterologous sequences (or unnatural moieties) beyond the LPXTG motif. SrtA has been used for many in vitro applications, including peptide–, protein–, and carbohydrate–peptide ligations.[106,107] In 2007, Ploegh and co-workers reported the first sortase modification of proteins on live cells.[108] In their report, an MHC H-2Kb protein was modified with a variety of oligoglycine probes containing biotin, fluorescein, tetramethylrhodamine, an aryl azide, and an ortho-nitrophenyl group. In later studies, Nagamune and co-workers used SrtA on live cells to label the extracellular C terminus of the membrane protein ODF with biotin and Alexa Fluor 488. The ligation of GFP to ODF was also demonstrated using SrtA.[109] The benefit of sortase tagging is that there is no observed limitation to the size of the modification introduced, thereby eliminating the need for two-step strategies. However, only C-terminal modifications are possible with this technique.
Cell-surface proteins have been selectively modified using phosphopantetheinyl transferases (PPtases) from E. coli (AcpS) and B. subtilis (Sfp). PPtases catalyze the addition of a CoA-activated phosphopantetheine group to the serine residue of an acyl or peptidyl carrier protein (Scheme 7, entry 7).[110] AcpS and Sfp do not load mammalian carrier proteins, and, therefore, these enzymes and their complementary bacterial carrier proteins can be used orthogonally within a mammalian system.[111] These enzymes are highly promiscuous and can introduce a variety of functionality, including biotin and cyanine dyes, into proteins.[112,113] PPtases were used to study the transport of the transferrin receptor 1[114] and yeast cell-wall protein Sag1.[115] In the latter study, a two-color labeling strategy was employed, which takes advantage of the pseudo-orthogonality of AcpS and Sfp. Sfp is not selective for the type of carrier protein it labels, while AcpS only modifies acyl carrier proteins. Thus, if a cell is exhaustively labeled with AcpS and then subjected to Sfp, two separate proteins may be selectively tagged and simultaneously studied.[115]
The PPtase system suffers from the same problems as GFP, in that the carrier proteins (80–120 residues) can often lead to substantial perturbations of the protein of interest. To overcome this limitation, Walsh and co-workers performed phage-display selections to obtain an 11-residue peptide (termed ybbR) that was selectively modified by Sfp.[116] Furthermore, phage-display screening resulted in two superior peptide sequences: one for Sfp and one for AcpS. The two new peptides (A1 and S6) both contain 12 residues and are modified more efficiently than ybbR. Additionally, these peptides display orthogonal reactivity to each other, as A1 is selective for modification by AcpS and S6 is selective for Sfp.[117] This makes the two-color labeling strategy originally demonstrated by Johnsson and co-workers[115] considerably more straightforward.
Johnsson and co-workers have developed a protein-labeling method that utilizes the human DNA repair protein O6-alkylguanine-DNA alkyltransferase (hAGT),[119] which repairs guanosine residues that are alkylated at the 6-oxo position by transferring the alkyl group to a resident cysteine.[118] These researchers found that when O6-benzylguanosine derivatives are introduced in cells, the benzyl group is readily transferred to hAGT. Thus, if hAGT is fused to a protein of interest, guanosine derivatives can be utilized to specifically label the desired protein (Scheme 7, entry 8).[119,120] This method has been termed “SNAP tag”. A recent adaptation, termed “CLIP tag”, employed an orthogonal enzyme that acts on modified cytosine residues.[121] Lastly, Promega has created a “HaloTag” where a protein of interest is fused to a bacterial haloalkane dehalogenase (DhaA) that has been mutated at the catalytic site to trap the covalent intermediate. The protein of interest can then be tagged using alkyl chloride probes (Scheme 7, entry 9).[122]
4. Bioorthogonal Reactions
Genetically encoded peptide tags have expanded the repertoire of proteins that can be probed in cellular systems. However, the other biomolecules—glycans, lipids, nucleic acids, and various metabolites—are not amenable to such genetically encoded tags. Instead, these biomolecules can be tagged by metabolic labeling with bioorthogonal chemical reporters, namely, functional groups that possess unique reactivity orthogonal to those of natural biomolecules.[123] The process entails two steps. First, cells (or organisms) are incubated with a metabolic precursor adorned with a unique functional group—the chemical reporter. The metabolite could be a monosaccharide for glycan labeling, a nucleoside for DNA labeling, an amino acid for protein labeling, or a fatty acid for lipid labeling. Once the chemical reporter is incorporated into the target biomolecule, it is treated in a second step with a probe molecule bearing complementary bioorthogonal functionality.
Exquisite selectivity of the chemical reporter and probe molecule is critical for execution of this method of biomolecule labeling, but equally important is the intrinsic kinetics of the chemical reaction. Most reactions used to selectively label biomolecules follow second-order kinetics, and consequently, their rates depend on the concentrations of the two reactive components and the second-order rate constant. While the concentrations of the labeled species can be controlled to some extent in in vitro settings, the labeled biomolecules are often at low concentrations in vivo. Biological labeling agents such as monoclonal antibodies typically bind their antigens with biomolecular rate constants that approach the diffusion limit (ca. 109M−1s−1).[124] Consequently, such reagents can be used at very low concentrations and still bind to their targets at reasonable rates. By contrast, most second-order chemical reactions have rate constants that are 8–15 orders of magnitude lower than this;[125] the reactions discussed in this section have rate constants ranging from 10−4 to 103M−1s−1.[126,127] These rate constants necessitate the use of relatively high concentrations (often high micromolar to millimolar) of secondary reagent when employing bioorthogonal chemical reactions in vivo; a parameter that may require consideration of the solubility and toxicity when designing the labeling reagent. This point highlights the importance of optimizing the intrinsic kinetics of the bioorthogonal reaction as a means of reducing the concentrations required for in vivo labeling.
In Section 4 we discuss the reactions developed for use in the bioorthogonal chemical reporter strategy, and then in Section 5 applications to specific biomolecule classes will be covered. The reactions include condensation of aldehydes and ketones with aminooxy and hydrazide probes (Section 4.1), the Staudinger ligation of triarylphosphines and azides (Section 4.2), and various reactions of azides and alkynes (Section 4.3). Bioorthogonal reactions involving alkenes are emerging (Section 4.4), although their use in complex biological systems is still on the horizon.
4.1. The Condensation of Ketones/Aldehydes with Amine Nucleophiles
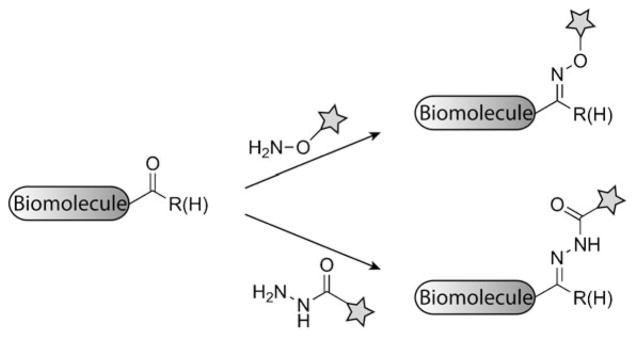
Ketones and aldehydes react with amine nucleophiles that are enhanced by the α effect. Prototypical examples are aminooxy and hydrazide compounds, which form oxime and hydrazone linkages, respectively, under physiological conditions (Scheme 8). While biological nucleophiles—amines, thiols, and alcohols—also react with ketones and aldehydes, the equilibrium in water generally favors the carbonyl compound.[128] Accordingly, ketones and aldehydes have a rich history in the field of protein modification.[100,129–131] These carbonyl compounds have not been widely employed for labeling biomolecules inside cells or within live organisms, in part because of competition with endogenous aldehdyes and ketones, including those in glucose and pyruvate (although notably, Schultz and co-workers reported the intracellular labeling of a ketone-functionalized protein).[132] However, aldehydes and ketones are absent from cell surfaces and in this environment they can serve as unique chemical reporters. For example, we demonstrated that certain keto sugars are metabolized by cells and integrated into cell-surface glycans, where they can be treated with aminooxy and hydrazide probes.[133,134] Paulson and co-workers introduced aldehydes into cell-surface sialic acid residues by mild periodate oxidation and then captured the modified glycoproteins by reaction with aminooxybiotin followed by streptavidin chromatography.[135] In this case, the use of aniline as a catalyst[135,136] accelerated the reaction under neutral conditions, an improvement over the typical acidic conditions used for oxime formation.[128] As described above, both biotin ligase[90] and aldehyde tags[100] have been used to label cell-surface proteins with ketones and aldehydes, respectively.
Scheme 8.
Bioorthogonal reactions of aldehydes/ketones. Aldehydes and ketones can condense with aminooxy compounds (top) or hydrazide compounds (bottom) to form stable oxime or hydrazone linkages, respectively.
4.2. The Staudinger Ligation of Azides and Triarylphosphines
Azide has proven to be a particularly powerful chemical reporter group. Unlike aldehydes and ketones, azide is totally absent from biological systems,[137] and it also possesses orthogonal reactivity to the majority of biological functional groups. Importantly, the azide group is small[138–140] and therefore only minimally perturbs a modified substrate. These favorable properties went unexploited in biological settings, however, until a suitable reactive partner for azide was developed. In 2000, azide made its debut as a chemical reporter group with the development of the Staudinger ligation.[141]
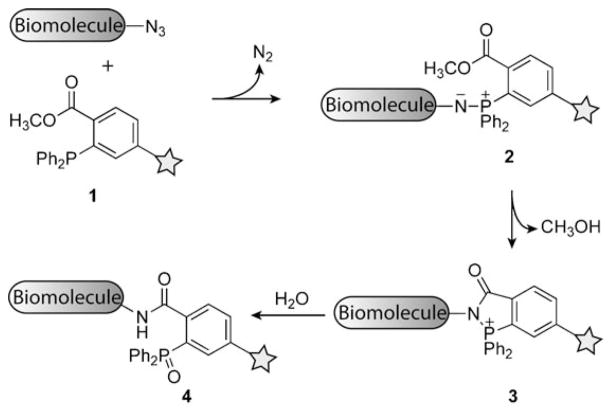
The Staudinger ligation is a modification of the classic Staudinger reduction of azides with triphenylphosphine.[142] As shown in Scheme 9, strategic placement of an ester group on one of the phosphine’s aryl substitutents (1) allows an intermediate aza-ylide (2) to undergo intramolecular formation of an amide bond (3). This step is a central feature of the reaction, because otherwise the aza-ylide intermediate would simply hydrolyze to afford the corresponding amine and phosphine oxide. However, hydrolysis of intermediate 3 produces a stable ligation product (4), which includes the phosphine oxide within its structure.
Scheme 9.
The Staudinger ligation of azides and triarylphosphines. Triarylphosphine 1 attacks the azido biomolecule, thereby releasing nitrogen from a four-coordinate transition state to yield aza-ylide 2, which undergoes intramolecular attack on the ester, extruding methanol, and resulting in bicycle 3. Upon hydrolysis, oxidation of the phosphine and formation of an amide bond occur to give ligation product 4.
The bioorthogonality of the reagents used in the Staudinger ligation warrants some discussion. Azides and phosphines have potential cross-reactivity with thiols and disulfides, respectively. Thiols are capable of reducing alkyl and aryl azides, especially under basic conditions, and dithiols such as dithiothreitol are particularly reactive in this regard.[143–145] Although the reduction of alkyl azides is quite slow at physiological pH, there is evidence that cytosolic glutathione can reduce the azide in 3′-azidothymidine.[146] We addressed this potential problem in the context of labeling glycans with azido sugars.[147] Jurkat cells bearing azido sugars in their cell-surface glycans were treated with a Staudinger ligation reagent or tris(2-carboxyethyl)phosphine (TCEP)—a trialkylphosphine known to readily reduce disulfides[148]—followed by an amine-reactive biotin probe. The quantity of amines on the cell surface was subsequently measured by incubation with a fluorescent avidin reagent followed by flow cytometry analysis. Cells labeled with azido sugar displayed slightly higher levels of cell-surface amines than unlabeled cells, which suggests that some azides had been reduced in situ. However, the quantity of cell-surface amines was significantly higher for cells treated with TCEP, thus indicating that the majority of the azides were not reduced by cellular thiols.
In principle, phosphines can reduce disulfide bonds.[148,149] However, triarylphosphines are generally not capable of reducing alkyl disulfides (namely, those found in biological systems) under physiological conditions.[150] We confirmed this observation in a biological setting by treating Jurkat cells with a triarylphosphine and quantifying the amount of free sulfhydryl groups on the cell surface by using a thiol-specific biotin reagent.[141] No increase in free sulfhydryl groups was observed. By contrast, TCEP produced a marked increase in free sulfhydryl groups.
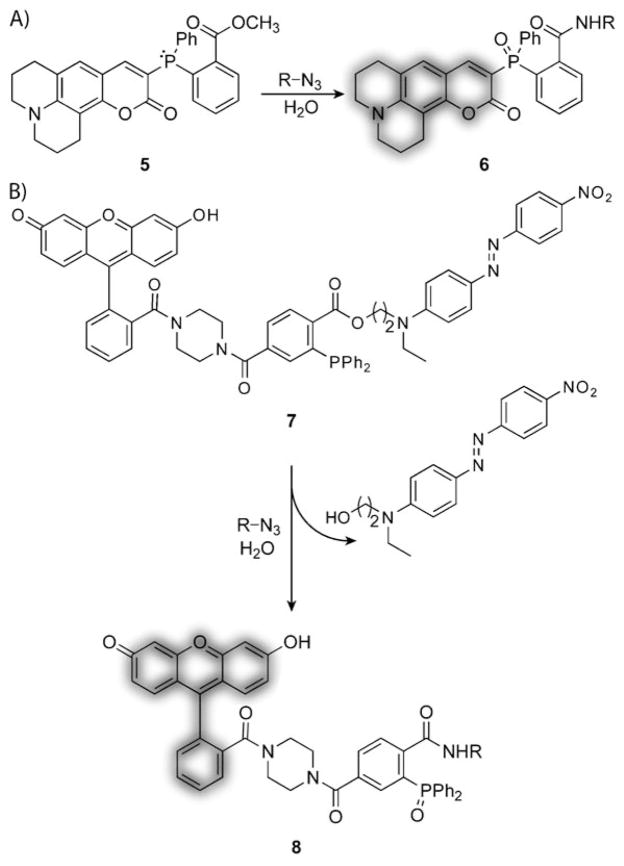
An appealing feature of the Staudinger ligation is that its mechanism lends itself to the invention of fluorogenic reagents for the real-time imaging of biomolecules. In 2003, Lemieux et al. reported a fluorogenic phosphine in which one of the aryl rings was replaced with a coumarin dye (5, Scheme 10A).[151] The fluorescence of 5 was quenched by the lone pair of electrons on the phosphorus atom. Phosphine oxidation during the course of the Staudinger ligation relieved the quenching effect, thereby producing a highly fluorescent biomolecule-bound product (6). One problem with this “smart” probe was background fluorescence caused by low levels of oxidation of the phosphine in air. To overcome this problem, we exploited another step of the Staudinger ligation—ester cleavage.[152] As shown in Scheme 10B, incorporation of a fluorescence resonance energy transfer (FRET) quenching group at the ester position in 7 provided an alternative means of fluorescence activation upon Staudinger ligation that was not sensitive to phosphine oxidation.
