Abstract
DNA interstrand cross-links (ICLs) are cytotoxic products of common anti-cancer drugs and cellular metabolic processes, whose mechanism(s) of repair remain poorly understood. In this study, we show that cross-link structure affects ICL repair in non-replicating reporter plasmids that contain a mispaired N4C-ethyl-N4C (C-C), N3T-ethyl-N3T (T-T), or N1I-ethyl-N3T (I-T) ICL. The T-T and I-T cross-links obstruct the hydrogen bond face of the base and mimic the N1G-ethyl-N3C ICL created by bis-chloroethylnitrosourea, whereas the C-C cross-link does not interfere with base pair formation. Host-cell reactivation (HCR) assays in human and hamster cells showed that repair of these ICLs primarily involves the transcription-coupled nucleotide excision repair (TC-NER) pathway. Repair of the C-C ICL was five-fold more efficient than repair of the T-T or I-T ICLs, suggesting the latter’s cross-links hinder lesion bypass following initial ICL unhooking. Luciferase expression from plasmids containing a C-C crosslink remnant on either the transcribed or non-transcribed strand increased in NER-deficient cells, indicating NER involvement occurs at a step prior to remnant removal, whereas expression from similar T-T remnant plasmids was inhibited in NER-deficient cells, demonstrating NER is required for remnant removal. Sequence analysis on repaired plasmids showed a high proportion of Cs inserted at the site of the T-T and I-T cross-links and HCR assays showed that Rev1 was likely responsible for these insertions. In contrast, both Cs and Gs were inserted at the C-C cross-link site and Rev1 was not required for repair, suggesting replicative or other translesion polymerases can bypass the C-C remnant.
Interstrand cross-links (ICLs) covalently link two bases on opposite strands of the DNA helix and can be formed by both endogenous and exogenous sources. ICLs are among the most cytotoxic DNA lesions to cells because they prevent the two DNA strands from separating, thereby inhibiting DNA replication and transcription. Additionally, DNA ICLs are cytotoxic products of bifunctional alkylating agents commonly used in cancer chemotherapy (1–3). If left unrepaired, ICLs signal cell death pathways. However, some cancer patients can become resistant to these types of chemotherapies through increased capacity for ICL repair (4–6). DNA ICLs also form endogenously as a result of reaction with lipid peroxidation byproducts (malondialdehyde, crotonaldehyde, and acrolein), nitric oxide, and abasic sites (7–12). Furthermore, lipid peroxidation products have been identified as one class of clinical biological markers for aging (13). A recent finding shows how long-term cancer survivors, who have received chemotherapeutic cross-linking agents, display symptoms of premature aging, a condition known as acquired premature progeroid syndrome (APPS) (13). Many proteins implicated in ICL repair have associated deficiency syndromes that display accelerated aging phenotypes, suggesting that ICLs left unrepaired contribute to aging phenotypes (14–16). Although ICL repair appears to be important for normal maintenance of genome integrity, enhanced ICL repair in tumor cells treated with cross-linking agents can become detrimental to the efficacy of cancer treatments.
Nucleotide excision repair (NER), homologous recombination, translesion synthesis, and many other repair pathways and proteins have been implicated in mammalian ICL repair, but the precise molecular mechanism(s) remains elusive (1, 3, 17). In E. coli and S. cerevisiae, NER is responsible for the initial release of the cross-link from one of the DNA strands (18–19). After release of the cross-link, a process termed “unhooking”, ICLs can be repaired in a homologous recombination-dependent manner when a non-damaged template is available in both prokaryotes and eukaryotes (19–20). Additionally, a recombination-independent repair pathway has been previously identified in E. coli (21–22), in S. cerevisiae (23–24), and in mammalian cells (25–28) that is required if a non-damaged template is not available, as is the case for the G1 phase of the cell cycle. This error-prone, recombination-independent pathway appears to utilize translesion synthesis to bypass the cross-link remnant in order to complete repair of the lesion; consequently, this pathway is likely to be essential for non-dividing cells, such as neurons, as well as in dividing cells during G1 phase of the cell cycle. Recent studies have suggested that cross-link structure and distortion levels affect how ICLs are recognized and repaired (21, 29–31). ICLs arising from various endogenous and exogenous sources have a variety of different structures. In the present study, we examined the effect of cross-links that obstruct the hydrogen bond face of a base on ICL repair in mammalian cells. Such obstruction could prevent DNA from serving as a faithful template during repair synthesis and thus affect overall ICL repair. We hypothesized that the location of the covalent linkage in the interstrand cross-link will dictate repair efficiencies and the involvement of repair proteins during replication-independent ICL repair in mammalian cells. To test this hypothesis, we prepared plasmid DNAs containing a single site-specific mispaired N4C-ethyl-N4C, mispaired N3T-ethyl-N3T, or mispaired N1I-ethyl-N3T ICL, whose structures are shown in Figure 1, and characterized their repair in mammalian cells. Plasmid DNAs were also prepared that contained either the C-C or T-T cross-link remnants placed on the transcribed strand or non-transcribed strand. These substrates allowed further investigation of the importance of lesion remnant removal in ICL repair outside of replication. Furthermore, repaired ICL containing plasmids were isolated from wild-type mammalian cells and analyzed by DNA sequencing to determine the effect of cross-link structure on lesion bypass. We found that ICLs that covalently block the hydrogen bond face of the base display significantly lower repair efficiencies than ICLs that do not interfere with the hydrogen bond face.
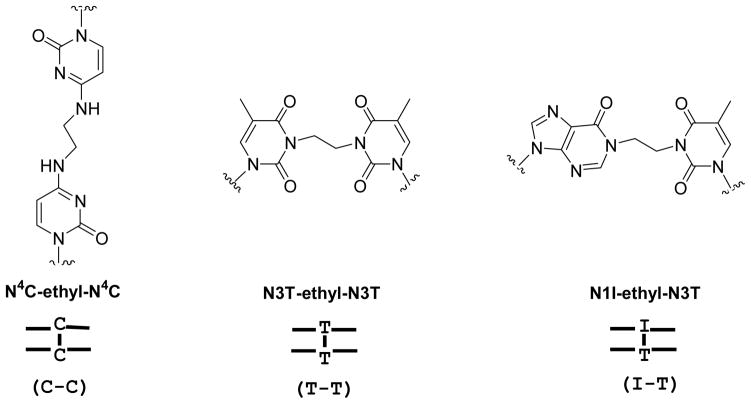
Figure 1.
Structures of the mismatch N4C-ethyl-N4C (C-C), N3T-ethyl-N3T (T-T), and N1I-ethyl-N3T (I-T) interstrand cross-links. The ethyl linker within the C-C cross-link does not hinder the Watson-Crick hydrogen bond face and consequently does not impede the ability of the cross-link to hydrogen bond correctly with guanine. Conversely, the ethyl linkers within the T-T and I-T cross-links block the Watson-Crick hydrogen bond face of the adducted base.
EXPERIMENTAL PROCEDURES
Materials
Protected deoxyribonucleoside 3′-O-phosphoramidites and oligonucleotide synthesis reagents were obtained from Glen Research, Inc. Polynucleotide kinase, T4 DNA ligase, E. coli DNA polymerase I (Klenow fragment), EcoRI, and BstXI were obtained from New England Biolabs, Inc. Van91I was obtained from Fermantas, Inc. Reactions employing these enzymes were carried out in buffer supplied by the manufacturer. All oligonucleotides were synthesized on an ABI DNA synthesizer (model 3400) and purified using high performance liquid chromatography (HPLC) on a Varian instrument using a 0.4 × 25 cm Dionex strong anion exchange (SAX) column. MALDI-TOF mass spectrometry was conducted on an Applied Biosystems Voyager mass spectrometer at the Mass Spectrometry/Proteomics Facility, Johns Hopkins School of Medicine, with support from a National Center for Research Resources shared instrumentation grant (1S10-RR14702). Phosphorimage screens were read on a Fuji Film FLA-7000 phosphorimager.
Substrate Preparation
The sequences of the non-damaged control, cross-linked, and monoadducted duplexes used to prepare the reporter plasmids are shown in Figure S1. Alkyl cross-linked duplexes containing a N4C-ethyl-N4C, N3T-ethyl-N3T, or N1I-ethyl-N3T ICL (see Figure 1) were synthesized, purified and characterized as previously described (32–34). The oligonucleotide containing the N4C-ethyl-N4C cross-link remnant (monoadduct) on the transcribed strand (t.s.) was prepared using a O4-triazole-U at the position of C followed by reaction with 5′-O-dimethoxytrityl-3′-O-t-butyldimethylsilyl-N4-(2-aminoethyl)deoxycytidine as previously reported (31, 34). Oligonucleotides containing a N4C-ethyl-N4C cross-link remnant on the non-transcribed strand (non t.s.) and a N3T-ethyl-N3T cross-link remnant on either the t.s. or non t.s. strand were prepared using cross-link phosphoramidites (32, 35), that contained 5′-O, 3′-O-bis tert-butyldimethylsilyl groups on the nucleoside lacking the β-cyanoethyl-N,N′-diisopropyl phosphoramidite group. The compositions of these oligonucleotides were confirmed by MALDI-TOF mass spectrometry: C-C t.s. m/z expected 3545, m/z observed 3544.4; C-C non t.s. m/z expected 3838, m/z observed 3837.94; T-T t.s. m/z expected 4441, m/z observed 4442.60; and T-T non t.s. m/z expected 4855, m/z observed 4856.30.
