Abstract
Kinetic analysis of the tissue factor (TF)-factor VIIa (FVIIa) binding interaction is helpful in investigating the structure-function relationships of TF-FVIIa. However, a wide variation exists among the reported binding affinities of FVIIa to TF, particularly when comparing KD values obtained from functional activity assays versus ligand binding studies. Surface plasmon resonance (SPR) technique was used frequently to investigate binding kinetics of FVIIa to TF in a lipid-free environment. In the present study we used TF embedded in a phospholipid bilayer for determining binding kinectis using SPR. The data revealed that FVIIa had a much higher binding affinity (>100-fold) for TF embedded in the phospholiid bilayer than TF in a lipid-free environment, approaching the KD values that were noted in the enzymatic activity assays. The present data suggest that SPR binding studies using TF embedded in phopsholipids is more appropritate for investigating how FVIIa (or FVIIa mutants/derivatives) may interact with TF in physiological settings.
Keywords: binding affinity, factor VIIa, phospholipid, surface plasmon resonance, tissue factor
Introduction
Factor VIIa (FVIIa) binding to tissue factor (TF) is a critical step in the initiation of blood coagulation. Kinetic analysis of the TF-FVIIa binding interaction is widely used to investigate the structure-function relationships of TF-FVIIa and to probe the effects of cellular alterations on TF-FVIIa function. However, studies of the TF-FVIIa binding interactions are complicated and a wide-range of dissociation equilibrium constants (10 pM to 90 nM) have been reported from various studies employing different experimental approaches (see ref [1]). In general, the measurement of FVIIa binding to TF in liposomes, performed by enzymatic activity assays, have yielded KD values in the picomolar range, whereas binding studies using cell-model systems or immobilized TF have yielded KD values in the nanomolar range. It is not entirely clear why such a wide variation exists in the reported binding affinities of FVIIa to TF.
One of the techniques commonly used to study the binding interactions of FVIIa with TF is surface plasmon resonance (SPR). SPR measurements on Biacore instruments have been used to investigate how specific alterations in FVIIa or TF affects the kinetics and affinity of their binding interaction [2–7]. Although these studies have yielded important insights into the structure-function relationships between TF and FVIIa, it is unclear whether the data obtained from these studies can be extrapolated to physiological settings as the calculated KD values by SPR were much higher (about 100-fold) than those obtained by fluoresence anisotropy or enzyme activity assays [1;8;9]. In the majority of SPR studies reported to date, soluble TF (TF1–219) was immobilized on a sensor chip using amine coupling chemistry. Alternatively, biotinylated FVIIa [2] or full-length TF [4] was immbolized on a sensor chip indirectly by capturing these molecules on either a streptavidin-derivitized surface or immobilized TF mAb, respectively. The reason for the wide differences in KD values obtained by SPR measurements or enzymatic activity are unclear, but could be the result of differences in TF preparations and/or the avaliablity of phospholipids or the substrate.
Tissue factor is an integral membrane glycoprotein and its activity is modulated by the surrounding phospholipid surface [10;11]. Although most of the binding energy of the TF-FVIIa complex comes from protein-protein interactions, the composition of the phospholipid bilayer or other membrane components surrounding TF may significantly influence its interaction with FVIIa either directly or indirectly by providing protein-membrane interactions between the FVIIa Gla domain and anionic phospholipids [12–14]. In the present study we have measured FVIIa binding interactions with TF embedded in a phospholipid bilayer and a lipid-free environment using SPR. The data demonstrate the influence of the phospholipid bilayer in FVIIa binding to TF.
Methods
The SPR studies were performed using a Biacore 3000. To measure FVIIa binding to TF in the lipid bilayer, recombinant TF containing transmembrane spanning domain (TF1–248) was first relipidated in either phosphatidylcholine (PC) alone or PC/phosphatidylserine (PS) (80:20) as described previously [15], with the exception that the TF:phospholipid ratio was 1:3000 (0.33 µM TF: 1 mM phospholipid). Control phospholipid vesicles were made in an identical fashion except that TF was omitted from the reconstitution. A lipophilic L1 chip (Biacore, Piscataway, NJ) was coated with PC (flow cell 1), TF/PC (flow cell 2), PC/PS (flow cell 3) and TF/PC/PS (flow cell 4) by injecting each preparation at a flow rate of 5 µl/min for 15 min followed by three 5 µl/min pulses of 10 nM NaOH to remove the unbound or loosely bound vesicles. Approximately 1900 RUs were immobilized in flow cell 1 and 1700 RUs were immobilized in all other flow cells. This represents about 35 pg of TF embedded in the lipid bilayer on flow cells 2 and 4. The chip was equilibrated overnight by passing running buffer (20 mM HEPES, 150 mM NaCl, 5 mM CaCl2, pH 7.4) over the chip at a flow rate of 5 µl/min. Kinetic analyses were performed at 37°C by flowing consecutively increasing concentrations of FVIIa (0 nM to 10 nM, in duplicates) in the running buffer containing 0.1% BSA over the sensor chip for 5 min (association time) followed by a 10 min dissociation period at a flow rate of 30 µl/min in parallel. Regeneration was performed with a 3-min pulse of 10 mM EDTA in HEPES buffer (20 mM HEPES, 150 mM NaCl, pH 7.4). FVIIa binding to TF in the lipid bilayer was determined by deducting the RU values noted in reference flow cells 1 and 3 from the RU values of flow cells 2 and 4, respectively (i.e., 2-1 and 4-3). Surface plasmon resonance data were fitted with a 1:1 binding model containing a mass transfer coefficient using BiaEvaluations software v 4.1 (Biacore).