Scheme 10.
Fluorogenic phosphines for the Staudinger ligation. A) Coumarin-based fluorogenic phosphine that becomes fluorescent upon phosphine oxidation. B) FRET-based fluorogenic phosphine that becomes fluorescent upon release of the quencher.
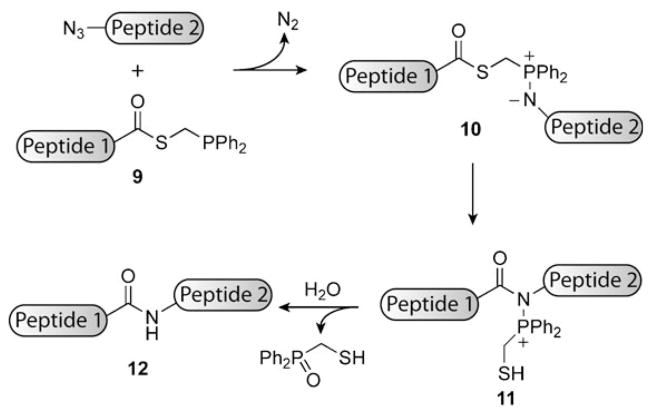
The Staudinger ligation has been adapted for applications beyond biomolecule labeling, most significantly for protein synthesis. “Traceless” versions of the Staudinger ligation have been developed to produce amide bonds without inclusion of the phosphine oxide moiety.[153] Raines and co-workers merged this concept with thioester chemistry (reminiscent of NCL) to develop a traceless Staudinger ligation for peptide coupling (Scheme 11).[154–156] This traceless Staudinger-mediated peptide coupling involves the attack of a peptide containing a C-terminal phosphinothioester (9) with an azide-labeled peptide. Attack of the phosphine in 9 on the azide results in iminophosphorane 10, which rearranges to 11. Hydrolysis of 11 facilitates the ligation of the two original peptides through formation of a native amide bond (12). Unlike the standard NCL process, the method of Raines and co-workers does not require the presence of a cysteine residue at the ligation site.[154,155] Additional reagents have been synthesized for the traceless Staudinger ligation, including an ester-linked version.[157] Optimization of the phosphine reagents for steric, electronic, and coulombic factors has also been performed.[158,159]
Scheme 11.
Peptide coupling by the traceless Staudinger ligation. The phosphine appended to peptide 1 through a C-terminal thioester (9) attacks the azide attached to peptide 2 in a manner analogous to the original Staudinger ligation to yield iminophosphorane 10, which rearranges to 11. Hydrolysis of 11 results in coupling product 12 plus a phosphine oxide by-product.
The exquisite bioorthogonality of azides and triarylphosphines has enabled the use of the Staudinger ligation for probing biomolecules in many cellular environments, as well as within living animals.[147,160] However, like any reaction, the Staudinger ligation is not without limitations. It suffers from relatively slow reaction kinetics, necessitating high concentrations of a triarylphosphine (>250 μM). A series of kinetic studies determined that the Staudinger ligation displays second-order kinetics in reactions with alkyl azides (k = 10−3M−1s−1), which indicates that the rate-determining step is the attack of the phosphine on the azide.[161] Unfortunately, all efforts to improve the kinetics of the Staudinger ligation by increasing the nucleophilicity of the phosphine reagents (such as by addition of electron-donating groups to its aryl substituents or replacement of aryl with alkyl substituents) also resulted in increased susceptibility to phosphine oxidation in air, which subverts the desired ligation.[162] As discussed above, the kinetics can be critical to the success of in vivo biomolecule labeling studies. Consequently, there has been considerable interest in developing faster azide reactions.
4.3. Reactions of Azides and Alkynes
An alternate mode of reactivity for the azide is its participation as a 1,3-dipole in a [3+2] cycloaddition with alkenes and alkynes. This reaction, first reported at the end of the 19th century,[163] has been proposed to proceed by a concerted cycloaddition since the 1950s, when Rolf Huisgen introduced the concept of 1,3-dipolar cycloadditions.[164] However, the high temperatures or pressures required to promote the cycloaddition of azides and most dipolarophiles are not compatible with living systems.[165] Nevertheless, the potential of this transformation, especially the cycloaddition of azides and alkynes to form aromatic triazole products (ΔG° ≈ −61 kcalmol−1),[166] was too great for it to be overlooked.[167]
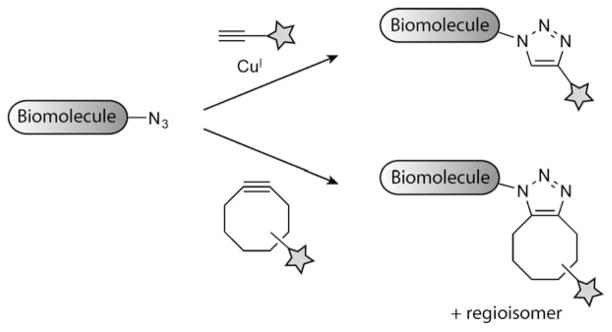
In separate efforts, the Sharpless and Meldal research groups discovered that the formal 1,3-dipolar cycloaddition of azides with terminal alkynes to produce 1,4-disubstituted 1,2,3-triazoles could be effectively catalyzed by copper(I) (Scheme 12).[168,169] This reaction, now termed the copper-catalyzed azide–alkyne 1,3-dipolar cycloaddition (CuAAC), takes advantage of the formation of a copper acetylide to activate terminal alkynes toward reaction with azides. The copper(I)-catalyzed cycloaddition proceeds roughly seven orders of magnitude faster than the uncatalyzed cycloaddition,[166] and the reaction can be further accelerated by the use of specific ligands for copper(I).[170,171] CuAAC has all the properties of a click reaction (including efficiency, simplicity, and selectivity), as defined by Sharpless and co-workers.[172] In fact, it has become the quintessential click reaction and is often refered to simply as “click chemistry”. CuAAC has gained widespread use in organic synthesis, combinatorial chemistry, polymer chemistry, materials chemistry, and chemical biology.[173–182] The formal cycloaddition between azides and terminal alkynes can also be catalyzed by ruthenium(II) to obtain 1,5-disubstituted 1,2,3-triazole products,[183] but this reaction is used far less frequently than CuAAC. The first report of CuAAC as a bioconjugation strategy was demonstrated by Finn and co-workers through the attachment of dyes to cowpea mosaic virus.[184]
Scheme 12.
Bioorthogonal [3+2] cycloadditions of azides and alkynes to form triazoles. Terminal alkynes are activated by CuI to undergo cycloaddition with azides under physiological conditions (top). Cyclooctynes react with azides through a strain-promoted [3+2] cycloaddition (bottom).
To date, the use of CuAAC in living systems has been hindered by the toxicity of copper(I).[185] Bacterial and mammalian cells as well as zebrafish embryos have been subjected to click chemistry conditions. E. coli expressing protein-associated azides have been labeled with 100 μM CuBr for 16 h and survived the initial labeling, but were no longer able to divide.[186,187] Similarly, mammalian cells can survive low concentrations (below 500 μM) of copper(I) for 1 hour.[162] However, considerable cell death[162] occurs when optimized CuAAC conditions that require 1 mM copper(I) are employed.[188] Zebrafish embryos exhibited a similar sensitivity to copper(I). When the embryos were treated with 1 mM CuSO4, 1.5 mM sodium ascorbate, and 0.1 mM tris(benzyltriazolylmethyl)amine ligand,[171] all the embryos were dead within 15 minutes.[162] Thus, as presently formulated, CuAAC is of limited use for labeling biomolecules in living systems.
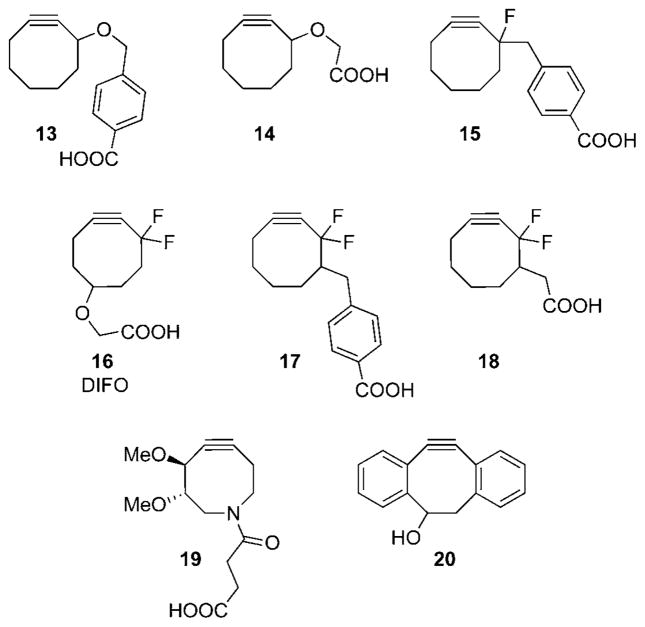
To improve upon the biocompatibility of the azide–alkyne cycloaddition, we sought to activate alkynes by a method other than metal catalysis, namely by ring strain (Scheme 12, bottom). The roots of the strain-promoted azide cycloadditions precede the Huisgen era and date back to when Alder and Stein discovered that dicyclopentadiene reacted considerably faster than cyclopentadiene in reactions with azides.[189,190] Studies on strained alkenes and alkynes continued through the 1960s, and during this time, Wittig and Krebs reported that cyclooctyne, the smallest stable cycloalkyne, reacted “like an explosion” when combined with phenylazide.[191] Building on this classic literature, we synthesized a biotin conjugate of cyclooctyne 13 and demonstrated that it labeled azides effectively within cell-surface glycans with no apparent cytotoxic effects.[192] This study laid the foundation for the investigation of a series of cyclooctynes (Scheme 13, 14–20) that enable the detection of azides in living systems through the strain-promoted [3 +2] cycloaddition.[193–196]
Scheme 13.
Cyclooctyne reagents for copper-free click reactions.
Still, the first-generation strain-promoted cycloaddition was no faster than the Staudinger ligation (k = 10−3M−1s−1)[192] and considerably slower than CuAAC. With the aim of improving the kinetics of the process, a series of compounds bearing electron-withdrawing fluorine atoms at the propargylic positions were investigated. The addition of one fluorine atom (cyclooctyne 15) modestly increased the rate (ca. 4-fold)[194] but a gem-difluoro group afforded a dramatic 60-fold enhancement (k = 10−1M−1s−1).[195] This difluorinated cyclooctyne (16, abbreviated DIFO) demonstrated comparable kinetics to CuAAC in biomolecule labeling experiments, which prompted us to dub its reaction with azides “copper-free click chemistry”. DIFO–fluorophore conjugates have now proven to be exceptional reagents for imaging azide-labeled biomolecules within complex biological systems, including live cells,[195] C. elegans,[162] and zebrafish embryos,[197] with very low background fluorescence. However, background labeling is sometimes observed in protein labeling experiments analyzed by Western blot, perhaps because of nonspecific hydrophobic interactions or as yet uncharacterized reactions with protein functionalities.[162]
Since the report of DIFO, a number of new cyclooctyne reagents have been synthesized. Two more synthetically tractable DIFO analogues 17 and 18 have been synthesized, and will hopefully make copper-free click chemistry more accessible to researchers in the area of chemical biology. We have also synthesized a more water-soluble azacyclooctyne (19) that was designed to improve other attributes of the cyclooctyne reagents such as pharmacokinetic properties.[196] Boons and co-workers have reported the use of dibenzocyclooctyn-ol 20 in copper-free click reactions.[198] This reagent is nontoxic, has reaction kinetics similar to DIFO, and is synthetically quite accessible. Recent theoretical studies by Houk and co-workers have provided frameworks in which cyclooctyne reactivity can be predicted and optimized.[199] This theoretical basis may enable the design of more reactive congeners for synthetic pursuit.
4.4. Bioorthogonal Reactions with Alkenes
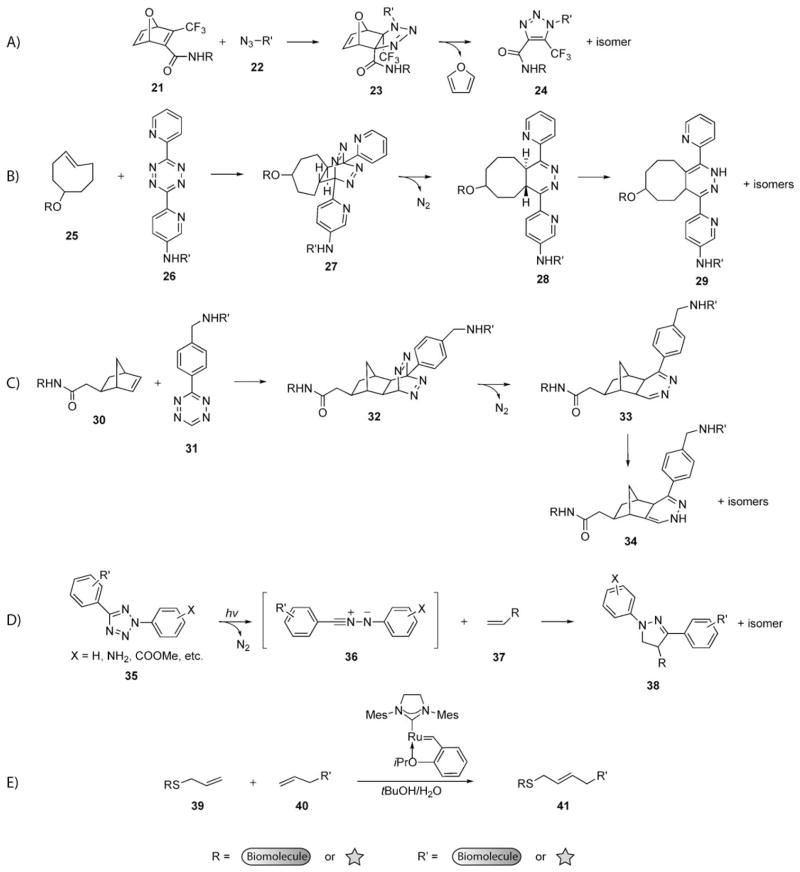
Alkenes have also been used with 1,3-dipoles and dienes in cycloadditions promoted by ring strain or light (Scheme 14). The product of an azide–alkene cycloaddition is a triazoline, which is relatively unstable compared to an aromatic triazole and is not advantageous for applications where a ligation product is desired.[167] Rutjes and co-workers circumvented this problem by using oxanorbornadienes containing electron-deficient olefins (21) as substrates. The oxanorbornadiene underwent a [3+2] cycloaddition with an azide (22) to produce 23, which then proceeded through a Diels–Alder reaction to extrude furan and yield triazole product 24 (Scheme 14A).[127,200] These reagents are straightforward to synthesize, but they display relatively slow reaction rates (k =10−4M−1s−1).
