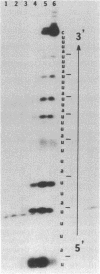
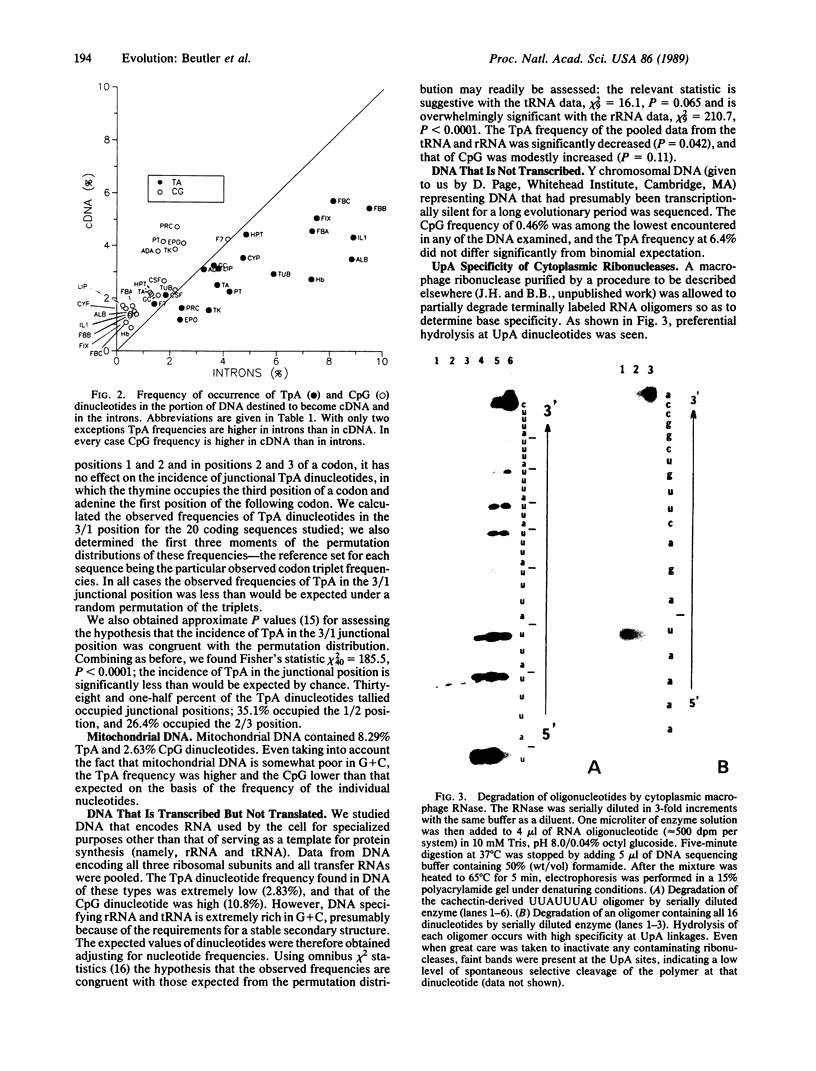
Abstract
Noting the scarcity of CpG dinucleotide in total genomic DNA derived from higher organisms and the scarcity of TpA dinucleotide in total genomic DNA derived from most life forms, we examined the distribution of these dinucleotides in sequences derived from functionally distinct types of human DNA, including mitochondrial DNA, intergenic DNA, intron DNA, and DNA destined to be represented in the cytoplasm as mRNA, tRNA, or rRNA. While CpG frequency has fallen to its lowest levels in DNA that is transcriptionally silent, TpA is most stringently excluded in DNA destined to be expressed as mRNA in the cytosol. This observation suggests that the selective pressures leading to the removal of CpG and TpA operate at different levels. With respect to TpA, dinucleotide scarcity may reflect a requirement for mRNA stability and may indicate the action of UpA-selective ribonucleases. We propose that, by reason of its instability, UpA must have been very rare in primordial RNA. Therefore, tRNA with the anticodon for this dinucleotide may have failed to evolve, making UpA the primordial doublet "stop" codon. The modern triplet code has faithfully conserved this arrangement in the two universal stop codons, UAA and UAG.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alff-Steinberger C. Codon usage in Homo sapiens: evidence for a coding pattern on the non-coding strand and evolutionary implications of dinucleotide discrimination. J Theor Biol. 1987 Jan 7;124(1):89–95. doi: 10.1016/s0022-5193(87)80254-0. [DOI] [PubMed] [Google Scholar]
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Cooper D. N., Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988 Feb;78(2):151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton R. A. Doublet frequencies in the DNA of genetic code limit organisms. J Mol Evol. 1973 Nov 27;2(4):293–302. doi: 10.1007/BF01654097. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Natural variation in the genetic code. Annu Rev Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- JOSSE J., KAISER A. D., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. VIII. Frequencies of nearest neighbor base sequences in deoxyribonucleic acid. J Biol Chem. 1961 Mar;236:864–875. [PubMed] [Google Scholar]
- Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985 May 5;183(1):1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- Lennon G. G., Fraser N. W. CpG frequency in large DNA segments. J Mol Evol. 1983;19(3-4):286–288. doi: 10.1007/BF02099976. [DOI] [PubMed] [Google Scholar]
- Nussinov R. Doublet frequencies in evolutionary distinct groups. Nucleic Acids Res. 1984 Feb 10;12(3):1749–1763. doi: 10.1093/nar/12.3.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Nearest neighbor nucleotide patterns. Structural and biological implications. J Biol Chem. 1981 Aug 25;256(16):8458–8462. [PubMed] [Google Scholar]
- Nussinov R. Strong doublet preferences in nucleotide sequences and DNA geometry. J Mol Evol. 1984;20(2):111–119. doi: 10.1007/BF02257371. [DOI] [PubMed] [Google Scholar]
- Page D. C., Mosher R., Simpson E. M., Fisher E. M., Mardon G., Pollack J., McGillivray B., de la Chapelle A., Brown L. G. The sex-determining region of the human Y chromosome encodes a finger protein. Cell. 1987 Dec 24;51(6):1091–1104. doi: 10.1016/0092-8674(87)90595-2. [DOI] [PubMed] [Google Scholar]
- SWARTZ M. N., TRAUTNER T. A., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem. 1962 Jun;237:1961–1967. [PubMed] [Google Scholar]
- Santibanez-Koref M., Reich J. G. Dinucleotide frequencies in different reading frame positions of coding bacterial DNA sequences. Biomed Biochim Acta. 1986;45(9):1105–1109. [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Youssoufian H., Kazazian H. H., Jr, Phillips D. G., Aronis S., Tsiftis G., Brown V. A., Antonarakis S. E. Recurrent mutations in haemophilia A give evidence for CpG mutation hotspots. 1986 Nov 27-Dec 3Nature. 324(6095):380–382. doi: 10.1038/324380a0. [DOI] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Leinfelder W., Böck A. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc Natl Acad Sci U S A. 1987 May;84(10):3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]