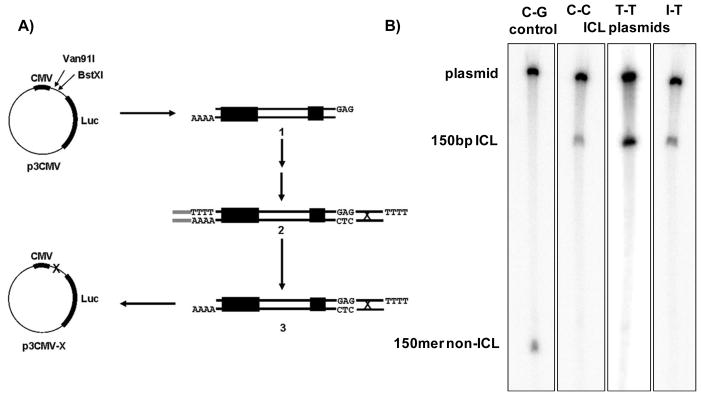
Cross-link-, cross-link remnant-, and non-cross-link-containing plasmids were prepared as outlined in Figure 2A. First, p3CMV, a 5.7 kb plasmid containing a CMV promoter and a firefly luciferase reporter gene (Luc) was generated by inserting a CMV promoter into the multicloning site of pGL3-Basic (Promega). Next, p3CMV was digested with Van91I and BstXI to produce linear plasmid DNA 2A.1, which was gel purified. An adaptor duplex, whose sequence includes part of the recognition site for BstXI was then ligated to the BstXI compatible end. A second ligation of a short cross-linked, monoadducted, or non-cross-linked linear duplex to the Van91I compatible end was carried at 0.4 μM to create 2A.2. The excess duplexes were removed after each ligation step using a MicroSpin S-400 spin column. Plasmid DNA was then digested with BstXI to cleave off the adaptor duplex and the resulting linear duplex 2A.3 was purified on a spin column to remove the cleaved adaptor oligonucleotide. The DNA was circularized by incubating 1nM 2A.3 with T4 DNA ligase. The resulting circular damaged plasmid DNA, p3CMV-X, was purified using cesium chloride-ethidium bromide density gradient centrifugation, which separates the circular plasmid from linear and nicked circular species. The purified plasmid was extracted with 1-butanol (saturated with 1X TE pH 8.0) to remove the ethidium bromide, ethanol precipitated, aliquoted and stored in 1X TE pH 8.0 at −20°C. For the plasmids containing an I-T ICL or cross-link lesion remnants, an additional EcoRI digestion was performed prior to the CsCl step in order to remove contaminating original plasmid. The resulting cross-linked plasmids contained a single site-specific ICL 31nt downstream of the end of the CMV promoter and 73nt upstream of the transcription start site of the firefly luciferase gene. To ensure that the plasmids contained the inserted duplex, the purified plasmids were incubated with EcoRI, whose recognition sequence is only present in the original insert of p3CMV. In the case of the cross-linked plasmids, the presence of the cross-link was confirmed by digesting the plasmid with HindIII which released a 150bp cross-linked fragment. The overhanging ends of this fragment and those of the linear plasmid DNA were filled in with radiolabeled nucleotides using the Klenow fragment of DNA polymerase I. The reaction products were analyzed on a 6% denaturing polyacrylamide gel run at 60°C described in ref. (30) and the results are shown in Figures 2B and S2. Purity of the cross-linked plasmids was additionally confirmed by directly electroporating each purified cross-linked plasmid into a repair-deficient E. coli uvrA recA mutant strain (AB2480).
Figure 2.
A) Scheme of cross-linked plasmid preparation. First, p3CMV was double digested with Van91I and BstXI to create the linear duplex, 2A.1. Next, an adaptor duplex was ligated onto the BstXI compatible end followed by ligation with the cross-linked duplex to form 2A.2. Blocking one end of the duplex prevented multiple ligations of the cross-linked duplex. Digestion by BstXI released the adaptor duplex to produce the cross-linked linear duplex, 2A.3. Finally, the linear cross-linked duplex was circularized under dilute conditions to form the single site-specific interstrand cross-linked plasmid, p3CMV-X. B) Characterization of interstrand cross-linked plasmids. Plasmids were digested with a restriction enzyme to release a 150bp fragment and the fragments were radiolabeled using the Klenow fragment of E. coli DNA polymerase I. The labeled fragments were then analyzed on a 6% gel under denaturing conditions.
Cell Culture
Wild-type CHO (AA8), UV135 (XPG), UV61 (CSB), GM15983 (XPC), and irs1SF (XRCC3), cells were generous gifts from Dr. Michael Seidman. The HeLa cells were a gift from Dr. Michael Matunis. The V79 cells were a gift from Dr. Les Hanakahi. Hec59 and Hec59 + Chr. 2 cells were gifts from Dr. Randy Legerski. HCT116 and HCT116 + Chr. 3 were gifts from Dr. Alan Clark. The MEF cell lines were kind gifts from Drs. Niels de Wind and Maki Moriya. The DT40 cell lines were a generous gift from Dr. Julian Sale. UV41 (XPF) cells were purchased from the American Type Culture Collection (ATCC).
Wild-type HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS). The human mutant cell line, GM15983 (XPC), was also grown in DMEM supplemented with 10% FBS. Wild-type V79 cells were grown in minimal essential medium (MEM) alpha supplemented with 10% FBS. Wild-type Chinese hamster ovary (CHO) cells (AA8) and their derived mutants, UV41 (XPF), UV135 (XPG), and UV61 (CSB), were grown in minimal essential medium (MEM) alpha supplemented with 10% FBS. The mismatch repair human mutant cell line HCT116 (hMLH1) and its complemented parental cell line HCT116 + Chr. 3 were also grown in minimal essential medium (MEM) alpha supplemented with 10% FBS. The mismatch human repair mutant cell line Hec59 (hMSH2) and its complemented parental cell line Hec59 + Chr. 2 were grown in DMEM/F12 supplemented with 10% FBS. Wild-type and REV1−/− MEFs were grown in DMEM supplemented with 10% FBS. DT40 wild-type and REV1−/− cells were grown in DMEM supplemented with 10% FBS and 3% chicken serum. All cells were grown in the presence of 100 units/mL of penicillin and 100 μg/mL of streptomycin. The complemented parental lines for the mismatch repair mutants: HCT116 + Chr. 3 and Hec59 + Chr. 2 were grown in the presence of 400μg/mL of G418.
Transfection and Host-Cell Reactivation Assays
Transient transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen) as recommended by the manufacturer. Cells seeded in 24-well plates at a density of 8.5–13 × 104 cells/well were co-transfected with 2.5 ng of either a non-damaged or damaged reporter plasmid DNA (p3CMV-X), 0.25ng of a Renilla non-damaged plasmid (pHRL-TK), and 212.5 ng of filler DNA (pGEM3Z). Serum-free media containing the DNA/liposome complex was replaced after 6 h with the cell line’s normal growth media. Cells were lysed 26 h after transfection, and luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega). The relative light units (RLU) were measured using a Turner Designs TD 20/20 luminometer. For the MEF cells, a reverse transfection method was performed whereby 2.5 × 105 cells in serum-free media were seeded in combination with the DNA/liposome complex described above. Serum-free media containing the DNA/liposome complex was replaced after 6 h with normal serum-containing growth media and the cells were lysed 26 h after transfection. For the DT40 cells, 2.5 × 105 cells in serum-containing media were seeded in combination with the DNA/liposome complex described above and the cells were lysed 26 h after transfection. At least three replicates were carried out for each transfection and the standard error was determined. For a given damaged plasmid in a given cell line, the percent repair efficiency was determined as the percent expression from the damaged plasmid relative to that of the corresponding non-damaged control plasmid.
