Abstract
U-insertion/deletion RNA editing of mitochondrial mRNAs in trypanosome mitochondria is mediated by a core complex (RECC) containing around 16–20 proteins which is linked to several other multiprotein complexes by RNA. There are two known subcomplexes in the RECC: the REL1 subcomplex which contains the REL1 RNA ligase, the MP63 zinc finger-containing protein and the REX2 U-specific 3'-5' exonuclease; and the REL2 subcomplex which contains the REL2 RNA ligase, the RET2 3' TUTase and the MP81 zinc finger-containing protein. In this study we have affinity isolated recombinant TAP-tagged Leishmania major RET2 and Leishmania tarentolae MP63, REL1 and REL2 proteins after expression in baculovirus-infected insect cells. Recombinant MP63 protein was found to stimulate several in vitro activities of recombinant REL1; these activities include autoadenylation, bridged ligation and even pre-cleaved gRNA-mediated U-insertion editing with RET2 which is in the REL2 subcomplex. There was no effect of recombinant MP63 on similar REL2 ligation activities. The specificity for REL1 is consistent with MP63 being a component of the REL1 subcomplex. These results suggest that in vivo the interaction of MP63 with REL1 may play a role in regulating the overall activity of RNA editing.
Keywords: RNA editing, trypanosomes, mitochondria, RECC, ligation
Introduction
Uridine insertion/deletion RNA editing in trypanosomatid mitochondria is a post-transcriptional process involving the specific insertion and deletion of uridine nucleotides into immature maxicircle transcripts thereby creating translatable mRNAs (Simpson et al. 2003). Guide RNAs (gRNAs) contain the sequence information for editing (Avila and Simpson 1995; Blum et al. 1990; Panigrahi et al. 2008; Pollard and Hajduk 1991; Shu and Stuart 1993; Sturm and Simpson 1990a, b; Yasuhira and Simpson 1995). Editing is mediated by several interacting multiprotein complexes. The core complex or RECC contains 15–20 proteins (Simpson et al. 2004; Stuart et al. 2005), sediments around 20–25S in glycerol gradients and migrates as a band of around 1 MDa in native (Peris et al. 1997) and blue native gels (Li et al. 2009; Osato et al. 2009; Peris et al. 1997). Low resolution 3D structures of the RECC particles from T. brucei and L. tarentolae were recently published (Golas et al. 2009; Li et al. 2009). The specific nomenclature suggestions for the editing complex and proteins which we recently proposed (Simpson et al. 2009) will be used in this paper.
Several of the RECC proteins have conserved motifs that suggest biochemical functions, and the functions of some of these proteins have been confirmed using recombinant proteins. These proteins have been given functional names replacing the operational names. These include the REL1 and REL2 RNA ligases (Gao et al. 2005), the REX1 and REX2 3'-5' U-specific exonucleases (Ernst et al. 2009; Kang et al. 2005; Rogers et al. 2007), the RET2 3' TUTase (Aphasizhev et al. 2003; Ernst et al. 2003), and the REN1, REN2 and REN3 endonucleases (Carnes et al. 2005, 2008; Kang et al. 2006; Panigrahi et al. 2008; Trotter et al. 2005).