Scheme 14.
Bioorthogonal reactions with alkenes. A) Reaction of oxanorbornadienes and azides to yield triazoles. B) Inverse-demand Diels–Alder reaction of dipyridyl tetrazines and trans-cyclooctenes. C) Inverse-demand Diels–Alder reaction of monoaryl tetrazines and norbornenes. D) Photoinduced 1,3-dipolar cycloaddition of tetrazoles and alkenes. E) Ruthenium-catalyzed cross-metathesis of two alkenes in water.
By contrast, the reaction of tetrazines with strained alkenes as reported by Fox and co-workers is very fast.[126] They reported the inverse-electron demand Diels–Alder reaction of trans-cyclooctene 25 with dipyridyltetrazine 26 to form ligation product 29 (Scheme 14B). The reaction proceeds via intermediate 27, which rapidly loses N2 to yield intermediate 28, which isomerizes to the final ligation product 29. This reaction proceeds very rapidly in water (k = 103M−1s−1). Independently, Hilderbrand and co-workers developed the reaction of norbornene 30 and tetrazine 31 (Scheme 14C).[201] This reaction proceeds by the same mechanism; however, it does not occur as rapidly as the tetrazine ligation of Fox and co-workers. It is important to note that normal-demand Diels–Alder reactions have been used in the context of bioconjugation reactions, but they often require dienophiles that are also Michael acceptors (for example, maleimides).[202–205] Thus, competing Michael additions from biological nucleophiles severely limit the selectivity of these reactions.
Lin and co-workers have developed a photochemical 1,3-dipolar cycloaddition between diaryl tetrazoles 35 and simple alkenes (Scheme 14D) to form pyrazolines.[206] The first report of the reaction required light with a wavelength of 302 nm to produce the nitrile–imine dipole 36. In an effort to reduce potential photodamage to living systems, Lin and coworkers have modified the aryl groups on the tetrazole so that the reaction can occur with irradiation at a wavelength of 365 nm.[207] This reaction does not require an activated alkene, which makes its application to living systems more facile, since techniques for metabolic labeling of biomolecules with simple alkenes are well precedented (see Section 5). In fact, alkene-containing proteins in E. coli have been modified with diaryl tetrazoles.[208] An additional advantage of this reaction is that the resulting pyrazoline cycloadducts (38) are fluorescent.
Chemoselective modification of alkenes by cross-metathesis is emerging as a bioorthogonal reaction. Ruthenium-catalyzed olefin metathesis is remarkably tolerant of functional groups and has been employed with biomolecule substrates for some time, usually in organic solvents.[209–212] Over the past few years, multiple research groups have sought to develop water-soluble olefin metathesis catalysts.[213–216] Cross-metathesis with these catalysts has been particularly challenging and only a few substrates have been successful. Recently, Davis and co-workers have modified proteins containing allyl sulfide groups (39) through cross-metathesis in a tert-butanol/water mixture by using the Hoveyda–Grubbs second-generation catalyst (Scheme 14E).[217] The high selectivity and functional-group tolerance of cross-metathesis, coupled with the ease of introduction of alkenes into biomolecules, render this technique well-poised for further application in biology.
5. Applications of Bioorthogonal Chemical Reactions
The bioorthogonal chemical reporter strategy requires integration of one reactive component into target biomolecules within cells or organisms. Proteins, glycans, lipids, and nucleic acids have all been adorned with bioorthogonal functional groups in a global or site-selective fashion.
5.1. Proteins
5.1.1. Metabolic Labeling (Residue-Specific Modification)
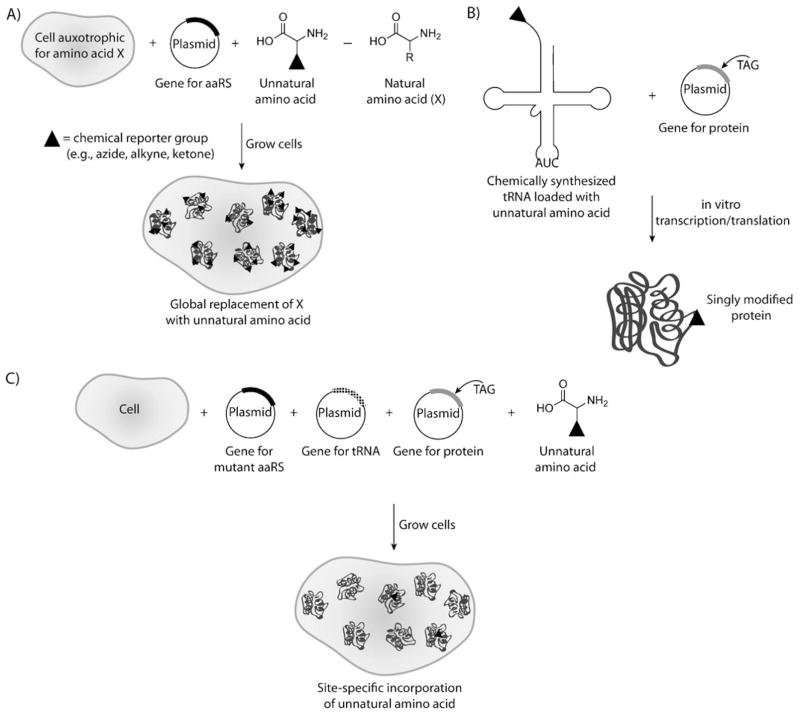
A straightforward method of introducing chemical reporters into cellular proteins, pioneered by Tirrell and co-workers, is to simply subject cells to an unnatural amino acid that is tolerated by the translational machinery, particularly the aminoacyl-tRNA synthetases (aaRSs; Scheme 15A).[218–221] The concept dates back to the 1950s, when methionine residues were shown to be replaced by their selenium analogues after the addition of selenomethionine to methionine-depleted growth media.[222] We now know that surrogates for methionine, leucine, tryptophan, or phenylalanine can be incorporated into proteins expressed in E. coli, although reports can be found for replacement of almost any amino acid with an unnatural derivative. The yields are optimal when the E. coli strain is rendered auxotrophic for the amino acid being targeted for replacement, and over-expression of the required aaRS can be helpful as well.
Scheme 15.
Incorporation of unnatural amino acids into proteins. A) Global incorporation of an unnatural amino acid using auxotrophic cell lines and promiscuous aminoacyl tRNA synthetases (aaRSs). B) Site-specific incorporation of an unnatural amino acid in vitro by using a transcription/translation system, a chemically synthesized tRNA loaded with the unnatural amino acid, and the gene for the protein of interest mutated at the site of modification. C) Site-specific incorporation of an unnatural amino acid in vivo using mutant tRNA and aaRS as well as the gene for the protein of interest mutated at the site of modification.
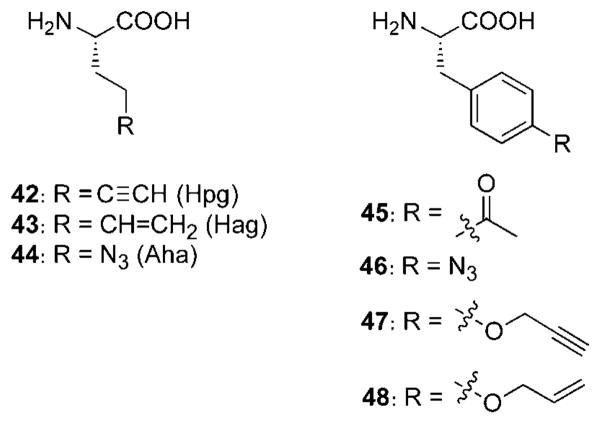
By using this method, a variety of bioorthogonal functional groups have been incorporated into proteins, both in E. coli and in mammalian cells. For example, the methionine surrogates homopropargylglycine (42, Hpg), homoallylglycine (43, Hag), and azidohomoalanine (44, Aha) were used to introduce alkynes, alkenes, and azides, respectively, in proteins with good efficiency (Scheme 16).[187,223] Hpg was employed to label newly synthesized proteins with an azidocoumarin dye by CuAAC in bacterial[224] and mammalian[225] systems. Similarly, Aha has been used to interrogate newly synthesized proteins by labeling with CuAAC,[186] Staudinger ligation,[226] and, more recently, with cyclooctyne probes.[227] The method has also been applied to proteomic analysis of newly synthesized proteins, a process termed bioorthogonal noncanonical amino acid tagging (BONCAT).[228] Moreover, Aha and Hpg have been used together to image two distinct protein populations simultaneously.[229] Azides and alkynes have also been installed within viruslike capsids by replacing methionine residues with Aha and Hpg.[230] Preliminary data indicate that allylcysteine can also be incorporated as a methionine surrogate and then potentially modified by cross-metathesis.[217]
Scheme 16.
Selected amino acids that have been site-specifically incorporated into proteins in vivo.
Some unnatural amino acids are too structurally dissimilar from their native relatives for recognition by natural aaRS enzymes. In such cases, the aaRSs can be mutated to accept the unnatural substrate, either by rational structure-based[231,232] or selection-based methods.[233] Such strategies have been used to obtain a mutant phenylalanine tRNA synthetase (ePheRS)[231] and incorporate a ketone-bearing amino acid para-acetyl-phenylalanine (45) into proteins.[231,234] The same ePheRS has also been used to incorporate para-azido-, para-bromo-, and para-iodophenyl-alanines into proteins.[235,236] The azide allowed for modification with phosphines or alkynes, while the halogen derivatives were modified by using palladium-catalyzed cross-coupling methods.[237,238]
5.1.2. Genetic Encoding (Site-Specific Modification)
Site-specific introduction of unnatural amino acids into proteins was first reported 20 years ago as an in vitro technique,[239,240] and it has since been extended, primarily through the work of Schultz and coworkers, to in vivo applications.[241] This method utilizes the codon UAG (the amber nonsense stop codon), which normally directs termination of protein synthesis, to instead encode an unnatural amino acid loaded onto a complementary tRNA. The tolerance of the ribosome for unnatural amino acids allows for incorporation into proteins during normal translation (Scheme 15B,C).
To integrate unnatural amino acids into proteins in an in vitro setting (Scheme 15B), the gene encoding the protein of interest is first mutated so that the amber stop codon is situated at the desired modification site, and all other amber stop codons are removed from the sequence. A tRNA is synthesized that contains a complementary anticodon as well as a covalently ligated unnatural amino acid at the 3′ end. The addition of this artificial tRNA and the gene encoding the mutated protein to an E. coli in vitro transcription/translation system generates the modified protein.[239] Many unnatural amino acids have been incorporated into proteins by using this technique; however, it suffers from low yields and considerable labor is involved in synthesizing the amino-acylated tRNA.[242] Despite these limitations, the in vitro technique has been used to study a variety of proteins. Residues in α helices and β sheets were replaced with ester analogues to explore the contributions of backbone amide bonds in protein structure.[243–245] In addition, the contribution of cation–π interactions to protein stability was analyzed by using unnatural amino acids.[246] A notable extension of the system has been microinjection of engineered mRNA and aminoacylated tRNA into Xenopus oocytes.[247,248] Dougherty, Lester, and co-workers have used this approach to study the mechanism of neuroreceptors through electrophysiology,[333] and to this end more than 100 unnatural amino acids have been incorporated into Xenopus ion channels. The in vitro method coupled with microinjection is ideally suited for these applications because of the very low levels of protein that can be detected using electrophysiology.
To overcome the limitations of the in vitro method, Schultz and co-workers have created an in vivo system for site-specific mutagenesis of unnatural amino acids (Scheme 15C). The breakthrough was the selection of orthogonal tRNA and aaRSs that recognized the amber stop codon and unnatural amino acid, respectively.[241] Expression of the corresponding genes in a heterologous host together with the gene encoding the desired protein with the amber mutation produced the modified protein. Typically, the cell-culture media is supplemented with the unnatural amino acid, but an E. coli strain engineered to produce para-aminophenylalanine was able to incorporate this “21st amino acid” by total biosynthesis.[249] The extension of in vivo unnatural amino acid mutagenesis to yeast[250,251] and mammalian cells[252–254] has also been achieved.
Dozens of unnatural amino acids have been incorporated into proteins by using the in vivo amber stop codon method; most are based on an aromatic (tyrosine or phenylalanine) core.[241] Amino acids containing azides (44, 46),[255] alkynes (42, 47),[256] ketones (45),[132,257] and alkenes (43, 48)[258] have all been incorporated for further reaction with bioorthogonal chemical reporters (Scheme 16). Other functional groups installed into proteins include anilines,[259,260] aryl halides,[261] boronic acids,[262] photoisomerizable and cross-linking groups,[263,264] post-translational modifications,[265,266] caged versions of amino acids to allow the masking of putatively important residues,[267–269] and even whole fluorophores.[270,271]
5.1.3. Activity-Based Labeling of Enzymes
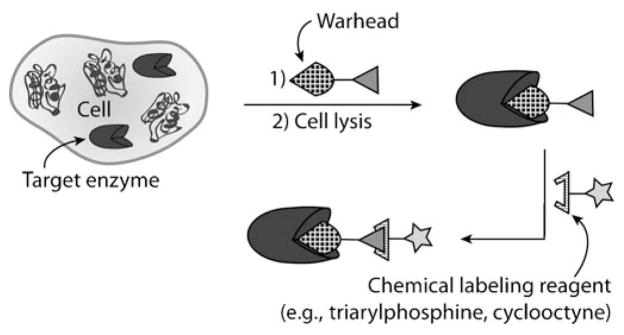
Activity-based protein profiling (ABPP) allows for the study of specific classes of enzymes based on their catalytic mechanism, often through the use of large molecules which contain a probe (fluorophore or biotin) conjugated to a functional group designed to react with residues in the target enzyme’s active site (a mechanism-based inhibitor). The bioorthogonal chemical reporter strategy has improved ABPP by eliminating the need for a large chemical probe during the covalent labeling process (Scheme 17).[272,273] In its initial form, ABPP made use of an electrophilic group (known as a warhead)[239] that covalently modifies the targeted class of enzymes and is conjugated to an affinity probe such as a biotin or a fluorophore. While useful for tagging active enzymes from cell lysates, such large probes often had poor pharmacokinetic properties that prevented their use in vivo.[274] Consequently, enzyme activities could not be studied in their native environments.
