Abstract
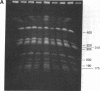
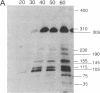
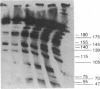
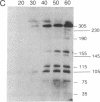
Cell division in Caulobacter crescentus yields progeny cells that differ with respect to cell structure and developmental program. Chromosome replication initiates in the daughter stalked cell but is repressed in the daughter swarmer cell until later in the cell cycle. To study cell-type-specific DNA initiation, chromosome replication was directly analyzed by pulsed-field gel electrophoresis. Analysis of Dra I restriction fragments of DNA taken at various times from synchronized cell cultures labeled with 2'-deoxy[3H]guanosine has allowed us to determine the origin of DNA replication, the rate and direction of fork movement, and the order of gene replication. The first labeled Dra I fragment to appear contains the site of replication initiation. Based on the correlation of the physical and genetic maps derived by Ely and Gerardot [Ely, B. & Gerardot, C. J. (1988) Gene 68, 323-333], the origin was localized to a 305-kilobase fragment containing the rrnA gene. Furthermore, the sequential replication through unmapped Dra I fragments has enabled us to localize their positions on the genome. The order of appearance of labeled restriction fragments revealed that the chromosome replicates bidirectionally at a fork movement rate of 21 kilobases per minute.
Full text
PDF
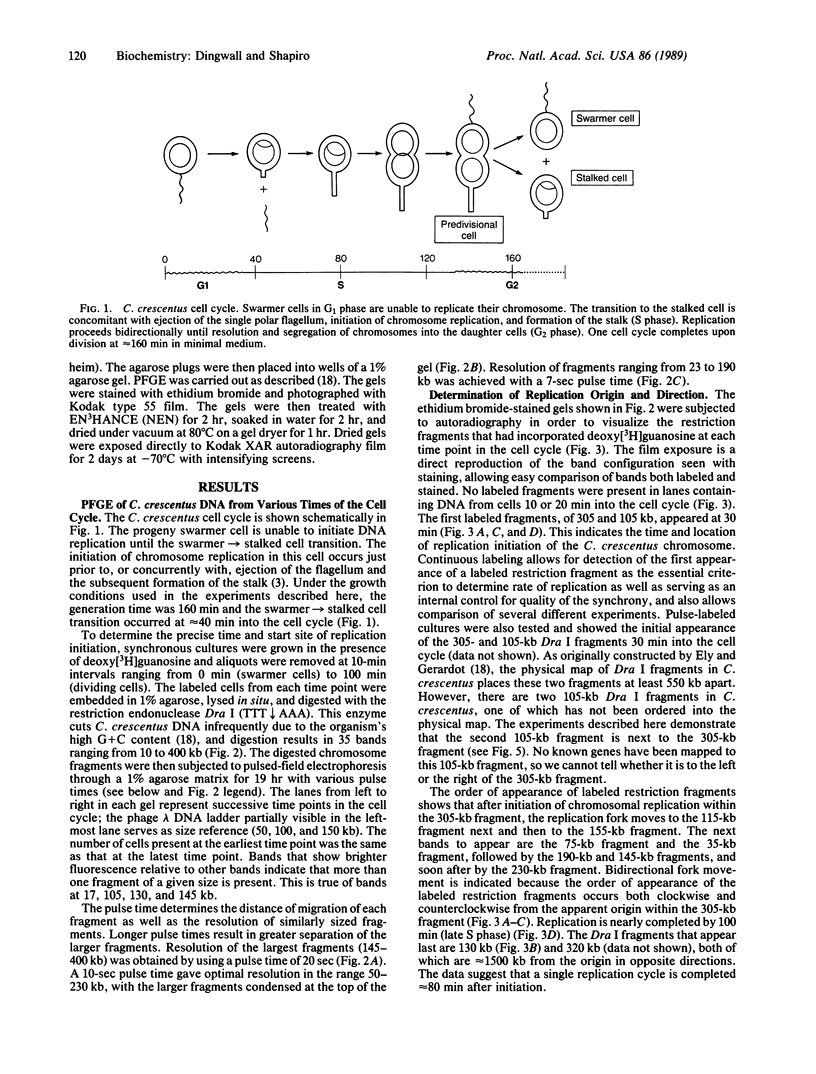
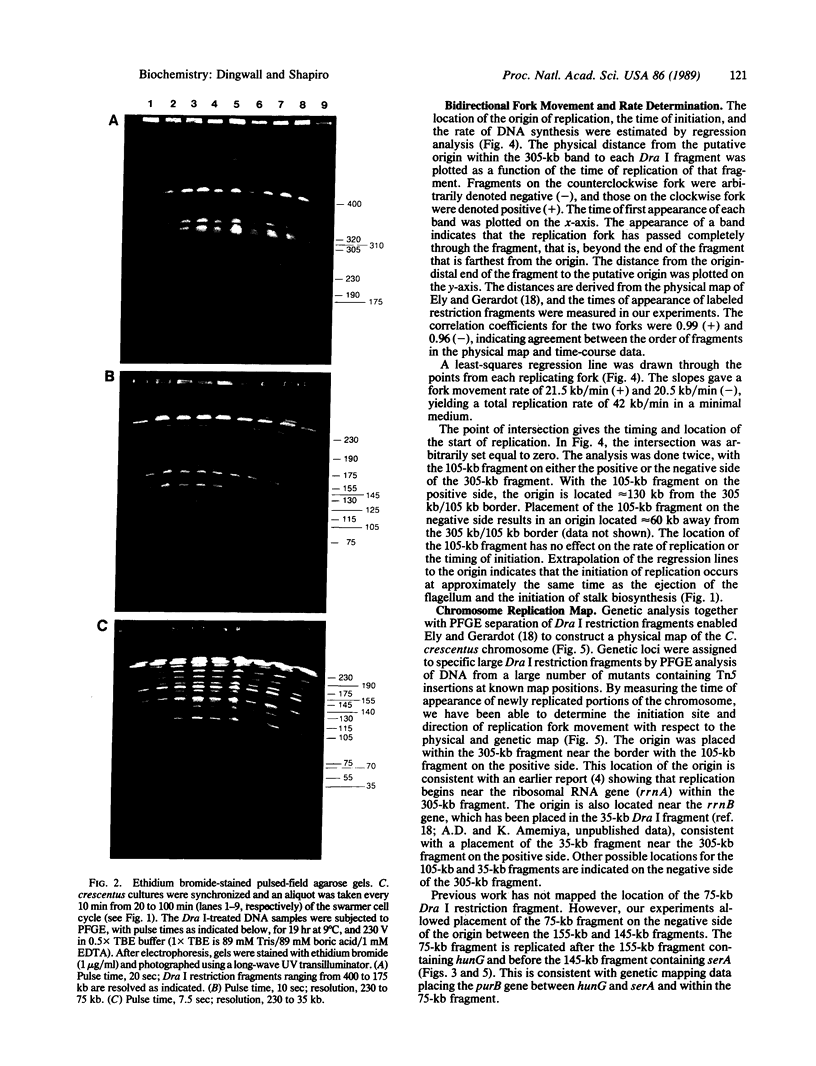
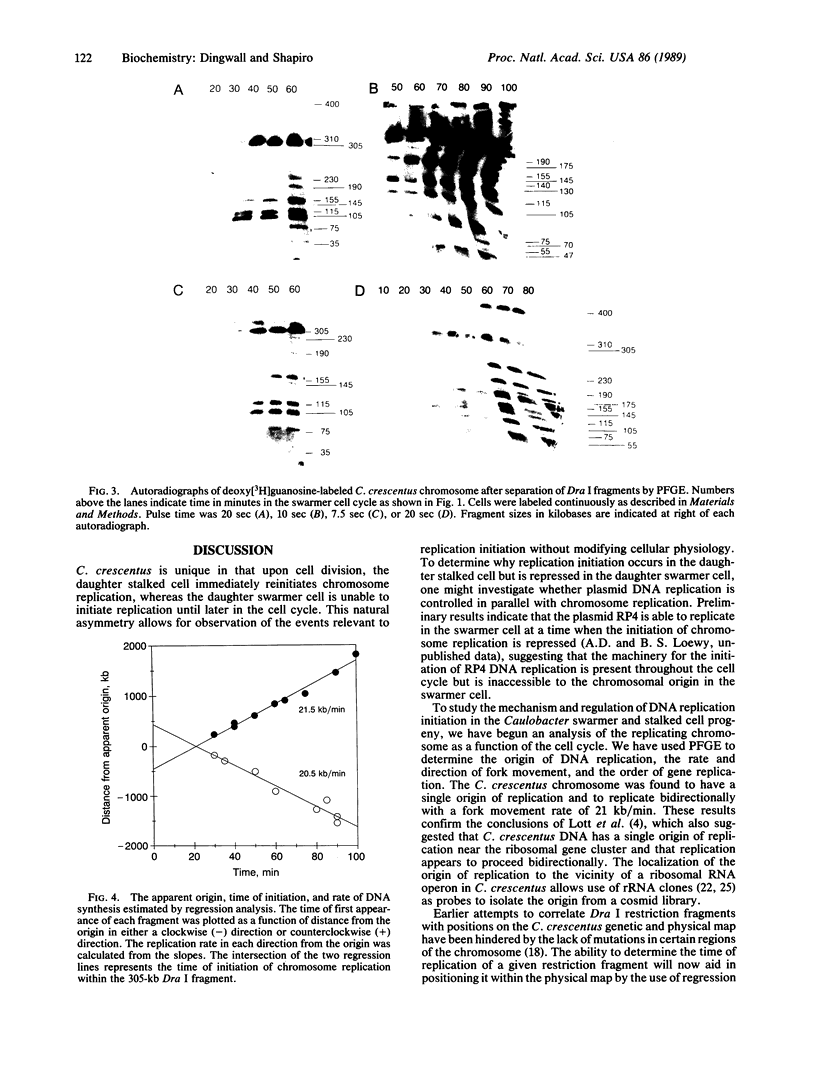