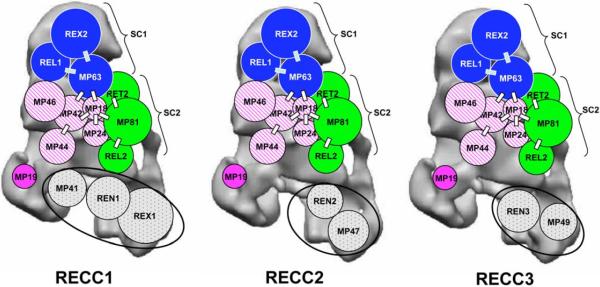
Interactions between RECC protein components have been studied by direct isolation, yeast two hybrid analysis, chemical cross-linking and subcomplex reconstitution with recombinant proteins (Aphasizhev et al. 2003; Schnaufer et al. 2003, 2009; Simpson et al. 2004; Stuart et al. 2005). Two subcomplexes have been identified: the REL1 subcomplex (SC1) contains REL1, MP63 and REX2, and the REL2 subcomplex (SC2) contains REL2, MP81 and RET2 (Aphasizhev et al. 2003; Schnaufer et al. 2003). Evidence for the interaction of these subcomplexes came from in vitro experiments showing that recombinant MP63 (rMP63) interacts not only with rREL1 and rREX2 as expected, but also with rREL2 and rMP81, which are components of the REL2 subcomplex (Kang et al. 2003; Schnaufer et al. 2003, 2009). Also, both REX2 and MP81 interact with MP18 (Schnaufer et al. 2003, 2009). Five proteins - MP24 (Salavati et al. 2006), MP18 (Tarun et al. 2008), MP44 (Wang et al. 2003), MP46 (Babbarwal et al. 2007) and MP42 (Guo et al. 2008) - were found to be involved in the stability of the RECC since down regulation of expression of these proteins in T. brucei produces disruptions of the complex, suggesting that these have extensive protein-protein interactions. A number of RECC proteins (MP81, MP63, MP46, MP42, MP41, and MP47) contain zinc finger motifs which are found in many regulatory proteins. We showed that disruption of the one of the two C2H2 motifs in MP63 in T. brucei led to a partial growth defect and a substantive breakdown of the RECC (Kang et al. 2003), suggesting a structural role for this motif. A model incorporating the known interactions of RECC proteins (Schnaufer et al. 2009) is shown in Figure 1.
Figure 1.
2D Model of RECC proteins within the 3D structure of the L. tarentolae RECC (Li et al. 2009). The areas are proportional to the molecular weights. Protein-protein interactions (Schnaufer et al. 2009) are indicated by bars. The SC1 and SC2 subcomplexes are indicated. The circled proteins are specific for each RECC subclass. Proteins whose removal causes disruption of the complex are indicated by crosshatching. The localization of REL1 has been established by tomography (Li et al. 2009) but the localization of other proteins is based solely on the known protein-protein interactions (Schnaufer et al. 2009) and otherwise is hypothetical. A single copy of each protein is assumed, but there are indications that some (e.g. REL1, MP63) may be present in more than one copy (Aphasizhev et al. 2003; Kang et al. 2003), but this must be resolved by further work.
In this paper we show that recombinant MP63 protein specifically stimulates several activities of recombinant REL1 RNA ligase in vitro and speculate on a possible in vivo regulatory role.
Results
Purification of Recombinant REL1 and REL2 Ligases, RET2 TUTase and MP63
TAP-tagged Lt REL1, Lt REL2 and Lt MP63 were overexpressed in insect cells using the Baculovirus expression system (Invitrogen), and affinity-purified using the standard TAP procedure (Puig et al. 2001). Lm RET2 was purified by binding to IgG agarose followed by Cellulose Phosphate chromatography. This step was used since this protein was not released from calmodulin-agarose with EGTA. Stained gels and Western analysis of the final protein preparations are shown in Figure 2 A, B. Recombinant REL1 and REL2 were purified to near homogeneity. The rREL1 had, in addition to the expected band at 50 kDa, several slowly migrating closely spaced minor bands, as has been observed previously (Gao et al. 2005) and the reason for which is unclear. These minor bands reacted with anti-CBP antibody and could be autoadenylated with α[32P]ATP (Fig. 2 B, C), suggesting that they are conformational isomers of REL1. The purified rRET2 was contaminated with two other minor bands, which were identified as insect hsp70 and β-tubulin by mass spectrometry analysis, and the purified MP63 was contaminated with one other protein, which was identified as β-tubulin.
Figure 2.
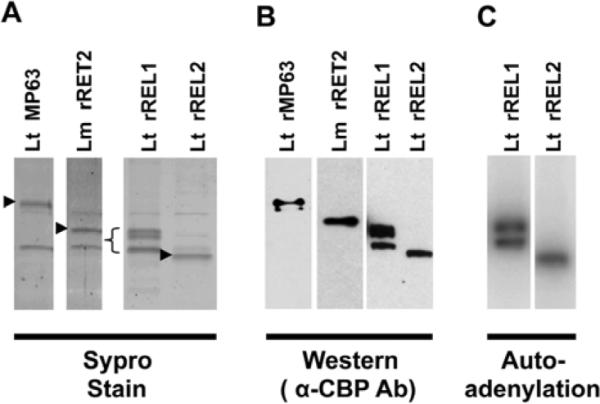
Isolation of CBP-tagged Lt REL1, Lt REL2 and Lt MP63 and Lm RET2 recombinant proteins. (A). Sypro-stained SDS gels of purified proteins. The bands marked with arrows represent the indicated proteins, as determined by mass spectrometry. (B). Western blot of the gels in (A) probed with α-CBP antibody. (C) Autoadenylation of rREL1 and rREL2. Note that all three bands show autoadenylation, indicating that they all contain enzymatically active REL1 ligase.