Scheme 17.
Activity-based protein profiling using a bioorthogonal chemical reporter. A warhead group is functionalized with a chemical reporter and introduced into cells. Following cell lysis, a chemical labeling reagent facilitates identification or visualization of the modified enzymes.
In 2003, the research groups of Cravatt and Overkleeft independently reported the application of bioorthogonal reactions to ABPP. Cravatt and co-workers used an azide-functionalized phenyl sulfonate that targeted serine hydrolases, while Overkleeft and co-workers used an azide-functionalized vinyl sulfone that targeted proteasomes.[188,275] The azide probes were introduced into live cells, where the target enzymes were covalently modified. Cell lysates were generated and subsequently treated with an alkyne-rhodamine probe through CuAAC ligation (Cravatt) or with a biotinylated triarylphosphine probe through a Staudinger ligation (Overkleeft). Cravatt and co-workers further showed that this two-step procedure allowed the extension of ABPP to live mice.[188] Since the original report, Cravatt and coworkers have used this strategy to profile breast cancer cells[276] and cytochromes P450,[277] and have also determined that the copper(I)-catalyzed labeling procedure is more effective when the alkyne is attached to the electrophilic trap and detection is performed with an azido probe.[276] Additionally, Ploegh and co-workers have used an azido-epoxide warhead, a triarylphosphine-biotin probe, and streptavidin-Alexa Fluor 647 for live-cell imaging of cathepsin proteases.[278]
5.2. Glycans
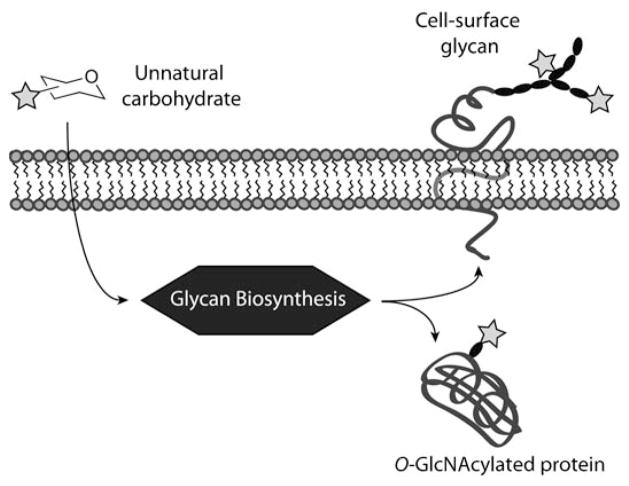
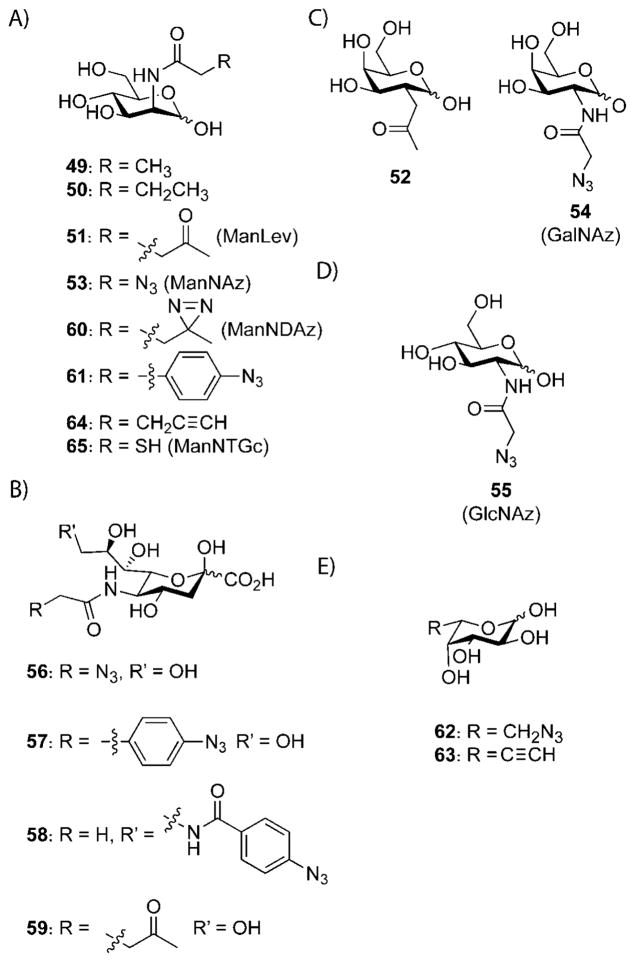
Proteins can be modified experimentally though genetics (amber stop codon methods and the addition of peptide sequences), biosynthesis (metabolic incorporation of unnatural amino acids using auxotrophs), and function (activity–based protein profiling). Many of these techniques are not applicable to glycans because these biopolymers do not have the genetic template and enzymatic activities that proteins can possess. However, glycans can be modified by metabolic labeling with biosynthetic precursors (Scheme 18).[279] To date, unnatural sialic acid (Sia), N-acetylgalactosamine (GalNAc), N-acetylglucosamine (GlcNAc), and fucose (Fuc) residues have been successfully incorporated into glycans through salvage pathways or, in the case of sialic acid, through de novo biosynthesis (Scheme 19). This technique, known as metabolic oligosaccharide engineering, was first employed by Reutter and co-workers to alter the acyl side chains on cell-surface sialic acids in live cells and animals using N-acetylmannosamine (ManNAc) derivatives 49 and 50 as the metabolic precursors.[280,281] These unnatural sialic acids were used to study host–virus interactions,[282] neuronal cell differentiation,[283] and polysialic acid.[284] Building upon these studies, we have exploited metabolic pathways to introduce bioorthogonal functional groups into sialylated glycans. Toward this end, a ketone-containing ManNAc derivative (51), known as N-levulinoylmannosamine (ManLev), was shown to be metabolized in a variety of cell lines.[133,285,286] The ketone groups were then treated with biotin hydrazide and detected by Western blot or flow cytometry. The latter method was also used to promote gene transfer and adenoviral uptake through the targeting of an anti-adenovirus antibody to ketone-containing glycans.[287]
Scheme 18.
Metabolic oligosaccharide engineering. Unnatural carbohydrate metabolites are taken up by cells and incorporated into cell-surface glycans and/or cytosolic O-GlcNAcylated proteins.
Scheme 19.
Unnatural carbohydrate metabolites for use in metabolic oligosaccharide engineering. A) N-Acetylmannosamine (ManNAc) metabolites. B) Sialic acid (Sia) metabolites. C) N-Acetylgalactosamine (GalNAc) metabolites. D) N-Acetylglucosamine (GlcNAc) metabolite. E) Fucose (Fuc) metabolites.
The GalNAc salvage pathway enzymes proved to be less tolerant than the sialic acid biosynthetic enzymes, as N-levulinoylgalactosamine (GalLev) was not significantly metabolized in mammalian cells. Ketone isostere 52 was synthesized to introduce ketone groups into GalNAc containing glycans.[134] This compound was successfully incorporated into glycans present in ldlD CHO cells, which lack the UDP-Gal/UDP-GalNAc 4-epimerase and cannot biosynthesize their own GalNAc.[288]
In 2000, we reported that azides could be incorporated into glycans containing sialic acid by treatment of cells with ManNAc derivative 53, N-azidoacetylmannosamine (ManNAz).[141] This report also marked the introduction of the Staudinger ligation, as the azidosialic acid residues were detected with triarylphosphine-biotin probes followed by flow cytometry analysis. Contrary to the levulinoyl derivatives, the azido derivatives could easily be extended to the GalNAc and GlcNAc salvage pathways (using GalNAz (54) and GlcNAz (55), respectively), thus facilitating the analysis of mucin-type O-linked glycans and O-GlcNAcylated proteins with the Staudinger ligation or strain-promoted [3+2] cycloaddition.[289–291] Additionally, we have shown that azides can be incorporated into glycans in mice and zebrafish by using ManNAz or GalNAz as metabolic precursors.[160,197,292] For in vivo studies, the azido glycans can be identified by either the Staudinger ligation[160] or copper-free click reaction.[195] Alternatively, they can be analyzed ex vivo by using any azide reaction.
Azides and other unnatural groups have also been incorporated into cell-surface sialic acid residues by using the sialic acid analogues directly. Modification of the 5-N-acyl and 9-OH positions of sialic acid are well tolerated, and numerous unnatural groups have been introduced therein to study sialic acid binding events.[293] Alkyl and aryl azides have been incorporated into cell-surface glycans through sialic acid precursors modified at the 5- and 9-positions (56–58).[294,295] Photo-cross-linking of sialic acid 58 was used to study the glycan ligands for CD22.[295] Tanaka and Kohler have performed similar photo-cross-linking studies using a diazirine-containing ManNAc derivative, 60 (ManNDAz).[296] Luchansky et al. employed sialic acids 56, 57, and 59 along with their corresponding ManNAc precursors 53, 61, and 51 to show that bypassing the first step of the sialic acid biosynthetic pathway often increases the yield of cell-surface glycans. Notably, the sialic acid biosynthetic pathway proved to be just as efficient as the salvage pathway for the incorporation of azides into sialic acid residues when ManNAz (53) and the corresponding azidosialic acid 56 were employed as substrates.[294]
Fucosylated glycans have been labeled with 6-azidofucose 62;[297,298] however, fucose 62 was found to be toxic in many mammalian cell lines and has not been extended into living animals. Wong and co-workers reported the use of 6-alkynyl fucose 63, which was less toxic than the azido analogue.[299] The Wong research group also successfully labeled cell-surface sialic acids with alkynyl ManNAc precursor 64,[299] and Wu and co-workers used the same metabolite to label cell-surface glycans in mice.[300] The degree of sialic acid metabolic labeling with alkynyl ManNAc is superior to that of ManNAz;[300] however, the alkyne must be detected by using CuAAC, which is not ideal for the detection of glycans in a living system, because of the toxicity of copper(I). Yarema and co-workers reported the metabolic labeling of sialic acids on stem cells with thio-ManNAc analogue 65 (ManNTGc), and showed that this unnatural glycan influenced their differentiation.[301]
The ability to metabolically label glycans with bioorthogonal functional groups has made these biopolymers amenable, for the first time, to molecular imaging. Toward this end, Chang et al. used triarylphosphine fluorophore conjugates to visualize azide-labeled glycans on cultured cells.[302] This same report demonstrated that two glycan subtypes could be imaged simultaneously by introduction of ketones into one glycan type and azides into another. Azide-labeled glycans were also imaged by the Staudinger ligation with fluorogenic phosphine 7.[152] Using the triarylphosphines, cell-surface glycans could be profiled; however, the inherently slow rate of the Staudinger ligation prevented more dynamic events from being observed. Subsequently, Baskin et al. performed glycan transport experiments on azide-labeled glycans with DIFO–fluorophore conjugates to gain insight into the rate and trajectory of internalization from the membrane.[195] DIFO–fluorophore conjugates have been used with great success to label glycans in developing zebrafish; this is the first example of molecular imaging of glycans in a live organism.[197] Spatiotemporal analysis of GalNAz-labeled glycans revealed dynamic aspects of mucin glycoproteins throughout the developmental program. Recently, alkyne-containing carbocyanine dyes (absorption in the near-IR region) have also been used to image azido glycans in mammalian cells by using CuAAC.[303]
There is growing interest in the use of chemical reporters embedded within glycans for proteomic analysis of glycosylation, an effort that may reveal new biomarkers of disease. For example, we have shown that cells treated with GalNAz incorporate the unnatural metabolite into mucin glycoproteins, and these can be identified by reaction with a phosphine-based affinity probe followed by capture, elution, trypsin digestion, and identification by mass spectrometry.[162] Through this procedure we discovered that GalNAz also labels O-GlcNAcylated proteins, presumably through the conversion of UDP-GalNAz into UDP-GlcNAz by the enzyme UDP-Glc/GalNAc C-4 epimerase. Zhao and coworkers analyzed O-GlcNAc-modified proteins of the cytosol and nucleus through metabolic labeling with GlcNAz followed by reaction with a phosphine probe, enrichment, and mass spectrometry identification.[304,305] In a similar way, Lemoine and co-workers analyzed the O-GlcNAc proteome.[306] By using the metabolic precursor alkynyl ManNAc (64), Wong and co-workers isolated sialylated glycoproteins by reacting lysates with an azido-biotin probe through CuAAC, capturing the tagged proteins on streptavidin-conjugated beads, and directly digesting the proteins off the beads with trypsin, which yielded an enriched sample ready for mass spectrometric analysis. By using this method, over 200 glycoproteins were identified, with the majority bearing sialylated N-linked glycans.[307] These precedents establish a platform for comparative glycoproteomic studies that seek to identify changes in glycoprotein abundance or in the structures of the pendant glycans that correlate with disease.
5.3. Lipids
Like glycans, lipids are secondary metabolites and post-translational modifications that cannot be directly studied using genetically encoded reporters. Bioorthogonal chemical reporters have proven particularly useful for probing lipids in cellular systems (Scheme 20A).
Scheme 20.
Unnatural metabolites containing bioorthogonal functional groups. A) Fatty acids and isoprenol metabolites for metabolic labeling of lipids. B) Nucleic acid metabolites for the incorporation of alkynes and azides into RNA and DNA.