Isolation of Plasmid DNA from Mammalian Cells and Lesion Bypass Analysis
For sequencing analysis, Lipofectamine complex containing 50–150ng of cross-linked plasmid (p3CMV-X) was transfected into HeLa cells seeded at a density of 15 × 105 cells per 60mm dish. the cells were lysed 26 h after transfection and plasmid DNA was recovered by a Hirt extraction method (36). The C-C ICL repaired plasmids were isolated from the nuclear fraction to reduce background colonies (see Results), but the other repaired ICL plasmids were isolated from a whole-cell lysate. Isolated plasmids were then electroporated into an ICL repair-deficient E. coli uvrA recA mutant strain (AB2480) and the transformed bacteria were grown on LB/amp plates. Plasmid DNA from the resulting colonies was isolated using a Qiagen Mini-Prep kit. The plasmid DNA was digested with EcoRI to ensure that the isolated plasmids contained the cross-link-containing insert sequence. The transcribed strands of the isolated repaired plasmids were then sequenced to determine the effect of cross-link structure on lesion bypass.
RESULTS
Structures of DNA Interstrand Cross-linked Substrates
The chemical structures of the mispaired N4C-ethyl-N4C (C-C), mispaired N3T-ethyl-N3T ICL (T-T), and mispaired N1I-ethyl-N3T (I-T) interstrand cross-links used in this study are shown in Figure 1. Each of the interstrand cross-links contains an ethyl linker that covalently tethers the two strands of the DNA duplex. The ethyl group of the N4C-ethyl-N4C ICL links the exocyclic amino groups of the cytosines, an arrangement that does not interfere with the Watson-Crick hydrogen bond face of the base pair (34–35). This ICL mimics alkyl cross-links that are formed by nitrogen mustards. In contrast, the ethyl linker of the T-T and I-T ICLs links ring nitrogens of the bases and consequently obstructs their Watson-Crick hydrogen bond faces (32–33). These ICLs mimic the N1G-ethyl-N3C ICL formed by the chemotherapeutic agent bis-chloroethylnitrosourea (BCNU).
Preparation and Characterization of Plasmids Containing a Single Site-Specific Cross-Linked or Cross-link Remnant
Short synthetic duplexes containing single site-specific cross-links or cross-link remnants were synthesized and purified as previously described (32–35). The sequences of these duplexes are shown in Figure S1. The duplexes were designed with overhanging ends complementary to restriction sites within the luciferase reporter plasmid, p3CMV. Non-damaged and damaged plasmid substrates were prepared by the method shown in Figure 2A. In this procedure, an adaptor duplex was first ligated to the linearized plasmid to prevent the cross-linked or remnant-containing duplex from subsequently ligating to both ends of the DNA. Because ligation of both the adaptor and damaged duplexes are bimolecular reactions, they were performed at moderately high plasmid DNA concentrations of 0.4 μM, which resulted in yields of 90% or greater. The adaptor duplex contained part of a restriction enzyme recognition sequence that permitted its cleavage to create a cross-link or cross-link remnant containing linear plasmid that could subsequently be circularized. The circularization reaction, a unimolecular reaction, was performed at low DNA concentrations, conditions that disfavor formation of multiple linear duplexes. We found that circularization reactions carried out at concentrations lower than 1.0 nM resulted in the least amount of multimerization. The resulting circular, cross-linked plasmid DNA, p3CMV-X, was purified by centrifugation using cesium chloride/ethidium bromide density gradients. This procedure was used to prepare up to 10 μg of damaged plasmid that contained a site-specific ICL or cross-link remnant and control plasmids that contained a non-damaged duplex.
The original plasmid, p3CMV, contained an EcoRI recognition site in the insert sequence that was removed with the BstXI and Van91I digestions. Each of the purified non-damaged and damaged plasmids was treated with EcoRI to ensure that they did not contain the original insert. No linearization was detected when the EcoRI treated plasmids were analyzed by agarose gel electrophoresis in the presence of SYBR Green, a result that confirmed the correctly inserted sequence was present in all plasmid substrates (data not shown).
The cross-linked plasmids were further characterized by first digesting them with the HindIII restriction enzyme. This treatment releases a 150 bp fragment that contains the cross-link. The products of this digestion were labeled by a Klenow fill-in reaction and then analyzed by electrophoresis on a denaturing polyacrylamide gel. As shown in Figures 2B and S2, two products were observed: the 150 bp cross-linked duplex, whose mobility is significantly slower than that of the corresponding 150 bp non-cross-linked duplex, and the much larger linear plasmid DNA. There were no detectable non-cross linked fragments in any of the cross-linked preparations. In addition, purified cross-linked plasmids were directly electroporated into a repair-deficient uvrA recA mutant strain (AB2480) in order to quantify the amount of contaminating non-cross-linked plasmid. All three of the cross-linked plasmid preparations had between 0.5–0.8% of the number of colonies compared to a non-damaged control plasmid when propagated in this strain. This result indicates that the purity for all of the purified cross-linked substrates is greater than 99%. As noted in the Methods section, an additional EcoRI digestion to remove contaminating non-cross-linked plasmid in the final sample was used in the preparation of the I-T ICL and the cross-link remnant plasmid preparations. This extra step did not significantly enhance the level of purity as the level of contaminating plasmid was similar for all three cross-links.
Effect of Cross-Link Structure on Replication-Independent ICL Repair in Mammalian Cells
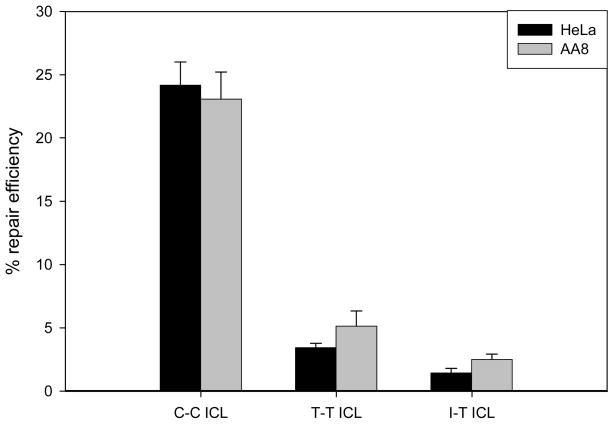
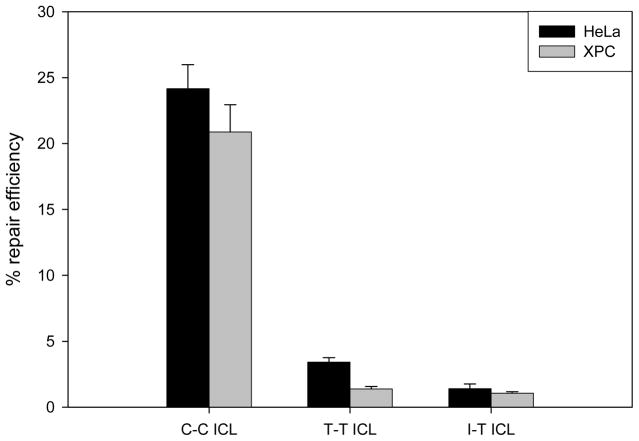
We have observed an NER-independent unhooking reaction that was responsive to the level of distortion of the ICL, but not the chemical structure (30). We then demonstrated that after unhooking, the T-T and I-T cross-links, which block the hydrogen bond face, are not bypassed by Klenow fragment of E. coli Pol I, T4 polymerase or polymerases present in Manley-type mammalian cell extracts, although the C-C cross-link was efficiently bypassed in all cases (31). To examine whether cross-link structure affects ICL repair in vivo, host-cell reactivation (HCR) assays were performed using non-replicating plasmids containing either the C-C, T-T, or I-T cross-links in wild-type human (HeLa) cells and in wild-type Chinese Hamster Ovary (CHO) cells. For each cell line, the percent repair efficiency was determined as the percent expression from the damaged plasmid relative to that of the corresponding non-damaged control plasmid.
The results of these experiments are shown in Figure 3. Expression of the luciferase gene from the plasmid containing the C-C ICL relative to that from the corresponding non-cross-linked control plasmid, a measure of cross-link repair efficiency, was approximately 24% in HeLa cells and 23% in CHO cells. These results, which are similar to those of Li and colleagues (25–26), who studied repair of psoralen and mitomycin C interstrand cross-links, demonstrate that both cell types are capable of repairing the interstrand cross-link under conditions that do not depend on replication of the plasmid or the presence of non-damaged donor plasmids. Repair was also observed for plasmids containing the T-T or I-T cross-links. However, the repair efficiencies for the T-T ICL in HeLa and CHO cells was 3.4% and 5.0%, respectively, and for the I-T ICL in HeLa and CHO cells was 1.4% and 2.4%, respectively. These data show that cross-link structure affects repair and that blocking the hydrogen bond face of the base significantly reduces the repair efficiency of DNA interstrand cross-links.
Figure 3.