Recombinant MP63 Specifically Stimulates the Autoadenylation Activity of rREL1
RNA ligase can be covalently labeled with α[32P]ATP due to the fact that the ligation reaction involves hydrolysis of ATP to AMP which remains covalently linked to the enzyme until donated to an RNA substrate. Both purified rREL1 and rREL2 showed in vitro autoadenylation activity (Fig. 2C). Addition of rMP63 increased the level of the autoadenylation activity of REL1 in a dose-dependent fashion (Fig. 3A) but had no effect on the level of REL2 activity (Fig. 3B).
Figure 3.
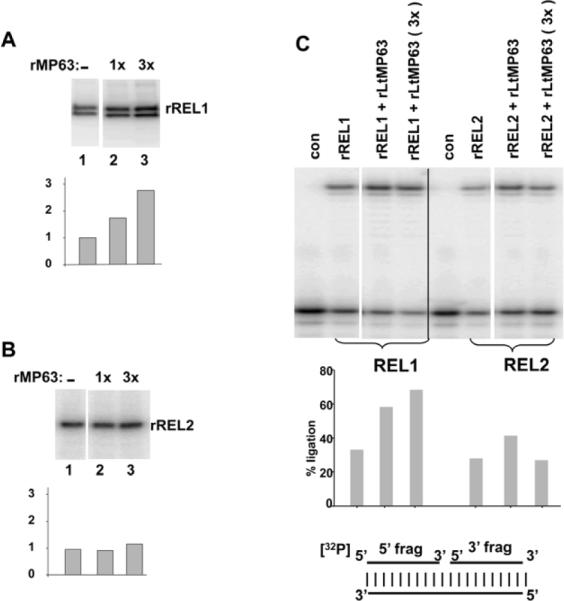
Stimulatory effect of rMP63 on autoadenylation and ligation activity of rREL1. (A) Autoadenylation of rREL1. 1X and 3X indicates 3 nM and 9 nM rMP63 (final concentration). This was repeated three times with an error range of ± 10% (B) Autoadenylation of rREL2. The small increase in REL2 activity in lane 3 is within the error range. (C) ligation of bridged nicked RNA substrate by rREL1 and rREL2. 1X MP63 indicates 1 nM protein. A diagram of the RNA substrate is shown below. The minor increase of REL2 activity in the rREL2+rMP63 (1X) lane was not reproducible.
Recombinant MP63 Specifically Stimulates the Bridged Ligation Activity of rREL1
Pre-cleaved editing is an in vitro reaction in which the mRNA is already cleaved at the editing site and the two fragments are bridged by the cognate gRNA, thereby avoiding the endonuclease requirement. Both rREL1 and rREL2 can efficiently ligate a bridged nicked substrate with similar Km values (Gao et al. 2005). Addition of rMP63 showed, however, a specific upregulation of bridged ligation activity of rREL1 and did not affect the bridged ligation activity of rREL2 (Fig. 3C).
Recombinant MP63 Specifically Stimulates the Pre-cleaved Editing Activity of rREL1
U-insertion editing has been proposed to be mediated by the REL2 subcomplex which contains the RET2 3' TUTase (Cruz-Reyes et al. 2002). Pre-cleaved gRNA-mediated U-insertion can be reconstituted in vitro with rRET2 plus either rREL1 or rREL2 (Fig. 4 A, B). We found that rMP63 increased the extent of the REL1-mediated pre-cleaved +1 U- and +2 U-insertion activities but had no effect on the rREL2-mediated pre-cleaved U-insertion activities (Fig. 4 A, B). This is consistent with the specificity of the effect of rMP63 on rREL1 in vitro.
Figure 4.