To measure FVIIa binding to soluble TF (sTF), TF1–219 was immobilized on a CM5 sensor chip using amine coupling chemistry according to the instructions provided by the manufacturer. Briefly, the carboxymethyl dextran surface of a CM5 chip was first activated with a mixture of 1-ethyl-3-(3-dimethylaminopropyl) carbamide (EDC) and N-hydroxysuccinamide (NHS). Ten µg/ml sTF was then passed over the activated surface of flow cell 2 at a rate 5 µl/min in acetate buffer (10 mM Na acetate, pH 5.0). Approximately 100 RU of sTF (i.e., 100 pg) was coupled to the chip. Excess (unreacted) sites were blocked with ethanolamine. Flow cell 1 was activated and immediately blocked with ethanolamine as a reference. Kinetic analysis was performed essentially as described above except FVIIa concentrations passed over the chip were required to be much higher (10 to 500 nM).
Results and Discussion
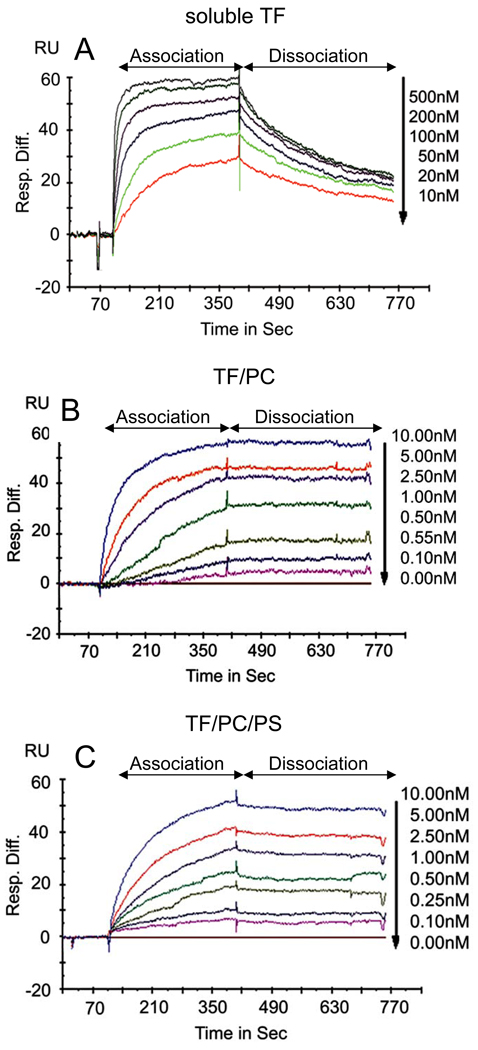
Although SPR analysis has been used frequently to analyze FVIIa binding kinetics to TF, these analyses were limited to TF in a lipid-free environment. To our knowledge, the present study was the first to use relipidated TF for the analysis of FVIIa binding to TF using SPR. Marked differences were noted between FVIIa binding to TF embedded in phospholipids versus FVIIa binding to TF in a lipid-free environment. As shown in Fig. 1, FVIIa bound to TF in a lipid bilayer much tighter than it bound to sTF. Analysis of FVIIa binding to sTF or full-length TF not inserted into a phospholipid bilayer in earlier SPR studies yielded values of kon = 0.2 to 4 × 105M−1s−1 and koff = 0.5 to 2 ×10−3 s−1 with a calculated KD of 2 to 25 nM [2–7]. The dissociation equilibrium binding constant obtained in the present study with sTF (KD of 11.6 nM; Table 1) is consistent with these reported values. FVIIa associates with TF inserted in the phospholipid bilayer at a rate 5 times faster than that observed for FVIIa binding to sTF. More importantly, FVIIa bound to TF in the lipid bilayer did not dissociate significantly over the time examined. Increasing the dissociation time to 30 min or longer did not alter the characteristics of the dissociation curve nor did increasing flow rates (data not shown). Thus, the TF-FVIIa complex on the phospholipid bilayer appears to be very stable. Although accurately calculating dissociation equilibrium constants, when there is little dissociation, is prone to some error, the approximately calculated KD values are helpful for comparative purposes. The dissociation equilibrium constant obtained by SPR in the present study (45 pM) is very similar to that obtained by enzymatic assays or fluorescence anisotropy [1;8;16]. The increased binding affinity of FVIIa to TF inserted in a phospholipid bilayer compared to TF in a lipid-free environment largely reflects the modulatory effect of lipids on the binding interaction and not on structural differences between sTF and full-length TF. This is supported by a binding analysis carried out with full-length TF immobilized on non-inhibitor antibody by SPR that yielded similar dissociation constants as measured with sTF [4]. It is interesting to note that in this study using SPR, FVIIa bound to TF embedded in a neutral lipid environment with a similar affinity to that observed for TF inserted in a phospholipid bilayer containing PS (Fig. 1 and Table 1). The similar binding kinetics observed with TF embedded in neutral lipids versus anionic phospholipids suggests that the FVIIa Gla domain interaction with anionic phospholipids contributes minimally to the binding energy of the TF-FVIIa complex. While this observation is in agreement with other published studies using recombinant