Cross-link structure affects interstrand cross-link repair in wild-type mammalian cells. The graph represents data obtained from host-cell reactivation assays employing plasmids containing a single site-specific C-C, T-T, or I-T ICL. Percent repair efficiency represents the relative level of luciferase expression from a damaged plasmid compared to that from a non-damaged control plasmid. Six replicates were performed and the error bars represent the standard error for each data point.
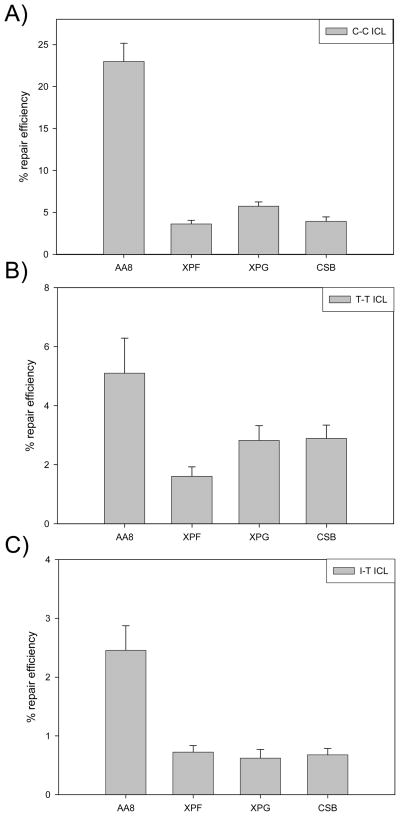
Requirement of NER in Replication-Independent ICL Repair is Independent of ICL Structure
We and others have previously shown, in vitro, that the NER pathway makes dual 5′ incisions removing an undamaged oligo 5′ of an ICL, but does not unhook the cross-link in mammalian cell extracts (30–31). In order to further examine the role of NER in replication-independent ICL repair in vivo, HCR assays were completed for the plasmids containing C-C, T-T, or I-T ICLs in wild-type CHO cells (AA8) and derived NER mutants, UV41 (XPF), UV135 (XPG), and UV61 (CSB), to determine if replication-independent repair is NER-dependent. As shown in Figure 4A, the repair efficiency of the C-C ICL was reduced from 23% in wild-type cells to 3.6%, 5.1%, and 4.0% in the XPF mutant, XPG mutant, and CSB mutants, respectively. Similar reductions were seen for the T-T and I-T cross-links. The repair efficiencies in the XPF mutant, XPG mutant, and CSB mutants were 1.6%, 2.8%, and 2.9%, respectively, for the T-T ICL (Figure 4B), and 0.7%, 0.6%, and 0.7%, respectively, for the I-T ICL (Figure 4C). These results show that transcription-coupled nucleotide excision repair (TC-NER) is required for repair of all three ICLs.
Figure 4.
Transcription coupled-nucleotide excision repair (TC-NER) is involved for interstrand cross-link repair in mammalian cells. The graph represents data obtained from host-cell reactivation assays employing plasmids containing a single site-specific C-C, T-T, or I-T ICL. Percent repair efficiencies of the C-C (A), T-T (B), and I-T ICL (C) interstrand cross-linked plasmids transfected into wild type CHO (AA8) and derived NER mutants: UV41 (XPF), UV135 (XPG), and UV61 (CSB) are shown and represent the relative level of luciferase expression from a damaged plasmid compared to that from a non-damaged control plasmid. Six replicates were performed and the error bars represent the standard error for each data point.
The nucleotide excision repair pathway consists of two subpathways, global genome repair (GG-NER) and TC-NER. Because the cross-links are positioned in the transcribed region between the CMV promoter and the luciferase reporter gene, the plasmids should undergo constitutive transcription and therefore it is not surprising that the TC-NER subpathway is involved in repair of these ICLs.
To determine if the GG-NER subpathway is also involved, we examined the involvement of XPC which, along with hHR23B, forms the initial damage recognition factor in GG-NER yet is not required for TC-NER. We carried out HCR assays in a human cell line deficient in XPC as shown in Figure 5. Although there was a slight reduction in the repair efficiency for the T-T ICL in the XPC-deficient cells, the repair efficiencies for the C-C and I-T interstrand cross-links were not significantly affected in XPC-deficient human cells relative to a human wild-type cell line (HeLa). As a result, TC-NER appears to be the dominant NER subpathway responsible for repair of ICLs when using the plasmid reporter system in vivo.
Figure 5.
Repair efficiencies of C-C, T-T and I-T interstrand cross-linked plasmids transfected into HeLa or XPC-deficient cells. The graph represents data obtained from host-cell reactivation assays that are biased toward transcription-coupled repair as the plasmids used contain a single site-specific C-C, T-T, or I-T ICL between a constitutive CMV promoter and a luciferase gene. The repair efficiencies represent the relative level of luciferase expression from a damaged plasmid compared to that from a non-damaged control plasmid. Six replicates were performed and the error bars represent the standard error for each data point.
Homologous Recombination and Mismatch Repair Are Not Required for Repair of Replication-Independent ICL Repair
To verify that homologous recombination is not involved in replication-independent interstrand cross-link repair, repair efficiencies for each cross-linked plasmid were analyzed in an XRCC3-deficient cell line that lacks a member of the RecA/Rad51-related protein family and cannot undergo homologous recombination. As demonstrated by the data shown in Figure S3, the repair efficiencies of each of the cross-linked plasmids in the XRCC3-deficient cell line, its wild-type parental rodent cell line V79, and wild-type rodent cell line AA8 were comparable. This result confirms that homologous recombination is not required for replication-independent ICL repair.
To examine whether the inherent mismatches within the C-C, T-T, and/or I-T ICLs elicit a mismatch repair response, HCR assays were carried out in two mismatch repair-deficient human cell lines, Hec59 and HCT116. The repair efficiencies of each cross-linked plasmid in the MSH2-deficient (Hec59) cell line was compared to its parental cell line, Hec59 complemented with chromosome 2, and to a human wild-type cell line (HeLa) as shown in Figure S4. There was no significant difference between the hMSH2-deficient and hMSH2-proficient cells, indicating neither MutSα nor MutSβ are involved in reactivation of the reporter plasmids, as the hMSH2 peptide is part of both proteins. Similarly, repair efficiencies were determined in a hMLH1-deficient (HCT116) human cell line and compared to those of its parental line, HCT116 complemented with chromosome 3. As shown in Figure S5, there was a slight decrease in efficiency of repair (14% in the wild type versus 9% in the mutant) for the C-C plasmid, but not for the T-T or I-T plasmids. To test if this minor involvement of hMLH1 in repair of the C-C ICL was caused by the mismatch or the structure of the cross-link, we examined the repair efficiencies of a plasmid that contained the same N4C-ethyl-N4C cross-link in a -CG- staggered sequence (see Figure S2) rather than in a mismatch context. The -CG- oriented ICL has the same chemical structure as the C-C mismatched ICL and has essentially no effect on the structure of the DNA helix (35). As shown in Figure S5, the reduction in repair efficiency was similar for both the -CG- and C-C ICLs indicating that the involvement of hMLH1 is not dependent on the C-C mismatch but rather is dependent on the structure of the N4C-ethyl-N4C lesion.
NER is Not Required for Cross-Link Remnant Removal of the C-C ICL
The HCR assay measures a final readout of luciferase expression, and many different steps in the ICL repair process must take place before the luciferase gene is expressed. For instance, unhooking and repair synthesis at a minimum must take place before allowing expression. It is not clear whether the cross-link remnant affects expression in the HCR assay and if NER is required for remnant removal. Because many steps are actually being monitored in the HCR assay, reduced luciferase expression from a cross-linked plasmid in a mutant cell line indicates the proteins involvement in repair, but the exact step(s) at which it is acting remains in question.