Myristoylation and palmitoylation are the two predominant forms of fatty acid acylation on proteins.[308] Both have been probed using azido lipid analogues in cell-culture systems. Ploegh and co-workers demonstrated that azide-labeled fatty acid 66 is converted in situ into its CoA analogue and transferred to sites of myristoylation on endogenous proteins. The longer chain fatty acids 67–69, by contrast, were attached to proteins at sites normally modified by palmitoylation.[309] N-myristoylation has also been studied in vitro and in E. coli by using azido- and alkynyl-fatty acids 70 and 71 that were metabolically incorporated and subsequently detected through CuAAC.[310,311] Berthiaume and co-workers have studied the role of myristoylation in apoptosis by using azido-fatty acid 66 and the Staudinger ligation.[312] The same research group has also used similar methods to identify new palmitoylated proteins in the liver.[313]
Unlike myristoylation and palmitoylation, farnesyl groups (as well as geranylgeranyl groups) are derived from isoprenoid diphosphates.[308] Proteins that have a C-terminal CAAX (A =aliphatic, X =Met, Ser, Phe) sequence are farnesylated on the cysteine residue by protein farnesyltransferases (PFtases).[314] A variety of diphosphate farnesyl precursors containing unnatural functional groups (for example, alkynes, azides, ketones) have been synthesized and used to modify proteins in vitro.[315–319] Zhao and co-workers have incorporated azides into farnesyl groups in an in vivo setting through the introduction of azidoisoprenol 72 as a metabolic precursor. An enrichment aided by the Staudinger ligation was performed, and new farnesylated proteins were identified by mass spectrometry. The authors also stated that this method was successful in labeling geranylgeranylated proteins.[320] Also noteworthy is that some research groups have used protein farnesylation as a method of enzyme-mediated site-specific modification of proteins (Section 3.3) by using the conserved sequence recognized by PFtases.[321]
Recently, Neef and Schultz have extended the bioorthogonal chemical reporter strategy to study the dynamics of phospholipids in bilayers.[322] Alkyne-modified phosphatidic acids 73–75 were synthesized and introduced into mammalian cells. The S-acetylthioethyl (SATE) groups facilitated penetration of the cell membrane and were cleaved in the cytosol by esterases. The terminal alkyne-functionalized phospholipid 73 and nonhydrolyzable version 74 were visualized on fixed cells by using CuAAC with an azidocoumarin dye. Cyclo-octyne lipid 75 enabled direct visualization of lipid bilayers on live cells by copper-free click reactions. This study marks the first integration of a cyclooctyne into a biomolecule for cell-based studies.
5.4. Nucleic Acids
Nucleic acids adorned with bioorthogonal functional groups have been synthesized and used for various purposes (Scheme 20B),[323] most notably as a method for imaging cells undergoing DNA synthesis and replication.[324] Salic and Mitchison have reported that 5-ethynyl-2′-deoxyuridine (76, EdU) can be metabolically incorporated into DNA during replication and subsequently detected with an azido fluorophore through CuAAC. The alkynes can be detected in live cells if a cell-permeable fluorophore is employed, although the viability of cells is compromised after exposure to copper. By using this procedure, newly synthesized DNA can be visualized quickly and with good sensitivity under milder conditions than the traditional method in which 5-bromo-2′-deoxyuridine (BrdU) and a corresponding antibody are used.[324,325] EdU is even effective for visualizing cells undergoing DNA synthesis in mice.[324] More recently, RNA synthesis has been imaged using 5-ethynyluridine (77, EU) and fluorescent azides in an analogous method.[326]
The azido analogue of EdU, 5-azido-2′-deoxyuridine (78, AdU), was also reported, but detection of AdU by CuAAC suffered from considerably higher background signal than labeling using EdU.[324] AdU was not the first case in which an azide was installed into a nucleic acid. Aryl azidonucleotides have been used as photo-cross-linking agents to study interactions between DNA or RNA and other biomolecules.[327] Additionally, Rajski and co-workers have synthesized S-adenosyl-L-methionine (SAM) derivatives with azides and alkynes to probe DNA methyltransferase activity.[328–330]
6. Future Perspectives
Within the last decade, bioorthogonal reactions have become essential tools for chemical biologists. They have opened up new avenues for biological investigation and produced fundamental discoveries in areas as diverse as protein biophysics, neurophysiology, developmental and stem cell biology, and cancer detection. The need still exists for additional orthogonal functional groups and improvements in the kinetics and selectivities of the reactions already at hand. Mining of the periodic table and classic organic literature are two avenues that are likely to bear interesting fruit. Group 15 has been particularly lucrative in terms of bioorthogonal reactions, and perhaps the larger elements within this group, bismuth and antimony, could be similarly beneficial. In addition, novel combinations of phosphorus and sulfur have yet to be explored. Pericyclic reactions seem poised for biological applications because their rates are often accelerated in polar solvents and their concerted mechanisms leave little room for interruption by biological nucleophiles and electrophiles. Also, biologically compatible forms of energy may be strategically employed to promote otherwise dormant reactions in living systems. Light has already been harnessed,[206] but focused ultrasound is another energy source that might be exploited for bioorthogonal chemistry in vivo.
The extension of chemical reporters to studies of small-molecule metabolites is another promising future direction. Cofactors and cholesterol seem to be prime targets for the addition of chemical reporters. Notably, the Burkart and Marquez research groups have already made progress in extending the bioorthogonal chemical reporter strategy to the analysis of pantetheine cofactors[331] and the mechanism of action of flavone-8-acetic acid, respectively.[332] The field is quite open to the merger of innovative chemical reactions with biological targets that have defied conventional research tools.
Acknowledgments
We thank J. M. Baskin, K. E. Beatty, K. W. Dehnert, J. C. Jewett, and S. T. Laughlin for critical reading of the manuscript.
Biographies
Ellen Sletten was born in New Hampshire (USA) in 1984. She obtained her BS in Chemistry from Stonehill College in 2006, where she worked in the laboratory of Prof. Louis J. Liotta on the synthesis of polyhydroxylated pyrrolizidines. She is currently pursuing her PhD at UC Berkeley under the direction of Prof. Carolyn Bertozzi. Her research focuses on the synthesis of cyclooctyne reagents for use in copper-free click reactions.

Prof. Carolyn Bertozzi is the T.Z. and Irmgard Chu Distinguished Professor of Chemistry and Professor of Molecular and Cell Biology at UC Berkeley, and Professor of Molecular and Cellular Biology at UCSF. She is also the Director of the Molecular Foundry at the Lawrence Berkeley National Laboratory and an Investigator of the Howard Hughes Medical Institute. She earned her AB in Chemistry from Harvard University in 1988 and obtained her PhD at UC Berkeley in 1993 with Prof. Mark Bednarski. She carried out postdoctoral research at UCSF with Prof. Steven Rosen and joined the faculty at UC Berkeley in 1996.

Contributor Information
Prof. Ellen M. Sletten, Department of Chemistry, University of California, Berkeley, CA 94720 (USA)
Carolyn R. Bertozzi, Email: crb@berkeley.edu, Departments of Chemistry and Molecular and Cell Biology and Howard Hughes Medical Institute, University of California and The Molecular Foundry, Lawrence Berkeley National Laboratory, Berkeley, CA 94720 (USA), Fax: (+1)510-643-2628
References
- 1.http://nobelprize.org/nobel_prizes/chemistry/web
- 2.Nienhaus GU. Angew Chem. 2008;120:9130. [Google Scholar]; Angew Chem Int Ed. 2008;47:8992. [Google Scholar]
- 3.Glazer AN. Annu Rev Biochem. 1970;39:101. doi: 10.1146/annurev.bi.39.070170.000533. [DOI] [PubMed] [Google Scholar]
- 4.Kaiser ET, Lawrence DS, Rokita SE. Annu Rev Biochem. 1985;54:565. doi: 10.1146/annurev.bi.54.070185.003025. [DOI] [PubMed] [Google Scholar]
- 5.Veronese FM, Pasut G. Drug Discovery Today. 2005;10:1451. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- 6.Ryan SM, Mantovani G, Wang XX, Haddleton DM, Brayden DJ. Expert Opin Drug Delivery. 2008;5:371. doi: 10.1517/17425247.5.4.371. [DOI] [PubMed] [Google Scholar]
- 7.Wu AM, Senter PD. Nat Biotechnol. 2005;23:1137. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 8.Francis MB. In: Chemical Biology. Schreiber SL, Kapoor T, Wess G, editors. Wiley-VCH; Weinheim: 2007. p. 593. [Google Scholar]
- 9.Tilley SD, Joshi NS, Francis MB. In: The Wiley Encyclopedia of Chemical Biology. Begley T, editor. Wiley-VCH; Weinheim: 2008. [Google Scholar]
- 10.Hermanson GT. Bioconjugate Techniques. 1. Academic Press; San Diego, CA: 1996. [Google Scholar]
- 11.Antos JM, Francis MB. Curr Opin Chem Biol. 2006;10:253. doi: 10.1016/j.cbpa.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Hackenberger CPR, Schwarzer D. Angew Chem. 2008;120:10182. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:10030. [Google Scholar]
- 13.McFarland JM, Francis MB. J Am Chem Soc. 2005;127:13490. doi: 10.1021/ja054686c. [DOI] [PubMed] [Google Scholar]
- 14.Bernardes GJL, Chalker JM, Errey JC, Davis BG. J Am Chem Soc. 2008;130:5052. doi: 10.1021/ja800800p. [DOI] [PubMed] [Google Scholar]
- 15.Dondoni A. Angew Chem. 2008;120:9133. [Google Scholar]; Angew Chem Int Ed. 2008;47:8995. [Google Scholar]
- 16.Triola G, Brunsveld L, Waldmann H. J Org Chem. 2008;73:3646. doi: 10.1021/jo800198s. [DOI] [PubMed] [Google Scholar]
- 17.Campos LM, Killops KL, Sakai R, Paulusse JMJ, Damiron D, Drockenmuller E, Messmore BW, Hawker CJ. Macromolecules. 2008;41:7063. [Google Scholar]
- 18.Wittrock S, Becker T, Kunz H. Angew Chem. 2007;119:5319. doi: 10.1002/anie.200700964. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2007;46:5226. [Google Scholar]
- 19.Jonkheijm P, Weinrich D, Koehn M, Engelkamp H, Christianen PCM, Kuhlmann J, Maan JC, Nuesse D, Schroeder H, Wacker R, Breinbauer R, Niemeyer CM, Waldmann H. Angew Chem. 2008;120:4493. doi: 10.1002/anie.200800101. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:4421. [Google Scholar]
- 20.Joshi NS, Whitaker LR, Francis MB. J Am Chem Soc. 2004;126:15942. doi: 10.1021/ja0439017. [DOI] [PubMed] [Google Scholar]
- 21.Kodadek T, Duroux-Richard I, Bonnafous JC. Trends Pharmacol Sci. 2005;26:210. doi: 10.1016/j.tips.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Meunier S, Strable E, Finn MG. Chem Biol. 2004;11:319. doi: 10.1016/j.chembiol.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 23.McFarland JM, Joshi NS, Francis MB. J Am Chem Soc. 2008;130:7639. doi: 10.1021/ja710927q. [DOI] [PubMed] [Google Scholar]
- 24.Romanini DW, Francis MB. Bioconjugate Chem. 2008;19:153. doi: 10.1021/bc700231v. [DOI] [PubMed] [Google Scholar]
- 25.Tilley SD, Francis MB. J Am Chem Soc. 2006;128:1080. doi: 10.1021/ja057106k. [DOI] [PubMed] [Google Scholar]
- 26.Antos JM, Francis MB. J Am Chem Soc. 2004;126:10256. doi: 10.1021/ja047272c. [DOI] [PubMed] [Google Scholar]
- 27.Dixon HBF. J Protein Chem. 1984;3:99. [Google Scholar]
- 28.Cennamo C, Carafoli B, Bonetti EP. J Am Chem Soc. 1956;78:3523. [Google Scholar]
- 29.Dixon HBF. Biochem J. 1964;92:661. doi: 10.1042/bj0920661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. Angew Chem. 2006;118:5433. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:5307. [Google Scholar]
- 31.Scheck RA, Dedeo MT, Lavarone AT, Francis MB. J Am Chem Soc. 2008;130:11762. doi: 10.1021/ja802495w. [DOI] [PubMed] [Google Scholar]
- 32.Geoghegan KF, Stroh JG. Bioconjugate Chem. 1992;3:138. doi: 10.1021/bc00014a008. [DOI] [PubMed] [Google Scholar]
- 33.Li XF, Zhang LS, Hall SE, Tam JP. Tetrahedron Lett. 2000;41:4069. [Google Scholar]
- 34.de La Figuera N, Fiol S, Fernandez JC, Forns P, Fernandez-Forner D, Albericio F. Synlett. 2006:1903. [Google Scholar]
- 35.Tam JP, Yu QT, Miao ZW. Biopolymers. 1999;51:311. doi: 10.1002/(SICI)1097-0282(1999)51:5<311::AID-BIP2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 36.Dawson PE, Muir TW, Clarklewis I, Kent SBH. Science. 1994;266:776. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 37.Wieland T, Bokelmann E, Bauer L, Lang HU, Lau H. Justus Liebigs Ann Chem. 1953;583:129. [Google Scholar]
- 38.Dawson PE, Kent SBH. Annu Rev Biochem. 2000;69:923. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 39.Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Proc Natl Acad Sci USA. 2002;99:8536. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki T, Otomo T, Oda N, Kyogoku Y, Uegaki K, Ito N, Ishino Y, Nakamura H. J Am Chem Soc. 1998;120:5591. [Google Scholar]