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett J. T., Rhodes C. S., Ferber D. M., Jenkins B., Kuhl S. A., Ely B. Construction of a genetic map for Caulobacter crescentus. J Bacteriol. 1982 Mar;149(3):889–896. doi: 10.1128/jb.149.3.889-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras I., Shapiro L., Henry S. Membrane phospholipid composition of Caulobacter crescentus. J Bacteriol. 1978 Sep;135(3):1130–1136. doi: 10.1128/jb.135.3.1130-1136.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen S. T., Newton A. Chromosome replication during development in Caulobacter crescentus. J Mol Biol. 1972 Mar 14;64(3):671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Gerardot C. J. Use of pulsed-field-gradient gel electrophoresis to construct a physical map of the Caulobacter crescentus genome. Gene. 1988 Sep 7;68(2):323–333. doi: 10.1016/0378-1119(88)90035-2. [DOI] [PubMed] [Google Scholar]
- Evinger M., Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977 Oct;132(1):294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold J., Bellofatto V., Shapiro L., Amemiya K. Organization and nucleotide sequence analysis of an rRNA and tRNA gene cluster from Caulobacter crescentus. J Bacteriol. 1985 Jul;163(1):155–166. doi: 10.1128/jb.163.1.155-166.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstetter C. E., Leonard A. C. Coordinate initiation of chromosome and minichromosome replication in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3489–3494. doi: 10.1128/jb.169.8.3489-3494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseleau-Petit D., Képès F., Peutat L., D'Ari R., Képès A. DNA replication initiation, doubling of rate of phospholipid synthesis, and cell division in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3701–3706. doi: 10.1128/jb.169.8.3701-3706.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Tohdoh N., Jiang X. W., Okazaki T. The distribution and properties of RNA primed initiation sites of DNA synthesis at the replication origin of Escherichia coli chromosome. Nucleic Acids Res. 1985 Oct 11;13(19):6847–6866. doi: 10.1093/nar/13.19.