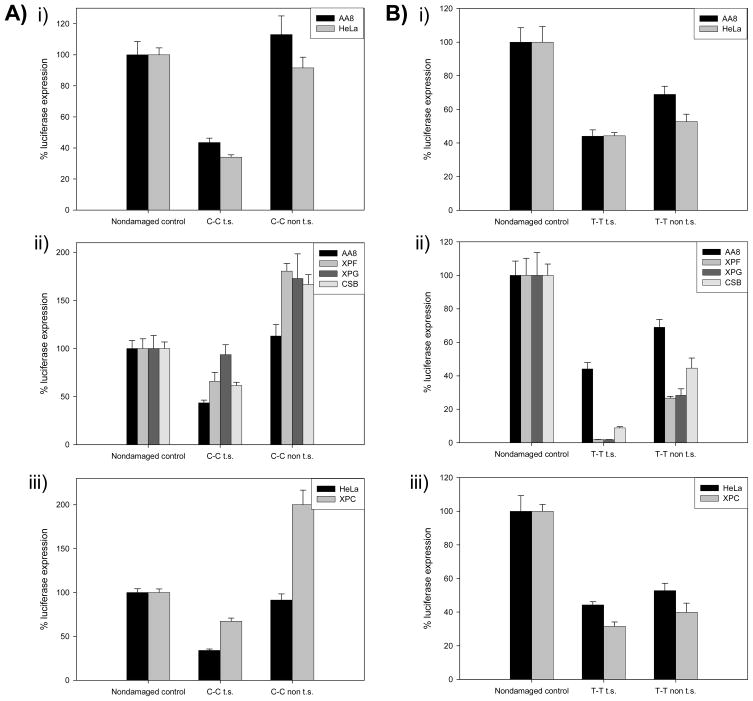
We have previously shown that the unhooked product we observe in mammalian cell extracts only contains a single tethered base (31). Therefore, these plasmids mimic an ICL repair intermediate that we have observed in mammalian extracts that has undergone unhooking and repair synthesis. To investigate whether NER is required for the cross-link remnant removal step of replication-independent ICL repair in cultured cells, plasmids containing a cross-link remnant lesion with a single tethered base were generated. Host-cell reactivation assays were first performed with plasmids containing a C-C cross-link remnant placed on either the transcribed or non-transcribed strand. As displayed in Figure 6Ai, when the C-C remnant was placed on the transcribed strand the efficiency of luciferase expression was 52% and 34% that of the non-damaged control plasmid in wild-type AA8 (CHO) and HeLa cells, respectively. In contrast, the plasmids containing the C-C cross-link remnant on the non-transcribed strand had relative expression efficiencies of 113% in AA8 and 92% in HeLa cells. These results suggest that in wild-type cells, transcription is only partially inhibited when the C-C remnant is placed on the transcribed strand and that it has essentially no effect when placed on the non-transcribed strand.
Figure 6.
Luciferase expression from plasmids containing a C-C (A) or T-T (B) cross-link remnant on the transcribed (t.s.) or non-transcribed (non t.s.) strand after transfection into wild-type HeLa or AA8 cells (panels Ai and Bi); XPF- (UV41), XPG- (UV135) or CSB- (UV61) deficient hamster cells (panels Aii and Bii); or XPC-deficient human cells (panels Aiii and Biii). The luciferase expression is given as the percent expression of a non-damaged control plasmid. Three replicates were performed and the error bars represent the standard error for each data point.
Host-cell reactivation assays were then carried out in hamster NER-deficient cell lines. As shown in Figure 6Aii, luciferase expression increased in the XPF, XPG and CSB mutants regardless of whether the C-C cross-link remnant was located on the transcribed or the non-transcribed strand of the plasmid. These results suggest that transcription past the N4C-ethyl-N4C cross-link remnant on is more efficient in the absence of an intact TC-NER complex.
To determine if GG-NER impedes transcription past the N4C-ethyl-N4C cross-link remnant, HCR assays were executed in human XPC-deficient cells. The results of this experiment, which are shown in Figure 6Aiii, suggest that the proteins involved in GG-NER pathway also act as impediments to transcription past a C-C remnant located on either strand of the plasmid.
Requirement of NER for T-T Cross-Link Remnant Removal is DNA Strand Dependent
Host-cell reactivation assays were next performed with plasmids containing a T-T cross-link remnant placed on either the transcribed or non-transcribed strand. As displayed in Figure 6Bi, when the T-T remnant was placed on the transcribed strand luciferase expression was 45% and 44% that of the non-damaged control in wild-type AA8 (CHO) and HeLa cells, respectively. The plasmids containing the T-T cross-linked remnant on the non-transcribed strand had slightly higher levels of luciferase expression, 69% and 53% in AA8 (CHO) and HeLa cells, respectively. These results indicate that the presence of a N3T-ethyl-N3T cross-link remnant on either the transcribed or non-transcribed strand partially impairs transcription in wild-type cells.
The T-T remnant-containing plasmids were subsequently assayed in the NER-deficient cell lines as displayed in Figure 6Bii. Luciferase expression in all three of the TC-NER mutants declined substantially for the plasmid containing the T-T remnant on the transcribed strand. Thus, it appears that a functional NER complex is essential for transcription past this T-T remnant when it is located on the transcribed strand. Surprisingly, luciferase expression was also reduced in the absence of intact NER machinery when the T-T remnant was placed on the non-transcribed strand. This result suggests that removal of the remnant from the non-transcribed strand, although not absolutely necessary for transcription, does facilitate transcription.
To determine if GG-NER is involved in T-T cross-link remnant removal, HCR assays were conducted in a human XPC-deficient cell line. The results of this experiment, which are shown in Figure 6Biii, show that the proteins involved in GG-NER pathway also appear to enhance transcription past the T-T remnant on either strand, although to a lesser extent than the proteins involved in TC-NER. This result suggests that in the HCR assay, TC-NER is the dominant NER subpathway required for transcription past a T-T cross-link remnant lesion placed on either DNA strand.
Taken together, the data suggest that the NER-dependence observed in replication-independent repair of the C-C ICL is due to an ICL repair step preceding the cross-link remnant removal step, and is not due to the cross-link remnant removal itself. Conversely, NER is required for T-T cross-link remnant removal. Accordingly, the dependence of NER for cross-link remnant removal depends on cross-link structure and on which strand the damage is located.
Cross-Link Structure Affects Lesion Bypass of Replication-Independent ICL Repair in Mammalian Cells
We next sequenced the repaired plasmids to determine the base composition at the site of the cross-links. Following transfection into HeLa cells, plasmids originally containing a single C-C, T-T, or I-T ICL were isolated from the cells using a modified Hirt procedure (36) and the recovered plasmids were electroporated into a repair-deficient E. coli uvrA recA mutant strain (AB2480). This strain was used in order to prevent bacterial repair of remaining unrepaired cross-linked plasmids transformed into E. coli, so that only repair from mammalian cells was monitored. Colonies were then isolated from the repair-deficient E. coli strain and analyzed by sequencing the DNA of the transcribed strand.
To validate that the observed repair was carried out by the HeLa cells and not by the bacterial cells, cross-linked plasmids were directly transformed into the AB2480 strain. The transformation efficiencies in these cells of the C-C, T-T, and I-T ICL plasmids compared to that of a non-damaged control plasmid were 2.7%, 0.8%, and 0.7%, respectively. Thirty colonies derived from each cross-linked plasmid were tested for their susceptibility to digestion by EcoRI in order to determine if they originated from contaminating plasmid that never contained the cross-link insert. Only the C-C ICL had colonies that were refractory to EcoRI, a result that suggests that the repair-deficient E. coli strain can only repair the C-C and not the T-T or I-T ICLs. Although the repair-deficient AB2480 strain showed repair of the C-C ICL plasmid, the repair was very low (~2.2%) of that compared to a non-damaged control. Twenty-four colonies that were refractory from the C-C ICL experiments were sequenced and all were repaired in an error-free manner. To reduce the background in the sequence analysis experiment for the C-C ICL plasmids, plasmids were isolated from the nuclear fraction. Because no direct repair occurred in the repair of the T-T and I-T ICLs in the bacterial mutant strain, the repair seen for these cross-links arose only from the mammalian cells.
Although no 1 bp deletions or mutations were seen at sites adjacent to the cross-link, large deletions (~1–2kB) were observed for all three cross-links. These deletions, which were seen in approximately 10% of the recovered repaired plasmids, were sequenced to determine the nature of the deletions. The deletions appeared to be random as they displayed no large regions of homology and/or small regions of homology near the deletion, a result that suggests a random end joining mechanism allowed the plasmid to still propagate in E. coli by keeping an intact ampicillin resistance gene and bacterial origin of replication.
The sequencing data is displayed in Table 1 and shows that of the 114 colonies tested, the C-C ICL was repaired in an error-free manner with approximately 60% Cs and 38% Gs inserted at the cross-link site on the transcribed strand. In contrast, the sequencing results show that in the case of the T-T ICL approximately 81% Cs were inserted at the site of the cross-link on the transcribed strand out of 104 colonies tested. Similarly, the I-T results showed approximately 94% Cs inserted at the site of the cross-link on the transcribed strand out of 84 colonies tested.
Table 1.
Sequence Analysisa
| C-C ICL | T-T ICL | I-T ICL | |
|---|---|---|---|
| C | 68 (60%) | 84 (81%) | 79 (94%) |
| G | 43 (38%) | 3 (3%) | 1 (1%) |
| A | 3 (3%) | 9 (9%) | 0 (0%) |
| T | 0 (0%) | 8 (8%) | 4 (5%) |
| Total colonies | 114 | 104 | 84 |
DNA sequence analysis of repaired reporter plasmids recovered after transfection into wild-type HeLa cells. Number reported represents colonies counted that contained the specific nucleotide at the site of the ICL on the transcribed strand of a repaired cross-linked plasmid.