- 41.Schwarzer D, Cole PA. Curr Opin Chem Biol. 2005;9:561. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 42.Muralidharan V, Muir TW. Nat Methods. 2006;3:429. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 43.Muir TW. Annu Rev Biochem. 2003;72:249. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 44.Muir TW, Sondhi D, Cole PA. Proc Natl Acad Sci USA. 1998;95:6705. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozawa T, Nogami S, Sato M, Ohya Y, Umezawa Y. Anal Chem. 2000;72:5151. doi: 10.1021/ac000617z. [DOI] [PubMed] [Google Scholar]
- 46.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Proc Natl Acad Sci USA. 1999;96:13638. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans TC, Martin D, Kolly R, Panne D, Sun L, Ghosh I, Chen LX, Benner J, Liu XQ, Xu MQ. J Biol Chem. 2000;275:9091. doi: 10.1074/jbc.275.13.9091. [DOI] [PubMed] [Google Scholar]
- 48.Giriat I, Muir TW. J Am Chem Soc. 2003;125:7180. doi: 10.1021/ja034736i. [DOI] [PubMed] [Google Scholar]
- 49.Yeo DSY, Srinivasan R, Uttamchandani M, Chen GYJ, Zhu Q, Yao SQ. Chem Commun. 2003:2870. doi: 10.1039/b309196a. [DOI] [PubMed] [Google Scholar]
- 50.Tsien RY. Annu Rev Biochem. 1998;67:509. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 51.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 52.Lippincott-Schwartz J, Patterson GH. Science. 2003;300:87. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 53.Shaner NC, Patterson GH, Davidson MW. J Cell Sci. 2007;120:4247. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 54.Andresen M, Schmitz-Salue R, Jakobs S. Mol Biol Cell. 2004;15:5616. doi: 10.1091/mbc.E04-06-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 56.Cao HS, Xiong YJ, Wang T, Chen BW, Squier TC, Mayer MU. J Am Chem Soc. 2007;129:8672. doi: 10.1021/ja070003c. [DOI] [PubMed] [Google Scholar]
- 57.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. J Am Chem Soc. 2002;124:6063. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 58.Stroffekova K, Proenza C, Beam KG. Pfluegers Arch. 2001;442:859. doi: 10.1007/s004240100619. [DOI] [PubMed] [Google Scholar]
- 59.Griffin BA, Adams SR, Jones J, Tsien RY. Methods Enzymol. 2000;327:565. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
- 60.Martin BR, Giepmans BNG, Adams SR, Tsien RY. Nat Biotechnol. 2005;23:1308. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 61.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. J Cell Biol. 2006;175:67. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffmann C, Gaietta G, Bunemann M, Adams SR, Oberdorff-Maass S, Behr B, Vilardaga JP, Tsien RY, Eisman MH, Lohse MJ. Nat Methods. 2005;2:171. doi: 10.1038/nmeth742. [DOI] [PubMed] [Google Scholar]
- 63.Roberti MJ, Bertoncini CW, Klement R, Jares-Erijman EA, Jovin TM. Nat Methods. 2007;4:345. doi: 10.1038/nmeth1026. [DOI] [PubMed] [Google Scholar]
- 64.Chaumont S, Khakh BS. Proc Natl Acad Sci USA. 2008;105:12063. doi: 10.1073/pnas.0803008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arhel N, Genovesio A, Kim KA, Miko S, Perret E, Olivo-Marin JC, Shorte S, Charneau P. Nat Methods. 2006;3:817. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- 66.Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Nat Chem Biol. 2007;3:779. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krishnan B, Gierasch LM. Chem Biol. 2008;15:1104. doi: 10.1016/j.chembiol.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Science. 2002;296:503. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 69.Taguchi Y, Shi Z-D, Ruddy B, Dorward DW, Greene L, Baron GS. Mol Biol Cell. 2008;20:233. doi: 10.1091/mbc.E08-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halo TL, Appelbaum J, Hobert EM, Balkin DM, Schepartz A. J Am Chem Soc. 2009;131:438. doi: 10.1021/ja807872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim KK, Escobedo JO, StLuce NN, Rusin O, Wong D, Strongin RM. Org Lett. 2003;5:5007. doi: 10.1021/ol035978q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Terpe K. Appl Microbiol Biotechnol. 2003;60:523. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 73.Guignet EG, Hovius R, Vogel H. Nat Biotechnol. 2004;22:440. doi: 10.1038/nbt954. [DOI] [PubMed] [Google Scholar]
- 74.Kapanidis AN, Ebright YW, Ebright RH. J Am Chem Soc. 2001;123:12123. doi: 10.1021/ja017074a. [DOI] [PubMed] [Google Scholar]
- 75.Lata S, Reichel A, Brock R, Tampe R, Piehler J. J Am Chem Soc. 2005;127:10205. doi: 10.1021/ja050690c. [DOI] [PubMed] [Google Scholar]
- 76.Goldsmith CR, Jaworski J, Sheng M, Lippard SJ. J Am Chem Soc. 2006;128:418. doi: 10.1021/ja0559754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valko M, Morris H, Cronin MTD. Curr Med Chem. 2005;12:1161. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 78.Hauser CT, Tsien RY. Proc Natl Acad Sci USA. 2007;104:3693. doi: 10.1073/pnas.0611713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ojida A, Honda K, Shinmi D, Kiyonaka S, Mori Y, Hamachi I. J Am Chem Soc. 2006;128:10452. doi: 10.1021/ja0618604. [DOI] [PubMed] [Google Scholar]
- 80.Honda K, Fujishima SH, Ojida A, Hamachi I. ChemBio-Chem. 2007;8:1370. doi: 10.1002/cbic.200700146. [DOI] [PubMed] [Google Scholar]
- 81.Honda K, Nakata E, Ojida A, Hamachi I. Chem Commun. 2006:4024. doi: 10.1039/b608684e. [DOI] [PubMed] [Google Scholar]
- 82.Nonaka H, Tsukiji S, Ojida A, Hamachi I. J Am Chem Soc. 2007;129:15777. doi: 10.1021/ja074176d. [DOI] [PubMed] [Google Scholar]
- 83.Nitz M, Franz KJ, Maglathlin RL, Imperiali B. ChemBio-Chem. 2003;4:272. doi: 10.1002/cbic.200390047. [DOI] [PubMed] [Google Scholar]
- 84.Martin LL, Sculimbrene BR, Nitz M, Imperiali B. QSAR Comb Sci. 2005;24:1149. [Google Scholar]
- 85.Franz KJ, Nitz M, Imperiali B. ChemBioChem. 2003;4:265. doi: 10.1002/cbic.200390046. [DOI] [PubMed] [Google Scholar]
- 86.Sculimbrene BR, Imperiali B. J Am Chem Soc. 2006;128:7346. doi: 10.1021/ja061188a. [DOI] [PubMed] [Google Scholar]
- 87.Fricker SP. Chem Soc Rev. 2006;35:524. doi: 10.1039/b509608c. [DOI] [PubMed] [Google Scholar]
- 88.Chen I, Ting AY. Curr Opin Biotechnol. 2005;16:35. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 89.Foley TL, Burkart MD. Curr Opin Chem Biol. 2007;11:12. doi: 10.1016/j.cbpa.2006.11.036. [DOI] [PubMed] [Google Scholar]
- 90.Chen I, Howarth M, Lin WY, Ting AY. Nat Methods. 2005;2:99. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 91.Howarth M, Takao K, Hayashi Y, Ting AY. Proc Natl Acad Sci USA. 2005;102:7583. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Slavoff SA, Chen I, Choi YA, Ting AY. J Am Chem Soc. 2008;130:1160. doi: 10.1021/ja076655i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin CW, Ting AY. J Am Chem Soc. 2006;128:4542. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sato H, Ikeda M, Suzuki K, Hirayama K. Biochemistry. 1996;35:13072. doi: 10.1021/bi952616k. [DOI] [PubMed] [Google Scholar]
- 95.Dutton A, Singer SJ. Proc Natl Acad Sci USA. 1975;72:2568. doi: 10.1073/pnas.72.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Nat Biotechnol. 2007;25:1483. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baruah H, Puthenveetil S, Choi YA, Shah S, Ting AY. Angew Chem. 2008;120:7126. doi: 10.1002/anie.200802088. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:7018. [Google Scholar]
- 98.Dierks T, Schmidt B, Borissenko LV, Peng JH, Preusser A, Mariappan M, von Figura K. Cell. 2003;113:435. doi: 10.1016/s0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 99.Carlson BL, Ballister ER, Skordalakes E, King DS, Breidenbach MA, Gilmore SA, Berger JM, Bertozzi CR. J Biol Chem. 2008;283:20117. doi: 10.1074/jbc.M800217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carrico IS, Carlson BL, Bertozzi CR. Nat Chem Biol. 2007;3:321. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 101.Dierks T, Lecca MR, Schlotterhose P, Schmidt B, von Figura K. EMBO J. 1999;18:2084. doi: 10.1093/emboj/18.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rush JS, Bertozzi CR. J Am Chem Soc. 2008;130:12240. doi: 10.1021/ja804530w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu P, Shui W, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR. Proc Natl Acad Sci USA. 2009;106:3000. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mazmanian SK, Liu G, Hung TT, Schneewind O. Science. 1999;285:760. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 105.Marraffini LA, DeDent AC, Schneewind O. Microbiol Mol Biol Rev. 2006;70:192. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Samantaray S, Marathe U, Dasgupta S, Nandicoori VK, Roy RP. J Am Chem Soc. 2008;130:2132. doi: 10.1021/ja077358g. [DOI] [PubMed] [Google Scholar]
- 107.Mao HY, Hart SA, Schink A, Pollok BA. J Am Chem Soc. 2004;126:2670. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 108.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Nat Chem Biol. 2007;3:707. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 109.Tanaka T, Yamamoto T, Tsukiji S, Nagamune T. ChemBio-Chem. 2008;9:802. doi: 10.1002/cbic.200700614. [DOI] [PubMed] [Google Scholar]
- 110.Johnsson N, George N, Johnsson K. ChemBioChem. 2005;6:47. doi: 10.1002/cbic.200400290. [DOI] [PubMed] [Google Scholar]
- 111.Joshi AK, Zhang L, Rangan VS, Smith S. J Biol Chem. 2003;278:33142. doi: 10.1074/jbc.M305459200. [DOI] [PubMed] [Google Scholar]
- 112.Yin J, Liu F, Li XH, Walsh CT. J Am Chem Soc. 2004;126:7754. doi: 10.1021/ja047749k. [DOI] [PubMed] [Google Scholar]
- 113.George N, Pick H, Vogel H, Johnsson N, Johnsson K. J Am Chem Soc. 2004;126:8896. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 114.Yin J, Lin AJ, Buckett PD, Wessling-Resnick M, Golan DE, Walsh CT. Chem Biol. 2005;12:999. doi: 10.1016/j.chembiol.2005.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vivero-Pol L, George N, Krumm H, Johnsson K, Johnsson N. J Am Chem Soc. 2005;127:12770. doi: 10.1021/ja0533850. [DOI] [PubMed] [Google Scholar]
- 116.Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, Walsh CT. Proc Natl Acad Sci USA. 2005;102:15815. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou Z, Cironi P, Lin AJ, Xu YQ, Hrvatin S, Golan DE, Silver PA, Walsh CT, Yin J. ACS Chem Biol. 2007;2:337. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- 118.Pegg AE. Mutat Res. 2000;462:83. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 119.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. Nat Biotechnol. 2003;21:86. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 120.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Proc Natl Acad Sci USA. 2004;101:9955. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gautier A, Juillerat A, Heinis C, Correa IR, Kindermann M, Beaufils F, Johnsson K. Chem Biol. 2008;15:128. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 122.Los GV, Zimprich C, McDougall MG, Karassina N, Learish R, Klaubert DH, Darzins A, Bulleit RF, Wood K. J Neurochem. 2005;94:15. [Google Scholar]
- 123.Prescher JA, Bertozzi CR. Nat Chem Biol. 2005;1:13. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 124.Voss EW. J Mol Recognit. 1993;6:51. doi: 10.1002/jmr.300060203. [DOI] [PubMed] [Google Scholar]
- 125.Turro NJ. Modern Molecular Photochemistry. Benjamin-Cummings; Menlo Park, CA: 1978. [Google Scholar]
- 126.Blackman ML, Royzen M, Fox JM. J Am Chem Soc. 2008;130:13518. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Berkel SS, Dirks ATJ, Debets MF, van Delft FL, Cornelissen JJLM, Nolte RJM, Rutjes FPJT. Chem-BioChem. 2007;8:1504. doi: 10.1002/cbic.200700278. [DOI] [PubMed] [Google Scholar]
- 128.Jencks WP. J Am Chem Soc. 1959;81:475. [Google Scholar]
- 129.Cornish VW, Hahn KM, Schultz PG. J Am Chem Soc. 1996;118:8150. [Google Scholar]
- 130.Rose K, Vizzavona J. J Am Chem Soc. 1999;121:7034. [Google Scholar]
- 131.Oshannessy DJ, Dobersen MJ, Quarles RH. Immunol Lett. 1984;8:273. doi: 10.1016/0165-2478(84)90008-7. [DOI] [PubMed] [Google Scholar]
- 132.Zhang ZW, Smith BAC, Wang L, Brock A, Cho C, Schultz PG. Biochemistry. 2003;42:6735. doi: 10.1021/bi0300231. [DOI] [PubMed] [Google Scholar]
- 133.Mahal LK, Yarema KJ, Bertozzi CR. Science. 1997;276:1125. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 134.Hang HC, Bertozzi CR. J Am Chem Soc. 2001;123:1242. doi: 10.1021/ja002962b. [DOI] [PubMed] [Google Scholar]
- 135.Zeng Y, Ramya TNC, Dirksen A, Dawson PE, Paulson JC. Nat Methods. 2009;6:207. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dirksen A, Hackeng TM, Dawson PE. Angew Chem. 2006;118:7743. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:7581. [Google Scholar]
- 137.Griffin RJ. Prog Med Chem. 1994;31:121. doi: 10.1016/s0079-6468(08)70020-1. [DOI] [PubMed] [Google Scholar]
- 138.Hendricks SB, Pauling L. J Am Chem Soc. 1925;47:2904. [Google Scholar]
- 139.Sidgwick NV, Sutton LE, Thomas W. J Chem Soc. 1933:406. [Google Scholar]
- 140.Knaggs IE. Nature. 1935;135:268. [Google Scholar]
- 141.Saxon E, Bertozzi CR. Science. 2000;287:2007. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 142.Staudinger H, Meyer J. Helv Chim Acta. 1919;2:635. [Google Scholar]