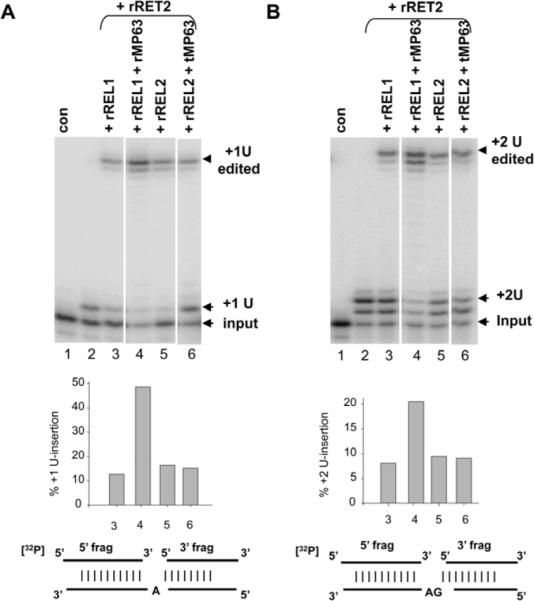
Stimulation of rRET2 + rREL1-mediated pre-cleaved U-insertion editing by rMP63. (A). +1U-insertion editing. A diagram of the RNA substrate is shown below. The histogram shows a quantitation of the lanes above each bar where % U-insertion = +1 U edited product/input (including the +1 U input) × 100. (B). +2U-insertion editing. See (A) for details.
Discussion
We have investigated the role of the MP63 zinc finger-containing protein in the REL1 subcomplex of the RECC using recombinant proteins in vitro. The autoadenylation activity and the bridged ligation activity of rREL1 were stimulated by the addition of rMP63. These effects were specific to REL1 since rMP63 had no affect on either the autoadenylation or bridged ligation activity of REL2, although both enzymes were previously shown to have similar Km values (Gao and Simpson 2003). The specificity of the effect of rMP63 on rREL1 is probably a function of the known in vitro interaction of REL1 and MP63 (Kang et al. 2003; Schnaufer et al. 2009), which are located in vivo in the same subcomplex (Fig. 1), although the mechanism is unclear.
A similar phenomenon has been previously reported for the MP81 protein from T. brucei, which is a component of the REL2 subcomplex (Schnaufer et al. 2003, 2009). Immunoprecipitated recombinant MP81 stimulated the activity of immunoprecipitated recombinant REL2 approximately 11 fold, but had no effect on the activity of recombinant REL1 (Schnaufer et al. 2003).
U-insertion editing, which requires the REN2 endonuclease for the initial cleavage (Carnes et al. 2005), has been proposed (Cruz-Reyes et al. 1998) to utilize the REL2 subcomplex and U-deletion editing, which requires the REN1 endonuclease (Trotter et al. 2005), the REL1 subcomplex. We examined the question whether rMP63, a component of the REL1 subcomplex, could affect in vitro editing mediated by rRET2, a component of the REL2 subcomplex. We first showed that pre-cleaved U-insertion editing could be reconstituted in vitro with just two recombinant enzymes, rRET2 and either rREL1 or rREL2. The addition of rMP63 to the rREL1-mediated U-insertion reaction increased the amount of the edited product for both +1 U editing and +2U editing. Again the stimulatory effect was specific for rREL1, although the extent of precleaved editing with rREL2 + rRET2 alone was equivalent to that of rREL1 + rRET2. Since there is no direct effect of rMP63 on the 3' TUTase activity of rRET2 (data not shown), it is likely that the stimulatory effect of rMP63 on U-insertion and ligation is the result of stimulation of autoadenylation activity of rREL1 by rMP63. This result is not surprising since down regulation of REL2 activity in T. brucei in vivo is not lethal (Gao and Simpson, 2003), suggesting that in vivo REL1 can substitute for REL2 in interacting with RET2 in U-insertion editing in spite of the localization in different subcomplexes (Fig. 1).
We showed previously that both zinc finger motifs of MP63 are required for stability of the RECC to different extents (Kang et al. 2003), suggesting that protein-protein interactions of MP63 with other RECC proteins may be involved with RECC stability. The specific stimulation of pre-cleaved U-insertion editing mediated by rREL1 and rRET2 reported in this paper may be due to the interaction of the rMP63 zinc finger motifs with rREL1 but this remains to be investigated. These results suggest that MP63 may have a regulatory role in the editing reaction in vivo by affecting the activity of the REL1 RNA ligase.