In order to verify that the E. coli strain AB2480 was itself not inserting Cs across from the T-T cross-link remnant that could hypothetically remain on the isolated plasmid, a plasmid containing a single T-T remnant on the non-transcribed strand was directly electroporated into AB2480. The transcribed strand of DNA isolated from 26 colonies was sequenced. All 26 colonies had an A placed at the site opposite the remnant. This result confirms that for both the T-T and I-T ICLs, the Cs were inserted by the HeLa cells and not by the repair-deficient bacterial cells.
Rev1 is Essential for Repair of ICLs that Block the Hydrogen Bond Face
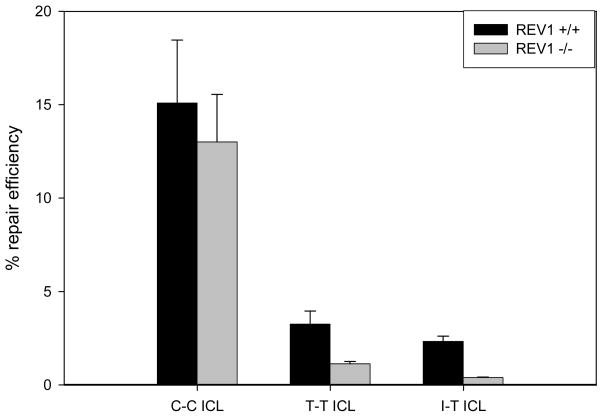
Because the sequencing data showed a high percentage of Cs inserted at the site of the T-T and I-T ICLs, we sought to determine if Rev1 was responsible. We first carried out HCR experiments in cells treated with anti-Rev1 siRNAs. The western blots displaying the results of the knockdown experiments are shown in Figure S6A. Although significant knockdown was achieved, essentially no effect on the repair of any of the three cross-links was observed. To determine if this lack of inhibition was due to insufficient knockdown of Rev1, REV1 knockout MEFs (37) and DT40 cells (38) were obtained and HCR assays were performed in these cells. As shown in Figure 7, the repair of the plasmid containing a C-C ICL relative to a non-damaged plasmid in wild-type MEF and REV1−/− MEF cells was approximately 15% and 13%, respectively. The relative repair percentage for the T-T ICL in wild-type MEF and REV1−/− MEF cells was 3.2% and 1.1%, respectively, and for the I-T ICL in wild-type MEF and REV1−/− MEF cells was 2.3% and 0.4%, respectively. Similar results were observed in REV1−/− DT40 cells and these results are shown in Figure S6B. These results clearly demonstrate that Rev1 is essential for bypass of cross-links that block the hydrogen bond face of the Watson-Crick base pair.
Figure 7.
Cross-link structure affects Rev1 involvement of lesion bypass in vivo. The graph represents repair efficiencies of C-C, T-T and I-T interstrand cross-linked plasmids transfected into wild-type (REV1+/+) and Rev1-deficient (REV1−/−) mouse embryonic fibroblast cell lines. Percent repair efficiency is the relative level of luciferase expression from a damaged plasmid compared to that from a non-damaged control plasmid. Six replicates were performed and the error bars represent the standard error for each data point.
DISCUSSION
DNA ICLs are one of the most deleterious lesions a cell can encounter, covalently linking the two strands of DNA in a double helix and completely blocking replication and transcription. These lesions are formed endogenously through byproducts of lipid peroxidation, such as malondialdehyde (7, 11–12) and many other endogenous sources (8–10, 39). Cytotoxic ICLs are also formed by many common chemotherapeutic bifunctional agents, such as cisplatin, mitomycin C, nitrosoureas, and nitrogen mustards (1, 3, 40). These cross-linking agents create cross-links of different structures in DNA, and our studies here further support the notion that they are likely repaired in a differential manner. For example, the cisplatin ICL, which links N7 atoms of G in a 5′-GC-3′ sequence, severely distorts the DNA helix (1, 3, 41), whereas mitomycin C creates relatively non-distorting ICLs by linking the G exocyclic amino groups of 5′-CG-3′ in the minor groove of DNA (1, 3, 42). Nitrosoureas, such as 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU), create ICLs between heterocyclic nitrogens N1G and N3C on the Watson-Crick hydrogen bond faces of a G-C base pair (3, 43). In contrast, nitrogen mustard ICLs, which reside in the DNA major groove, primarily link the N7 atoms of guanines in a 5′-GNC-3′sequence and thus are not expected to interfere with the Watson-Crick hydrogen bonding potential of guanine to the complementary cytosine (1, 44). Two of the three ICLs used in our study, T-T and I-T, are close mimics of the BCNU ICL where the cross-link blocks the Watson-Crick hydrogen bond face. The third ICL, C-C, mimics alkyl ICLs that do not block the Watson-Crick hydrogen bond face and are thus similar to those formed by nitrogen mustards. These cross-link mimics were used in this study and demonstrated that the location of the covalent linkage of the cross-link structure affects ICL repair.
Duplexes containing a single site-specific ICL were synthesized using automated phosphoramidite chemistry (32–34) and were used to construct cross-linked plasmids to study the repair of defined, structurally different ICLs in mammalian cells. We have shown previously that ICLs that differ structurally are not repaired in the same manner in vitro, as evidenced by the observation that ICL structure affects repair synthesis in mammalian whole-cell extracts (31). For example, of the three cross-links, C-C, T-T and I-T, studied, only the C-C cross-link was bypassed during the repair synthesis step in HeLa or CHO whole-cell extracts (31).
When the cross-linked plasmids were transfected into wild-type mammalian cells, luciferase expression from the C-C ICL plasmid was approximately 5-fold higher than that from either the T-T or the I-T ICL plasmid (see Figure 3). It appears then, that cross-link structure affects repair efficiency, and similar to our previously published results in mammalian extracts (31), blocking the Watson-Crick hydrogen bond face interferes with the repair process. A similar conclusion was reached by Loechler and colleagues who presented evidence for a recombination-independent ICL repair pathway in E. coli, and provided data suggesting specific pathways may repair ICLs differently due to the nature of the cross-link structure (21).
It should be noted that our cross-linked plasmids lack a mammalian origin of replication. Therefore, repair of the cross-link would not be expected to involve replication- or recombination-dependent pathways. To verify this, we showed that luciferase expression from the C-C, T-T or I-T plasmids was essentially the same in wild-type and XRCC3-deficient hamster cells (see Figure S3). The latter cells lack a member of the RecA/Rad51-related protein family, which is required for homologous recombination. Considering that a homologous non-damaged donor plasmid was not provided in these experiments, this result is consistent with what would be expected for replication-independent repair in a G1 type context. The repair efficiency for the C-C ICL plasmid, approximately 25%, is comparable to the efficiencies previously reported by Li and coworkers for recombination-independent repair of psoralen and mitomycin C cross-linked plasmids in human and hamster cells (25–26).
Because the C-C, T-T, and I-T plasmids contain mismatched base-pairs it seemed possible that the mismatched repair (MMR) pathway could be involved in their repair. However transfection of these plasmids into human cell lines deficient in hMLH1 or hMSH2, proteins required for MMR, did not adversely affect luciferase expression, suggesting that the MMR pathway is not involved in the repair of these lesions (see Figures S4 and S5). This result is consistent with the observation by Muniandy et al. that ICL repair is independent of MSH2 during G1 phase of the mammalian cell cycle (28).
In contrast, a significant decrease in repair efficiency was observed when the C-C, T-T, and I-T plasmids were transfected into hamster cell lines deficient in the NER endonucleases XPF or XPG (see Figure 4). Similar decreases were seen when the cross-linked plasmids were transfected into hamster cells deficient in CSB, a protein required for transcription-coupled NER (TC-NER). However, HCR assays performed in human XPC-deficient cells showed that XPC, which is involved in the initial recognition step of global genome repair NER (GG-NER), is not required for repair of the C-C and I-T ICLs, although it does appear to be involved in repair of the T-T ICL (see Figure 5). This difference may reflect the involvement of GG-NER at the level of the cross-link remnant removal, as XPC was involved for transcription across a T-T cross-link remnant when placed on either the transcribed or non-transcribed strand (see Figure 6Biii), whereas transcription from a plasmid containing the C-C remnant on either strand did not involve XPC. Therefore, the TC-NER subpathway appears to be the dominant subpathway required for the NER-dependent ICL repair of the C-C and I-T cross-links. This bias towards transcription-coupled repair is not unexpected as the cross-linked plasmids contain a strong CMV promoter and are constitutively transcribed, therefore this result does not rule out participation of GG-NER in ICL repair outside of a transcribed region. A similar dependence on the TC-NER pathway was observed by Li and colleagues for the repair of a psoralen cross-linked plasmid (25). However, they also observed that in the case of a mitomycin C ICL, both the GG- and TC-NER pathways were equally involved in repair of this lesion (26). Our experiments and those of Li et al., further emphasize that cross-link structure plays an important role in determining the pathway by which this type of lesion is repaired.