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lother H., Messer W. Promoters in the E. coli replication origin. Nature. 1981 Nov 26;294(5839):376–378. doi: 10.1038/294376a0. [DOI] [PubMed] [Google Scholar]
- Lott T., Ohta N., Newton A. Order of gene replication in Caulobacter crescentus; use of in vivo labeled genomic DNA as a probe. Mol Gen Genet. 1987 Dec;210(3):543–550. doi: 10.1007/BF00327210. [DOI] [PubMed] [Google Scholar]
- Messer W. Initiation of DNA replication in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3395–3399. doi: 10.1128/jb.169.8.3395-3399.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Seiki M., Yoshikawa H. Replication origin region of Bacillus subtilis chromosome contains two rRNA operons. J Bacteriol. 1983 Apr;154(1):50–57. doi: 10.1128/jb.154.1.50-57.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Newton A. Isolation and mapping of ribosomal RNA genes of Caulobacter crescentus. J Mol Biol. 1981 Dec 5;153(2):291–303. doi: 10.1016/0022-2836(81)90279-5. [DOI] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci O., Rickert M. Duplication of Escherichia coli during inhibition of net phospholipid synthesis. J Bacteriol. 1985 Apr;162(1):374–382. doi: 10.1128/jb.162.1.374-382.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Shapiro L. Generation of polarity during Caulobacter cell differentiation. Annu Rev Cell Biol. 1985;1:173–207. doi: 10.1146/annurev.cb.01.110185.001133. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Zyskind J. W., Smith D. W. The bacterial origin of replication, oriC. Cell. 1986 Aug 15;46(4):489–490. doi: 10.1016/0092-8674(86)90873-1. [DOI] [PubMed] [Google Scholar]