- 143.Cartwright IL, Hutchinson DW, Armstrong VW. Nucleic Acids Res. 1976;3:2331. doi: 10.1093/nar/3.9.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Staros JV, Bayley H, Standring DN, Knowles JR. Biochem Biophys Res Commun. 1978;80:568. doi: 10.1016/0006-291x(78)91606-6. [DOI] [PubMed] [Google Scholar]
- 145.Reardon JE, Crouch RC, St John Williams L. J Biol Chem. 1994;269:15999. [PubMed] [Google Scholar]
- 146.Handlon AL, Oppenheimer NJ. Pharm Res. 1988;5:297. doi: 10.1023/a:1015926720740. [DOI] [PubMed] [Google Scholar]
- 147.Saxon E, Luchansky SJ, Hang HC, Yu C, Lee SC, Bertozzi CR. J Am Chem Soc. 2002;124:14893. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- 148.Burns JA, Butler JC, Moran J, Whitesides GM. J Org Chem. 1991;56:2648. [Google Scholar]
- 149.Dmitrenko O, Thorpe C, Bach RD. J Org Chem. 2007;72:8298. doi: 10.1021/jo071271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Humphrey RE, McCrary AL, Webb RM. Talanta. 1965;12:727. [Google Scholar]
- 151.Lemieux GA, de Graffenried CL, Bertozzi CR. J Am Chem Soc. 2003;125:4708. doi: 10.1021/ja029013y. [DOI] [PubMed] [Google Scholar]
- 152.Hangauer MJ, Bertozzi CR. Angew Chem. 2008;120:2428. doi: 10.1002/anie.200704847. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:2394. [Google Scholar]
- 153.Soellner MB, Nilsson BL, Raines RT. J Am Chem Soc. 2006;128:8820. doi: 10.1021/ja060484k. [DOI] [PubMed] [Google Scholar]
- 154.Nilsson BL, Kiessling LL, Raines RT. Org Lett. 2000;2:1939. doi: 10.1021/ol0060174. [DOI] [PubMed] [Google Scholar]
- 155.Nilsson BL, Kiessling LL, Raines RT. Org Lett. 2001;3:9. doi: 10.1021/ol006739v. [DOI] [PubMed] [Google Scholar]
- 156.Soellner MB, Tam A, Raines RT. J Org Chem. 2006;71:9824. doi: 10.1021/jo0620056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Saxon E, Armstrong JI, Bertozzi CR. Org Lett. 2000;2:2141. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 158.Tam A, Soellner MB, Raines RT. Org Biomol Chem. 2008;6:1173. doi: 10.1039/b802336k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tam A, Raines RT. Bioorg Med Chem. 2009;17:1055. doi: 10.1016/j.bmc.2008.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Prescher JA, Dube DH, Bertozzi CR. Nature. 2004;430:873. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 161.Lin FL, Hoyt HM, Van Halbeek H, Bergman RG, Bertozzi CR. J Am Chem Soc. 2005;127:2686. doi: 10.1021/ja044461m. [DOI] [PubMed] [Google Scholar]
- 162.Bertozzi CR. unpublished results. [Google Scholar]
- 163.Michael A. J Prakt Chem. 1893;48:94. [Google Scholar]
- 164.Huisgen R. Angew Chem. 1963;75:604. [Google Scholar]; Angew Chem Int Ed Engl. 1963;2:565. [Google Scholar]
- 165.Hartzel LW, Benson FR. J Am Chem Soc. 1954;76:667. doi: 10.1021/ja01171a501. [DOI] [PubMed] [Google Scholar]
- 166.Himo F, Lovell T, Hilgraf R, Rostovtsev VV, Noodleman L, Sharpless KB, Fokin VV. J Am Chem Soc. 2005;127:210. doi: 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
- 167.Padwa A. 1,3-Dipolar Cycloaddition Chemistry. 1. Vol. 1. Wiley-Interscience Publication; New York: 1984. [Google Scholar]
- 168.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem. 2002;114:2708. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2002;41:2596. [Google Scholar]
- 169.Tornoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 170.Rodionov VO, Presolski SI, Diaz DD, Fokin VV, Finn MG. J Am Chem Soc. 2007;129:12705. doi: 10.1021/ja072679d. [DOI] [PubMed] [Google Scholar]
- 171.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org Lett. 2004;6:2853. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 172.Kolb HC, Finn MG, Sharpless KB. Angew Chem. 2001;113:2056. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2001;40:2004. [Google Scholar]
- 173.Lutz JF. Angew Chem. 2007;119:1036. [Google Scholar]; Angew Chem Int Ed. 2007;46:1018. [Google Scholar]
- 174.Sharpless KB, van der Eycken E. QSAR Comb Sci. 2007;26:1110. See entire issue. [Google Scholar]
- 175.Wu P, Fokin VV. Aldrichimica Acta. 2007;40:7. [Google Scholar]
- 176.Tron GC, Pirali T, Billington RA, Canonico PL, Sorba G, Genazzani AA. Med Res Rev. 2008;28:278. doi: 10.1002/med.20107. [DOI] [PubMed] [Google Scholar]
- 177.Fokin VV. ACS Chem Biol. 2007;2:775. doi: 10.1021/cb700254v. [DOI] [PubMed] [Google Scholar]
- 178.Fournier D, Hoogenboom R, Schubert US. Chem Soc Rev. 2007;36:1369. doi: 10.1039/b700809k. [DOI] [PubMed] [Google Scholar]
- 179.Bock VD, Hiemstra H, van Maarseveen JH. Eur J Org Chem. 2005:51. [Google Scholar]
- 180.Kolb HC, Sharpless KB. Drug Discovery Today. 2003;8:1128. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 181.Breinbauer R, Kohn M. ChemBioChem. 2003;4:1147. doi: 10.1002/cbic.200300705. [DOI] [PubMed] [Google Scholar]
- 182.Dondoni A. Chem Asian J. 2007;2:700. doi: 10.1002/asia.200700015. [DOI] [PubMed] [Google Scholar]
- 183.Zhang L, Chen XG, Xue P, Sun HHY, Williams ID, Sharpless KB, Fokin VV, Jia GC. J Am Chem Soc. 2005;127:15998. doi: 10.1021/ja054114s. [DOI] [PubMed] [Google Scholar]
- 184.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J Am Chem Soc. 2003;125:3192. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 185.Wolbers F, ter Braak P, Le Gac S, Luttge R, Andersson H, Vermes I, van den Berg A. Electrophoresis. 2006;27:5073. doi: 10.1002/elps.200600203. [DOI] [PubMed] [Google Scholar]
- 186.Link AJ, Tirrell DA. J Am Chem Soc. 2003;125:11164. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
- 187.Link AJ, Vink MKS, Tirrell DA. J Am Chem Soc. 2004;126:10598. doi: 10.1021/ja047629c. [DOI] [PubMed] [Google Scholar]
- 188.Speers AE, Adam GC, Cravatt BF. J Am Chem Soc. 2003;125:4686. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 189.Alder K, Stein G. Justus Liebigs Ann Chem. 1933;501:1. [Google Scholar]
- 190.Alder K, Stein G. Justus Liebigs Ann Chem. 1931;485:211. [Google Scholar]
- 191.Wittig G, Krebs A. Chem Ber. 1961;94:3260. [Google Scholar]
- 192.Agard NJ, Prescher JA, Bertozzi CR. J Am Chem Soc. 2004;126:15046. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 193.Codelli JA, Baskin JM, Agard NJ, Bertozzi CR. J Am Chem Soc. 2008;130:11486. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem Biol. 2006;1:644. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 195.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc Natl Acad Sci USA. 2007;104:16793. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sletten EM, Bertozzi CR. Org Lett. 2008;10:3097. doi: 10.1021/ol801141k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ning XH, Guo J, Wolfert MA, Boons GJ. Angew Chem. 2008;120:2285. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:2253. [Google Scholar]
- 199.Ess DH, Jones GO, Houk KN. Org Lett. 2008;10:1633. doi: 10.1021/ol8003657. [DOI] [PubMed] [Google Scholar]; Schoenebeck F, Ess DH, Jones GO, Houk KN. J Am Chem Soc. 2009;131:8121. doi: 10.1021/ja9003624. [DOI] [PubMed] [Google Scholar]
- 200.van Berkel SS, Dirks AJ, Meeuwissen SA, Pingen DLL, Boerman OC, Laverman P, van Delft FL, Cornelissen JJLM, Rutjes FPJT. ChemBioChem. 2008;9:1805. doi: 10.1002/cbic.200800074. [DOI] [PubMed] [Google Scholar]
- 201.Devarah NK, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2297. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.de Araujo AD, Palomo JM, Cramer J, Seitz O, Alexandrov K, Waldmann H. Chem Eur J. 2006;12:6095. doi: 10.1002/chem.200600148. [DOI] [PubMed] [Google Scholar]
- 203.de Araujo AD, Palomo JM, Cramer J, Kohn M, Schroder H, Wacker R, Niemeyer C, Alexandrov K, Waldmann H. Angew Chem. 2006;118:302. doi: 10.1002/anie.200502266. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:296. [Google Scholar]
- 204.Sun XL, Yang LC, Chaikof EL. Tetrahedron Lett. 2008;49:2510. doi: 10.1016/j.tetlet.2008.02.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Steven V, Graham D. Org Biomol Chem. 2008;6:3781. doi: 10.1039/b807843b. [DOI] [PubMed] [Google Scholar]
- 206.Song W, Wang Y, Qu J, Madden MM, Lin Q. Angew Chem. 2008;120:2874. doi: 10.1002/anie.200705805. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:2832. [Google Scholar]
- 207.Wang Y, Hu WJ, Song W, Lim RKV, Lin Q. Org Lett. 2008;10:3752. doi: 10.1021/ol801350r. [DOI] [PubMed] [Google Scholar]
- 208.Song W, Wang Y, Lin Q. J Am Chem Soc. 2008;130:9654. doi: 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]
- 209.Miller SJ, Grubbs RH. J Am Chem Soc. 1995;117:5855. [Google Scholar]
- 210.Clark TD, Ghadiri MR. J Am Chem Soc. 1995;117:12364. [Google Scholar]
- 211.Blackwell HE, Grubbs RH. Angew Chem. 1998;110:3469. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 1998;37:3281. [Google Scholar]
- 212.Maynard HD, Okada SY, Grubbs RH. J Am Chem Soc. 2001;123:1275. doi: 10.1021/ja003305m. [DOI] [PubMed] [Google Scholar]
- 213.Hong SH, Grubbs RH. J Am Chem Soc. 2006;128:3508. doi: 10.1021/ja058451c. [DOI] [PubMed] [Google Scholar]
- 214.Jordan JP, Grubbs RH. Angew Chem. 2007;119:5244. doi: 10.1002/anie.200701258. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2007;46:5152. [Google Scholar]
- 215.Binder JB, Blank JJ, Raines RT. Org Lett. 2007;9:4885. doi: 10.1021/ol7022505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gulajski L, Michrowska A, Naroznik J, Kaczmarska Z, Rupnicki L, Grela K. ChemSusChem. 2008;1:103. doi: 10.1002/cssc.200700111. [DOI] [PubMed] [Google Scholar]
- 217.Lin YA, Chalker JM, Floyd N, Bernardes GJL, Davis BGI. J Am Chem Soc. 2008;130:9642. doi: 10.1021/ja8026168. [DOI] [PubMed] [Google Scholar]
- 218.Link AJ, Mock ML, Tirrell DA. Curr Opin Biotechnol. 2003;14:603. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 219.Budisa N. Engineering the Genetic Code: Expanding the Amino Acid Repertoire for the Design of Novel Protiens. Wiley-VCH; Weinheim: 2006. [Google Scholar]
- 220.Hendrickson TL, de Crecy-Lagard V, Schimmel P. Annu Rev Biochem. 2004;73:147. doi: 10.1146/annurev.biochem.73.012803.092429. [DOI] [PubMed] [Google Scholar]
- 221.Budisa N. Angew Chem. 2004;116:6586. doi: 10.1002/anie.200300646. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2004;43:6426. [Google Scholar]
- 222.Cowie DB, Cohen GN. Biochim Biophys Acta. 1957;26:252. doi: 10.1016/0006-3002(57)90003-3. [DOI] [PubMed] [Google Scholar]
- 223.van Hest JCM, Kiick KL, Tirrell DA. J Am Chem Soc. 2000;122:1282. [PubMed] [Google Scholar]
- 224.Beatty KE, Xie F, Wang Q, Tirrell DA. J Am Chem Soc. 2005;127:14150. doi: 10.1021/ja054643w. [DOI] [PubMed] [Google Scholar]
- 225.Beatty KE, Liu JC, Xie F, Dieterich DC, Schuman EM, Wang Q, Tirrell DA. Angew Chem. 2006;118:7524. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:7364. [Google Scholar]
- 226.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Proc Natl Acad Sci USA. 2002;99:19. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Link AJ, Vink MKS, Agard NJ, Prescher JA, Bertozzi CR, Tirrell DA. Proc Natl Acad Sci USA. 2006;103:10180. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Proc Natl Acad Sci USA. 2006;103:9482. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Beatty KE, Tirrell DA. Bioorg Med Chem. 2008;18:5995. doi: 10.1016/j.bmcl.2008.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Strable E, Prasuhn DE, Udit AK, Brown S, Link AJ, Ngo JT, Lander G, Quispe J, Potter CS, Carragher B, Tirrell DA, Finn MG. Bioconjugate Chem. 2008;19:866. doi: 10.1021/bc700390r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kast P, Hennecke H. J Mol Biol. 1991;222:99. doi: 10.1016/0022-2836(91)90740-w. [DOI] [PubMed] [Google Scholar]
- 232.Ibba M, Kast P, Hennecke H. Biochemistry. 1994;33:7107. doi: 10.1021/bi00189a013. [DOI] [PubMed] [Google Scholar]
- 233.Kothakota S, Mason TL, Tirrell DA, Fournier MJ. J Am Chem Soc. 1995;117:536. [Google Scholar]
- 234.Datta D, Wang P, Carrico IS, Mayo SL, Tirrell DA. J Am Chem Soc. 2002;124:5652. doi: 10.1021/ja0177096. [DOI] [PubMed] [Google Scholar]
- 235.Sharma N, Furter R, Kast P, Tirrell DA. FEBS Lett. 2000;467:37. doi: 10.1016/s0014-5793(00)01120-0. [DOI] [PubMed] [Google Scholar]
- 236.Kirshenbaum K, Carrico IS, Tirrell DA. ChemBioChem. 2002;3:235. doi: 10.1002/1439-7633(20020301)3:2/3<235::AID-CBIC235>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 237.Kodama K, Fukuzawa S, Nakayama H, Kigawa T, Sakamoto K, Yabuki T, Matsuda N, Shirouzu M, Takio K, Tachibana K, Yokoyama S. ChemBioChem. 2006;7:134. doi: 10.1002/cbic.200500290. [DOI] [PubMed] [Google Scholar]
- 238.Kodama K, Fukuzawa S, Nakayama H, Sakamoto K, Kigawa T, Yabuki T, Matsuda N, Shirouzu M, Takio K, Yokoyama S, Tachibana K. ChemBioChem. 2007;8:232. doi: 10.1002/cbic.200600432. [DOI] [PubMed] [Google Scholar]
- 239.Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. Science. 1989;244:182. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 240.Bain JD, Glabe CG, Dix TA, Chamberlin AR, Diala ES. J Am Chem Soc. 1989;111:8013. [Google Scholar]
- 241.Wang L, Schultz PG. Angew Chem. 2005;117:34. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2005;44:34. [Google Scholar]
- 242.Ellman J, Mendel D, Anthony-Cahill S, Noren CJ, Schultz PG. Methods Enzymol. 1991;202:301. doi: 10.1016/0076-6879(91)02017-4. [DOI] [PubMed] [Google Scholar]
- 243.Ellman JA, Mendel D, Schultz PG. Science. 1992;255:197. doi: 10.1126/science.1553546. [DOI] [PubMed] [Google Scholar]