The precise involvement of NER in mammalian cross-link repair has yet to be determined. Studies in E. coli and S. cerevisiae have indicated that NER acts to initiate unhooking of ICLs (1, 21, 23, 45). A second round of NER is thought to remove the remaining cross-link remnant (1). Therefore, it is plausible that a cross-link remnant on the non-excised strand could serve as substrate for NER, which is capable of detecting a broad range of DNA lesions (46).
In order to determine whether NER is required for the cross-link remnant removal step, plasmids were constructed in which a C-C or T-T remnant was placed on either the transcribed or non-transcribed strand. Luciferase expression, which serves as a measure of transcription past the cross-link remnant, decreased approximately 60% when the C-C remnant was placed on the transcribed strand, whereas placing the remnant on the non-transcribed strand had essentially no effect in wild-type human and hamster cells (see Figure 6A). When these plasmids were transfected into cells deficient in TC-NER proteins, the level of expression actually increased indicating that NER impedes transcription past the C-C remnant when located on either the transcribed or non-transcribed strand. The results also suggest that the presence of the C-C remnant may destabilize the DNA thus facilitating unwinding of the double helix and resulting in increased luciferase expression compared to that of a non-damaged substrate.
In contrast to the behavior of the C-C remnant, NER appears to be essential for transcription past the T-T remnant when it is located on the transcribed strand (see Figure 6B). Most likely the presence of the linker attached to the hydrogen bond face of the T prevents RNA polymerase from inserting a base opposite the lesion thus inhibiting transcription. Consistent with this interpretation is our previous observation that DNA polymerases are able to copy past a C-C remnant but not past a T-T remnant when these lesions are placed on the template strand (31). Interestingly, transcription is also partially dependent on NER when the T-T remnant is located on the non-transcribed strand. These results are reminiscent of the inhibitory effects of G4 DNA and a 1,3 cisplatin intrastrand cross-link on transcription when these modifications are present on the non-transcribed strand (47–48). The results of the HCR experiments for the C-C, T-T, and I-T cross-linked plasmids in NER-deficient rodent cells clearly illustrate the involvement of TC-NER pathway in the repair of these ICLs. The residual repair seen in the NER-deficient cells is most likely due to low levels of contaminating non-damaged plasmid. The observation that NER was required for repair of the C-C ICL but not the C-C cross-link remnant indicates that NER is acting at an early step in the repair process. However, it is still not clear if NER itself actually unhooks the ICL or if instead it acts as a signal for repair of the cross-links. We have previously reported an ICL unhooking activity in mammalian cell extracts that is NER-independent. However, unlike the reporter plasmids used in the present study, the linear substrates used in the unhooking studies did not undergo transcription (30). We and others have observed, in vitro, that the NER apparatus makes dual incisions on the 5′-side of the ICL (30–31, 49). However, these dual 5′-incision do not result in cross-link unhooking in mammalian whole-cell extracts (30). It remains possible that these incisions occur in vivo and signal further processing of the cross-link as originally suggested by Sancar and coworkers (49). It is also possible that auxiliary factors present in vivo that are not active in Manley-based mammalian cell extracts enable NER to unhook the ICL. Alternatively, transcription itself and a stalled polymerase may provide a structure that is conducive to NER unhooking in vivo. Further investigation is needed to dissect the individual ICL repair steps in vivo in order to delineate the role of NER in ICL repair.
Sequence analysis was performed on the DNA of plasmids recovered from wild-type HeLa cells. The transcribed strand was sequenced because previous studies have shown that removal of intrastrand and interstrand lesions by TC-NER preferentially occurs on the transcribed strand (25–26, 50–51). In the case of the C-C ICL, the transcribed strands of the recovered plasmids contained approximately equal amounts of Cs (~40%) and Gs (~60%) inserted at the site of the cross-link (see Table 1). In contrast, in the case of the T-T and I-T ICLs, a higher proportion of Cs (>80%) were inserted at the cross-link site of the transcribed strand of the recovered plasmids. These results suggested that Rev1, a Y-family translesion polymerase, might be primarily responsible for bypassing the T-T and I-T cross-link remnants left on the non-transcribed after cross-link unhooking. This hypothesis seemed plausible because blockage of the hydrogen bond face of the cross-link remnant would not be expected to affect the dCMP transferase activity of Rev1, whose activity is DNA template-independent (52). Furthermore, Rev1 and Rev3 have previously been implicated in repair of plasmids containing psoralen and mitomycin C ICLs (27). Consistent with these expectations, the repair efficiency for the T-T and I-T ICLs was found to be significantly reduced in the REV1−/− MEF (37) and REV1−/− DT40 (38) cells compared to their parental wild-type cells, whereas repair of the C-C ICL was only slightly affected (see Figures 7 and S6B). This result suggests that Rev1 is a critical component of replication-independent ICL repair and is likely responsible for the high rate of C insertions seen for the T-T and I-T ICLs. Rev1, however, does not appear to be essential for repair of the C-C ICL, which suggests that cross-link structure dictates the involvement of translesion polymerases.
Although Rev1 appears to not be required for replication-independent repair of the C-C ICL, a large proportion of Cs were inserted at the lesion on the transcribed of plasmids recovered from wild-type cells. The initial cross-link unhooking step is expected to occur primarily on the transcribed strand during TC-NER (25–26), which leaves the C-C remnant on the non-transcribed strand. We have previously observed error-free bypass of a C-C remnant in the repair synthesis step in whole-cell extracts (31), and consequently subsequent bypass of this lesion by a replicative polymerase would be expected to result in insertion of a G in transcribed strand. The significant amounts of C inserted opposite the C-C lesion therefore suggests that Rev1 was still recruited to the site of the cross-link and was involved in bypass of this lesion in wild-type cells. The lack of an inhibitory effect on C-C ICL repair seen in the REV1 knockout cell lines most likely means that Rev1 is not absolutely required for bypass of this lesion. Thus, it appears that either the replicative polymerase is able to faithfully bypass the C-C remnant or other lesion bypass polymerases can compensate for the absence of Rev1.
It is becoming increasingly apparent that interstrand cross-link structure plays an important role in the repair of this type of DNA lesion and is thus an important parameter that should be considered when studying the molecular mechanisms of repair of both endogenously and exogenously formed cross-links (53). Such understanding could facilitate the design of more potent cross-linking chemotherapeutic treatments and lead to the identification of DNA repair protein targets that could be used to increase the efficacy of treatments for cancer, premature aging syndromes, and other diseases related to ICL repair.
Supplementary Material
Acknowledgments
We thank Dr. Michael Seidman for his many thoughtful comments and suggestions regarding this manuscript. This research was supported by grants from the National Cancer Institute (CA082785, P.S.M. and CA016783, S. Ludeman), the Natural Sciences and Engineering Research Council (NSERC, C.J.W.) of Canada, and the Canada Research Chair (CRC, C.J.W.) program. E.M.H. was supported by a training grant from the National Cancer Institute (T32 CA09110) and M.B.S was supported by a predoctoral fellowship from the American Heart Association (103527).
Abbreviations
- ICL
interstrand cross-link
- NER
nucleotide excision repair
- TC-NER
transcription-coupled NER
- GG-NER
global genome NER
- TE
Tris/EDTA
- nt
nucleotide
- bp
base pair
- HCR
host-cell reactivation
- t.s
transcribed strand
- non t.s
non-transcribed strand
Footnotes
This research was supported by grants from the National Cancer Institute (CA082785, CA016783, and T32CA09110), the Natural Sciences and Engineering Research Council (NSERC) of Canada, the Canada Research Chair program, and the American Heart Association (103527).