Methods
Cell culture and plasmid constructions
T. brucei 29-13 procyclic cells (from G. Cross, Rockefeller University, New York), which carry integrated T7 RNA polymerase and tetracycline repressor, were cultured as described (Gao et al. 2005).
Recombinant proteins expression in insect cells using the baculovirus system
Cloning and expression of the Lt REL1 maltose-binding protein fusion was described previously (Gao et al. 2005). Monoclonal antibody against Lt REL1 was prepared by the Caltech Monoclonal Facility and tested against L. tarentolae mitochondrial extract. Lt REL2, Lm RET2 and Lt MP63 were cloned into pET161/GW/D-TOPO as His-tagged proteins. The overexpressed proteins were purified with Talon metal affinity resin (Clontech). Polyclonal antibodies against Lt REL2, Lm RET2 and Lt MP63 were prepared by Pacific Immunologies (Ramona, CA). Anticalmodulin-binding peptide (CBP) polyclonal antibody was purchased from Upstate Cell Signaling Solutions (Charlottesville, VA). For expression in the Baculovirus system, Lt REL1, Lt REL2, Lt MP63 and Lm RET2 with C-terminal TAP tags were inserted into the BamHI-EcoRI sites of the pFastBac plasmid (Invitrogen). All proteins except Lm RET2 were purified by binding to IgG agarose followed by TEV release and binding to CBP agarose resin followed by EGTA release. Lm rRET2 irreversibly bound to the CBP agarose and was therefore purified by cellulose phosphate chromatography (Sigma) after release from IgG agarose. The purified recombinant proteins were stored in aliquots at −70 °C.
Ligation and precleaved editing assays
The following RNA substrates were chemically synthesized (Dharmacon, Lafayette, CO; IDT DNA, Coralville, IA) and gel-purified: 5' fragment, 5' GCACUACACGAUAAAUAUAAAAAG-3'; 5'-UU fragment, 5'-GCACUACACGAUAAAUAUAAAAAGUU-3'; 3' fragment, 5'-AACAUUAUGCUUCUUddC-3'; +2 Br RNA (AG Br), 5'-AAGAAGCAUAAUGUUAGCUUUUUAUAUUUAUCGUGUAGUCddG-3'; +1 Br RNA (A Br), 5'-AAGAAGCAUAAUGUUACUUUUUAUAUUUAUCGUGUAGUCddG-3' 0 brRNA, 5'-AAGAAGCAUAAUGUUCUUUUUAUAUUUAUCGUGUAGUCddG-3'.
The 5' fragment RNAs were 5'-phosphorylated with T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP. Complementary RNAs were annealed in 20 μl of 50 mM Tris·HCl (pH 7.5)/2.5 mM MgCl2/20 mM KCl/60 μg/ml BSA/1 mM DTT/30 μM ATP. UTP (1 mM) was added for the U-insertion assay. The reactions were incubated at 27 °C for 45 min for the ligation assay and 90 min for the RNA editing assay, and stopped by addition of three volumes of ethanol at −20 °C. The pellets were redissolved in 80% formamide/1 mM EDTA/50 mM Tris borate (pH 8.3) and electrophoresed in a 15% polyacrylamide-urea sequencing gel. The gel was dried and exposed to a PhosphorImager cassette. The signals were quantitated using IMAGEQUANT software (Molecular Dynamics).
Acknowledgements
We acknowledge the assistance of Martina Nebohacova and Agda Simpson in screening the mouse monoclonal sera for anti-REL1 antibody. We thank Ruslan Aphasizhev for cloning the RET2 gene, and Xuedong Kang for cloning the MP63 gene and characterizing the anti-MP63 antibody. This work was partially supported by NIH grant AI09102 to LS.