- 244.Koh JT, Cornish VW, Schultz PG. Biochemistry. 1997;36:11314. doi: 10.1021/bi9707685. [DOI] [PubMed] [Google Scholar]
- 245.Chapman E, Thorson JS, Schultz PG. J Am Chem Soc. 1997;119:7151. [Google Scholar]
- 246.Ting AY, Shin I, Lucero C, Schultz PG. J Am Chem Soc. 1998;120:7135. [Google Scholar]
- 247.Dougherty DA. Chem Rev. 2008;108:1642. doi: 10.1021/cr078207z. [DOI] [PubMed] [Google Scholar]
- 248.Dougherty DA. Curr Opin Chem Biol. 2000;4:645. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- 249.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. J Am Chem Soc. 2003;125:935. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]
- 250.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang ZW, Schultz PG. Science. 2003;301:964. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 251.Young TS, Ahmad I, Brock A, Schultz PG. Biochemistry. 2009;48:2643. doi: 10.1021/bi802178k. [DOI] [PubMed] [Google Scholar]
- 252.Liu WS, Brock A, Chen S, Chen SB, Schultz PG. Nat Methods. 2007;4:239. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 253.Kohrer C, Xie L, Kellerer S, Varshney U, RajBhandary UL. Proc Natl Acad Sci USA. 2001;98:14310. doi: 10.1073/pnas.251438898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ye SX, Kohrer C, Huber T, Kazmi M, Sachdev P, Yan ECY, Bhagat A, RajBhandary UL, Sakmar TP. J Biol Chem. 2008;283:1525. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 255.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. J Am Chem Soc. 2002;124:9026. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 256.Deiters A, Schultz PG. Bioorg Med Chem Lett. 2005;15:1521. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 257.Wang L, Zhang ZW, Brock A, Schultz PG. Proc Natl Acad Sci USA. 2003;100:56. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zhang ZW, Wang L, Brock A, Schultz PG. Angew Chem. 2002;114:2964. [Google Scholar]; Angew Chem Int Ed. 2002;41:2840. [Google Scholar]
- 259.Santoro SW, Wang L, Herberich B, King DS, Schultz PG. Nat Biotechnol. 2002;20:1044. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- 260.Carrico ZM, Romanini DW, Mehl RA, Francis MB. Chem Commun. 2008:1205. doi: 10.1039/b717826c. [DOI] [PubMed] [Google Scholar]
- 261.Wang L, Xie JM, Deniz AA, Schultz PG. J Org Chem. 2003;68:174. doi: 10.1021/jo026570u. [DOI] [PubMed] [Google Scholar]
- 262.Brustad E, Bushey ML, Lee JW, Groff D, Liu W, Schultz PG. Angew Chem. 2008;120:8344. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:8220. [Google Scholar]
- 263.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Proc Natl Acad Sci USA. 2002;99:11020. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Bose M, Groff D, Xie JM, Brustad E, Schultz PG. J Am Chem Soc. 2006;128:388. doi: 10.1021/ja055467u. [DOI] [PubMed] [Google Scholar]
- 265.Zhang ZW, Gildersleeve J, Yang YY, Xu R, Loo JA, Uryu S, Wong CH, Schultz PG. Science. 2004;303:371. doi: 10.1126/science.1089509. [DOI] [PubMed] [Google Scholar]
- 266.Liu CC, Schultz PG. Nat Biotechnol. 2006;24:1436. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 267.Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Nat Chem Biol. 2007;3:769. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- 268.Wu N, Deiters A, Cropp TA, King D, Schultz PG. J Am Chem Soc. 2004;126:14306. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- 269.Deiters A, Groff D, Ryu YH, Xie JM, Schultz PG. Angew Chem. 2006;118:2794. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:2728. [Google Scholar]
- 270.Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. Proc Natl Acad Sci USA. 2006;103:9785. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Wang JY, Xie JM, Schultz PG. J Am Chem Soc. 2006;128:8738. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 272.Speers AE, Cravatt BF. ChemBioChem. 2004;5:41. doi: 10.1002/cbic.200300721. [DOI] [PubMed] [Google Scholar]
- 273.Uttamchandani M, Li JQ, Sun H, Yao SQ. ChemBioChem. 2008;9:667. doi: 10.1002/cbic.200700755. [DOI] [PubMed] [Google Scholar]
- 274.Evans MJ, Cravatt BF. Chem Rev. 2006;106:3279. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 275.Ovaa H, van Swieten PF, Kessler BM, Leeuwenburgh MA, Fiebiger E, van den Nieuwendijk AMCH, Galardy PJ, van der Marel GA, Ploegh HL, Overkleeft HS. Angew Chem. 2003;115:3754. doi: 10.1002/anie.200351314. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2003;42:3626. [Google Scholar]
- 276.Speers AE, Cravatt BF. Chem Biol. 2004;11:535. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 277.Wright AT, Cravatt BF. Chem Biol. 2007;14:1043. doi: 10.1016/j.chembiol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hang HC, Loureiro J, Spooner E, van der Velden AWM, Kim YM, Pollington AM, Maehr R, Starnbach MN, Ploegh HL. ACS Chem Biol. 2006;1:713. doi: 10.1021/cb600431a. [DOI] [PubMed] [Google Scholar]
- 279.Keppler OT, Horstkorte R, Pawlita M, Schmidts C, Reutter W. Glycobiology. 2001;11:11R. doi: 10.1093/glycob/11.2.11r. [DOI] [PubMed] [Google Scholar]
- 280.Kayser H, Zeitler R, Kannicht C, Grunow D, Nuck R, Reutter W. J Biol Chem. 1992;267:16934. [PubMed] [Google Scholar]
- 281.Keppler OT, Stehling P, Herrmann M, Kayser H, Grunow D, Reutter W, Pawlita M. J Biol Chem. 1995;270:1308. doi: 10.1074/jbc.270.3.1308. [DOI] [PubMed] [Google Scholar]
- 282.Herrmann M, von der Lieth CW, Stehling P, Reutter W, Pawlita M. J Virol. 1997;71:5922. doi: 10.1128/jvi.71.8.5922-5931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Schmidt C, Stehling P, Schnitzer J, Reutter W, Horstkorte R. J Biol Chem. 1998;273:19146. doi: 10.1074/jbc.273.30.19146. [DOI] [PubMed] [Google Scholar]
- 284.Mahal LK, Charter NW, Angata K, Fukuda M, Koshland DE, Bertozzi CR. Science. 2001;294:380. doi: 10.1126/science.1062192. [DOI] [PubMed] [Google Scholar]
- 285.Yarema KJ, Mahal LK, Bruehl RE, Rodriguez EC, Bertozzi CR. J Biol Chem. 1998;273:31168. doi: 10.1074/jbc.273.47.31168. [DOI] [PubMed] [Google Scholar]
- 286.Jacobs CL, Goon S, Yarema KJ, Hinderlich S, Hang HC, Chai DH, Bertozzi CR. Biochemistry. 2001;40:12864. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- 287.Lee JH, Baker TJ, Mahal LK, Zabner J, Bertozzi CR, Wiemer DF, Welsh MJ. J Biol Chem. 1999;274:21878. doi: 10.1074/jbc.274.31.21878. [DOI] [PubMed] [Google Scholar]
- 288.Kingsley DM, Kozarsky KF, Hobbie L, Krieger M. Cell. 1986;44:749. doi: 10.1016/0092-8674(86)90841-x. [DOI] [PubMed] [Google Scholar]
- 289.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. Proc Natl Acad Sci USA. 2003;100:9116. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Hang HC, Yu C, Kato DL, Bertozzi CR. Proc Natl Acad Sci USA. 2003;100:14846. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Laughlin ST, Agard NJ, Baskin JM, Carrico IS, Chang PV, Ganguli AS, Hangauer MJ, Lo A, Prescher JA, Bertozzi CR. Methods Enzymol. 2006;415:230. doi: 10.1016/S0076-6879(06)15015-6. [DOI] [PubMed] [Google Scholar]
- 292.Dube DH, Prescher JA, Quang CN, Bertozzi CR. Proc Natl Acad Sci USA. 2006;103:4819. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Oetke C, Brossmer R, Mantey LR, Hinderlich S, Isecke R, Reutter W, Keppler OT, Pawlita M. J Biol Chem. 2002;277:6688. doi: 10.1074/jbc.M109973200. [DOI] [PubMed] [Google Scholar]
- 294.Luchansky SJ, Goon S, Bertozzi CR. ChemBioChem. 2004;5:371. doi: 10.1002/cbic.200300789. [DOI] [PubMed] [Google Scholar]
- 295.Han S, Collins BE, Bengtson P, Paulson JC. Nat Chem Biol. 2005;1:93. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 296.Tanaka Y, Kohler JJ. J Am Chem Soc. 2008;130:3278. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- 297.Rabuka D, Hubbard SC, Laughlin ST, Argade SP, Bertozzi CR. J Am Chem Soc. 2006;128:12078. doi: 10.1021/ja064619y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Proc Natl Acad Sci USA. 2006;103:12371. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Proc Natl Acad Sci USA. 2007;104:2614. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Angew Chem. 2009;121:4090. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:4030. [Google Scholar]
- 301.Sampathkumar SG, Li AV, Jones MB, Sun ZH, Yarema KJ. Nat Chem Biol. 2006;2:149. doi: 10.1038/nchembio770. [DOI] [PubMed] [Google Scholar]
- 302.Chang PV, Prescher JA, Hangauer MJ, Bertozzi CR. J Am Chem Soc. 2007;129:8400. doi: 10.1021/ja070238o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Shao FW, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2487. doi: 10.1021/bc800417b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Nandi A, Sprung R, Barma DK, Zhao YX, Kim SC, Falck JR, Zhao YM. Anal Chem. 2006;78:452. doi: 10.1021/ac051207j. [DOI] [PubMed] [Google Scholar]
- 305.Sprung R, Nandi A, Chen Y, Kim SC, Barma D, Falck JR, Zhao YM. J Proteome Res. 2005;4:950. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 306.Gurcel C, Vercoutter-Edouart AS, Fonbonne C, Mortuaire M, Salvador A, Michalski JC, Lemoine J. Anal Biochem. 2008;390:2089. doi: 10.1007/s00216-008-1950-y. [DOI] [PubMed] [Google Scholar]
- 307.Hanson SR, Hsu TL, Weerapana E, Kishikawa K, Simon GM, Cravatt BF, Wong CH. J Am Chem Soc. 2007;129:7266. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Resh MD. Nat Chem Biol. 2006;2:584. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 309.Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. J Am Chem Soc. 2007;129:2744. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 310.Heal WP, Wickramasinghe SR, Leatherbarrow RJ, Tate EW. Org Biomol Chem. 2008;6:2308. doi: 10.1039/b803258k. [DOI] [PubMed] [Google Scholar]
- 311.Heal WP, Wickramasinghe SR, Bowyer PW, Holder AA, Smith DF, Leatherbarrow RJ, Tate EW. Chem Commun. 2008:480. doi: 10.1039/b716115h. [DOI] [PubMed] [Google Scholar]
- 312.Martin DO, Vilas GL, Prescher JA, Rajaiah G, Falck JR, Bertozzi CR, Berthiaume LG. FASEB J. 2008;22:797. doi: 10.1096/fj.07-9198com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kostiuk MA, Corvi MM, Keller BO, Plummer G, Prescher JA, Hangauer MJ, Bertozzi CR, Rajaiah G, Falck JR, Berthiaume LG. FASEB J. 2008;22:721. doi: 10.1096/fj.07-9199com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Reiss Y, Stradley SJ, Gierasch LM, Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 1991;88:732. doi: 10.1073/pnas.88.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Rose MW, Xu JH, Kale TA, O’Doherty G, Barany G, Distefano MD. Biopolymers. 2005;80:164. doi: 10.1002/bip.20239. [DOI] [PubMed] [Google Scholar]
- 316.Labadie GR, Viswanathan R, Poulter CD. J Org Chem. 2007;72:9291. doi: 10.1021/jo7017747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Xu JH, DeGraw AJ, Duckworth BP, Lenevich S, Tann CM, Jenson EC, Gruber SJ, Barany G, Distefano MD. Chem Biol Drug Des. 2006;68:85. doi: 10.1111/j.1747-0285.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 318.Gauchet C, Labadie GR, Poulter CD. J Am Chem Soc. 2006;128:9274. doi: 10.1021/ja061131o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Nguyen UTT, Cramer J, Gomis J, Reents R, Gutierrez-Rodriguez M, Goody RS, Alexandrov K, Waldmann H. Chem-BioChem. 2007;8:408. doi: 10.1002/cbic.200600440. [DOI] [PubMed] [Google Scholar]
- 320.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng JK, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao YM. Proc Natl Acad Sci USA. 2004;101:12479. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Duckworth BP, Zhang ZY, Hosokawa A, Distefano MD. ChemBioChem. 2007;8:98. doi: 10.1002/cbic.200600340. [DOI] [PubMed] [Google Scholar]
- 322.Neef AB, Schultz C. Angew Chem. 2009;121:1526. doi: 10.1002/anie.200805507. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:1498. [Google Scholar]
- 323.Gramlich PME, Wirges S, Manetto A, Carell T. Angew Chem. 2008;120:8478. [Google Scholar]; Angew Chem Int Ed. 2008;47:8350. [Google Scholar]
- 324.Salic A, Mitchison TJ. Proc Natl Acad Sci USA. 2008;105:2415. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Gratzner HG. Science. 1982;218:474. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- 326.Jao CY, Salic A. Proc Natl Acad Sci USA. 2008;105:15779. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Sylvers LA, Wower J. Bioconjugate Chem. 1993;4:411. doi: 10.1021/bc00024a001. [DOI] [PubMed] [Google Scholar]
- 328.Comstock LR, Rajski SR. Nucleic Acids Res. 2005;33:1644. doi: 10.1093/nar/gki306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Weller RL, Rajski SR. Org Lett. 2005;7:2141. doi: 10.1021/ol0504749. [DOI] [PubMed] [Google Scholar]
- 330.Comstock LR, Rajski SR. J Org Chem. 2004;69:1425. doi: 10.1021/jo035485z. [DOI] [PubMed] [Google Scholar]
- 331.Meier JL, Mercer AC, Rivera H, Burkart MD. J Am Chem Soc. 2006;128:12174. doi: 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- 332.Malolanarasimhan K, Lai CC, Kelly JA, Iaccarino L, Reynolds D, Young HA, Marquez VE. Bioorg Med Chem. 2005;13:2717. doi: 10.1016/j.bmc.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 333.Lummis SCR, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Nature. 2005;438:248. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]; Xiu X, Puskar NL, Shanata JAP, Lester HA, Dougherty DA. Nature. 2009;458:534. doi: 10.1038/nature07768. [DOI] [PMC free article] [PubMed] [Google Scholar]