Supporting Information Available: Figure S1 shows the sequences of non-damaged, cross-linked, and cross-link remnant containing duplexes used to construct the reporter plasmids. Figure S2 illustrates the structure of a N4C-ethyl-N4C interstrand cross-link placed in a -CG- sequence and characterization of a -CG- cross-linked plasmid. Figure S3 displays repair efficiencies of ICLs transfected into wild type and XRCC3-deficient cells. Figure S4 and S5 show repair efficiencies of ICLs transfected into wild-type and mismatch repair-deficient cells. Figure S6 demonstrates Rev1 siRNA knockdown in HeLa cells and repair efficiencies of ICLs transfected into wild-type and REV1−/− DT40 cells. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 3.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 4.Friedman HS, Colvin OM, Kaufmann SH, Ludeman SM, Bullock N, Bigner DD, Griffith OW. Cyclophosphamide resistance in medulloblastoma. Cancer Res. 1992;52:5373–5378. [PubMed] [Google Scholar]
- 5.Dong Q, Johnson SP, Colvin OM, Bullock N, Kilborn C, Runyon G, Sullivan DM, Eaton J, Bigner DD, Nahta R, Marks J, Modrich P, Friedman HS. Multiple DNA repair mechanisms and alkylator resistance in the human medulloblastoma cell line D-283 Med (4-HCR) Cancer Chemother Pharmacol. 1999;43:73–79. doi: 10.1007/s002800050865. [DOI] [PubMed] [Google Scholar]
- 6.Panasci L, Xu ZY, Bello V, Aloyz R. The role of DNA repair in nitrogen mustard drug resistance. Anticancer Drugs. 2002;13:211–220. doi: 10.1097/00001813-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Summerfield FW, Tappel AL. Detection and measurement by high-performance liquid chromatography of malondialdehyde crosslinks in DNA. Anal Biochem. 1984;143:265–271. doi: 10.1016/0003-2697(84)90662-6. [DOI] [PubMed] [Google Scholar]
- 8.Caulfield JL, Wishnok JS, Tannenbaum SR. Nitric oxide-induced interstrand cross-links in DNA. Chem Res Toxicol. 2003;16:571–574. doi: 10.1021/tx020117w. [DOI] [PubMed] [Google Scholar]
- 9.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J Am Chem Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 10.Sczepanski JT, Jacobs AC, Greenberg MM. Self-promoted DNA interstrand cross-link formation by an abasic site. J Am Chem Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- 11.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA cross-links induced by alpha,beta-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niedernhofer LJ, Daniels JS, Rouzer CA, Greene RE, Marnett LJ. Malondialdehyde, a product of lipid peroxidation, is mutagenic in human cells. J Biol Chem. 2003;278:31426–31433. doi: 10.1074/jbc.M212549200. [DOI] [PubMed] [Google Scholar]
- 13.Grillari J, Katinger H, Voglauer R. Contributions of DNA interstrand cross-links to aging of cells and organisms. Nucleic Acids Res. 2007;35:7566–7576. doi: 10.1093/nar/gkm1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 15.Neveling K, Bechtold A, Hoehn H. Genetic instability syndromes with progeroid features. Z Gerontol Geriatr. 2007;40:339–348. doi: 10.1007/s00391-007-0483-x. [DOI] [PubMed] [Google Scholar]
- 16.Schumacher B, Garinis GA, Hoeijmakers JH. Age to survive: DNA damage and aging. Trends Genet. 2008;24:77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole RS, Sinden RR. Repair of cross-linked DNA in Escherichia coli. Basic Life Sci. 1975;5B:487–495. doi: 10.1007/978-1-4684-2898-8_10. [DOI] [PubMed] [Google Scholar]
- 19.Lehoczky P, McHugh PJ, Chovanec M. DNA interstrand cross-link repair in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2007;31:109–133. doi: 10.1111/j.1574-6976.2006.00046.x. [DOI] [PubMed] [Google Scholar]
- 20.Sladek FM, Munn MM, Rupp WD, Howard-Flanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J Biol Chem. 1989;264:6755–6765. [PubMed] [Google Scholar]
- 21.Berardini M, Mackay W, Loechler EL. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry. 1997;36:3506–3513. doi: 10.1021/bi962778w. [DOI] [PubMed] [Google Scholar]
- 22.Zietlow L, Bessho T. DNA polymerase I-mediated translesion synthesis in RecA-independent DNA interstrand cross-link repair in E. coli. Biochemistry. 2008;47:5460–5464. doi: 10.1021/bi702343y. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. Embo J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHugh PJ, Sarkar S. DNA interstrand cross-link repair in the cell cycle: a critical role for polymerase zeta in G1 phase. Cell Cycle. 2006;5:1044–1047. doi: 10.4161/cc.5.10.2763. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen X, Jun S, O’Neal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J Biol Chem. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- 28.Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J Biol Chem. 2009;284:27908–27917. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noll DM, Webba da Silva M, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS. Structure, flexibility, and repair of two different orientations of the same alkyl interstrand DNA cross-link. Biochemistry. 2005;44:6764–6775. doi: 10.1021/bi050014n. [DOI] [PubMed] [Google Scholar]
- 30.Smeaton MB, Hlavin EM, McGregor Mason T, Noronha AM, Wilds CJ, Miller PS. Distortion-dependent unhooking of interstrand cross-links in mammalian cell extracts. Biochemistry. 2008;47:9920–9930. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smeaton MB, Hlavin EM, Noronha AM, Murphy SP, Wilds CJ, Miller PS. Effect of Cross-Link Structure on DNA Interstrand Cross-Link Repair Synthesis. Chem Res Toxicol. 2009;22:1285–1297. doi: 10.1021/tx9000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilds CJ, Noronha AM, Robidoux S, Miller PS. Mispair-aligned N3T-alkyl-N3T interstrand cross-linked DNA: synthesis and characterization of duplexes with interstrand cross-links of variable lengths. J Am Chem Soc. 2004;126:9257–9265. doi: 10.1021/ja0498540. [DOI] [PubMed] [Google Scholar]
- 33.Wilds CJ, Xu F, Noronha AM. Synthesis and characterization of DNA containing an N1-2′-deoxyinosine-ethyl-N3-thymidine interstrand cross-link: a structural mimic of the cross-link formed by 1,3-bis-(2-chloroethyl)-1-nitrosourea. Chem Res Toxicol. 2008;21:686–695. doi: 10.1021/tx700422h. [DOI] [PubMed] [Google Scholar]
- 34.Noll DM, Noronha AM, Miller PS. Synthesis and characterization of DNA duplexes containing an N(4)C-ethyl-N(4)C interstrand cross-link. J Am Chem Soc. 2001;123:3405–3411. doi: 10.1021/ja003340t. [DOI] [PubMed] [Google Scholar]
- 35.Noronha AM, Noll DM, Wilds CJ, Miller PS. N(4)C-ethyl-N(4)C cross-linked DNA: synthesis and characterization of duplexes with interstrand cross-links of different orientations. Biochemistry. 2002;41:760–771. doi: 10.1021/bi011610u. [DOI] [PubMed] [Google Scholar]
- 36.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 37.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. Embo J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edfeldt NB, Harwood EA, Sigurdsson ST, Hopkins PB, Reid BR. Solution structure of a nitrous acid induced DNA interstrand cross-link. Nucleic Acids Res. 2004;32:2785–2794. doi: 10.1093/nar/gkh606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferguson LR, Pearson AE. The clinical use of mutagenic anticancer drugs. Mutat Res. 1996;355:1–12. doi: 10.1016/0027-5107(96)00019-x. [DOI] [PubMed] [Google Scholar]
- 41.Fichtinger-Schepman AM, van der Veer JL, den Hartog JH, Lohman PH, Reedijk J. Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: formation, identification, and quantitation. Biochemistry. 1985;24:707–713. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]
- 42.Tomasz M, Lipman R, Chowdary D, Pawlak J, Verdine GL, Nakanishi K. Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA. Science. 1987;235:1204–1208. doi: 10.1126/science.3103215. [DOI] [PubMed] [Google Scholar]
- 43.Tong WP, Kirk MC, Ludlum DB. Formation of the cross-link 1-[N3-deoxycytidyl),2-[N1-deoxyguanosinyl]ethane in DNA treated with N,N′-bis(2-chloroethyl)-N-nitrosourea. Cancer Res. 1982;42:3102–3105. [PubMed] [Google Scholar]
- 44.Brookes P, Lawley PD. The reaction of mustard gas with nucleic acids in vitro and in vivo. Biochem J. 1960;77:478–484. doi: 10.1042/bj0770478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saffran WA, Ahmed S, Bellevue S, Pereira G, Patrick T, Sanchez W, Thomas S, Alberti M, Hearst JE. DNA repair defects channel interstrand DNA cross-links into alternate recombinational and error-prone repair pathways. J Biol Chem. 2004;279:36462–36469. doi: 10.1074/jbc.M402323200. [DOI] [PubMed] [Google Scholar]
- 46.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 47.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tornaletti S, Patrick SM, Turchi JJ, Hanawalt PC. Behavior of T7 RNA polymerase and mammalian RNA polymerase II at site-specific cisplatin adducts in the template DNA. J Biol Chem. 2003;278:35791–35797. doi: 10.1074/jbc.M305394200. [DOI] [PubMed] [Google Scholar]
- 49.Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 51.Mellon I, Bohr VA, Smith CA, Hanawalt PC. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 53.Hlavin EM, Smeaton MB, Miller PS. Initiation of DNA Interstrand Cross-link Repair in Mammalian Cells. Environmental and Molecular Mutagenesis. 2010 doi: 10.1002/em.20559. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.