Abbreviations
- RECC
RNA Editing Core Complex
- rProtein
recombinant protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila H, Simpson L. Organization and complexity of minicircle-encoded guide RNAs from Trypanosoma cruzi. RNA. 1995;1:939–947. [PMC free article] [PubMed] [Google Scholar]
- Babbarwal VK, Feck M, Ernst NL, Schnaufer A, Stuart K. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei. RNA. 2007;13:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci USA. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Rusche LN, Piller KJ, Sollner-Webb B. T. brucei RNA editing: adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol Cell. 1998;1:401–409. doi: 10.1016/s1097-2765(00)80040-4. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J, Zhelonkina AG, Huang CE, Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. MolCell Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst NL, Panicucci B, Carnes J, Stuart K. Differential functions of two editosome exoUases in Trypanosoma brucei. RNA. 2009;15:947–957. doi: 10.1261/rna.1373009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst NL, Panicucci B, Igo RP, Jr, Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3' terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Gao G, Simpson AM, Kang X, Rogers K, Nebohacova M, Li F, Simpson L. Functional complementation of Trypanosoma brucei RNA in vitro editing with recombinant RNA ligase. Proc Natl Acad Sci USA. 2005;102:4712–4717. doi: 10.1073/pnas.0500553102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J Biol Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- Golas MM, Böhm C, Sander B, Effenberger K, Brecht M, Stark H, Göringer HU. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 2009;28:766–778. doi: 10.1038/emboj.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ernst NL, Stuart KD. The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei. Mol Cell Biol. 2008;28:6939–6953. doi: 10.1128/MCB.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Gao G, Rogers K, Falick AM, Zhou S, Simpson L. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc Natl Acad. Sci USA. 2006;103:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc Natl Acad Sci USA. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Falick AM, Nelson RE, Gao G, Rogers K, Aphasizhev R, Simpson L. Disruption of the zinc finger motifs in the Leishmania tarentolae LC-4 (=TbMP63) L-complex editing protein affects the stability of the L-complex. J Biol Chem. 2004;279:3893–3899. doi: 10.1074/jbc.M310185200. [DOI] [PubMed] [Google Scholar]
- Li F, Ge P, Hu W, Atanasov A, Rogers K, Guo Q, Osato D, Falick AM, Zhou H, Simpson L. Structure of the core editing complex (L-Complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc Natl Acad Sci USA. 2009;106:12306–12310. doi: 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato D, Rogers K, Guo Q, Li F, Richmond G, Klug F, Simpson L. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: in search of the editosome. RNA. 2009;15:1338–1344. doi: 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Ogata Y, Zikova A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2008;9:434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris M, Simpson AM, Grunstein J, Liliental JE, Frech GC, Simpson L. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol Biochem Parasitol. 1997;85:9–24. doi: 10.1016/s0166-6851(96)02795-8. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Hajduk SL. Trypanosoma equiperdum minicircles encode three distinct primary transcripts which exhibit guide RNA characteristics. Mol Cell Biol. 1991;11:1668–1675. doi: 10.1128/mcb.11.3.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rogers K, Gao G, Simpson L. Uridylate-specific 3' 5'-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J Biol Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- Salavati R, Ernst NL, O'Rear J, Gilliam T, Tarun S, Jr, Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Wu M, Park YJ, Nakai T, Deng J, Proff R, Hol WG, Stuart KD. A protein-protein interaction map of trypanosome ~20S editosomes. J Biol Chem. 2009 doi: 10.1074/jbc.M109.059378. (published online Dec 14, 2009), doi: 10.1074/jbc.M109.059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu HH, Stuart K. A Trypanosoma brucei minicircle encodes the same gRNAs as do minicircles of T.equiperdum ATCC 30019 and T.evansi type-A minicircles. Nucleic Acids Res. 1993;21:2951–2951. doi: 10.1093/nar/21.12.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Aphasizhev R, Lukes J, Cruz Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: A proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Sturm NR, Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990a;61:879–884. doi: 10.1016/0092-8674(90)90198-n. [DOI] [PubMed] [Google Scholar]
- Sturm NR, Simpson L. Partially edited mRNAs for cytochrome b and subunit III of cytochrome oxidase from Leishmania tarentolae mitochondria: RNA editing intermediates. Cell. 1990b;61:871–878. doi: 10.1016/0092-8674(90)90197-m. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Schnaufer A, Ernst NL, Proff R, Deng J, Ho W, Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Wang B, Ernst NL, Palazzo SS, Panigrahi AK, Salavati R, Stuart K. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhira S, Simpson L. Minicircle-encoded guide RNAs from Crithidia fasciculata. RNA. 1995;1:634–643. [PMC free article] [PubMed] [Google Scholar]