lipid-anchored TF [1;9], it is in contrast to studies using native brain-derived TF, where enzymatic activity studies showed a reduced affinity of FVIIa for TF embedded in neutral lipids [8]. The reasons for this discrepancy are currently unclear but may involve numerous factors, not the least of which are the presence of homodimers in the brain-derived material [8], as well as the differences in TF density on the lipid membrane that was used in these various studies. In light of marked differences in FVIIa binding interaction with TF in phospholipid-free environment vs. surrounding phospholipids, caution should be exercised in extrapolating the binding data obtained in phospholipid-free systems to biological systems where TF is embedded in a phospholipid membrane.
Fig 1. Sensograms of FVIIa binding to soluble TF or TF in a lipid bilayer.
Varying concentrations of FVIIa were passed through flow cells of a CM5 sensor chip coated with soluble TF (sTF) or an ethanolamine-blocked reference (A) or a lipophilic L1 sensor chip coated with PC, PC/PS, PC-TF or PC/PS-TF (B and C) at a flow rate of 30 µl/min for 5 min at 37°C (association time) followed by calcium-containing buffer for 10 min under the same flow conditions (dissociation time). The sensograms shown in the figure represent FVIIa binding to TF, derived by subtracting the values obtained with reference cells (ethanol amine, PC or PC/PS coated surfaces) from cells containing soluble TF, PC/TF or PC/PS-TF coated surfaces, respectively. The data were fitted to a 1:1 binding model containing a mass transfer coefficient using Biaevaluations software v 4.1 (Biacore). The concentrations of FVIIa used are specified in the figure.
Table 1. FVIIa binding to TF: Rate constants measured by surface Plasmon resonance.
The values represent mean ± SEM of 3 to 4 independent experiments.
| Binding site | kon (M−1s−1) | koff (s−1) | KD (M) |
|---|---|---|---|
| sTF | 4.75 ± 1.03 × 105 | 5.92 ± 0.72 × 10−3 | 1.16 ± 0.05 × 10−8 |
| TF/PC | 2.73 ± 0.94 × 106 | 2.34 ± 0.43 × 10−4 | 4.57 ± 1.09 × 10−11 |
| TF/PC/PS | 2.58 ± 0.72 × 106 | 2.33 ± 1.50 × 10−4 | 4.74 ± 0.93 × 10−11 |
It is more biologically relevant to use cells naturally expressing TF to characterize FVIIa interaction with TF. We and others have used radiolabeled FVIIa to investigate interaction of FVIIa with TF on cell surfaces [17–20]. These studies indicated that the KD for the interaction of cell-surface TF with FVIIa is in the 1 to 10 nM range. However, none of these measurements were true equilibrium measurements as these studies depended on the separation of free FVIIa from cell-bound FVIIa, which required multiple washings. This could dissociate a significant fraction of FVIIa bound to cell-surface TF and deviate from true equilibrium measurements, and thus introduce an error in determining KD accurately. This could explain why KD values determined from radioligand binding studies were much higher than those obtained from enzymatic titrations in functional activity assays [16;21]. Furthermore, the existence of TF in at least two different forms, cryptic and active, on cell surfaces, whose proportions and properties are ill-defined at present [10;22], makes it impractical to use cell-model systems for determining true affinity of FVIIa to TF. The present methodology where TF is embedded in a defined phospholipid composition and use of SPR to analyze the binding interactions overcomes some of the limitations that exist in measuring FVIIa-TF binding interactions using cell and other model systems. Although other methods, such as sedimentation equilibrium and pressure-dependent fluorescence anisotropy are also capable of measuring interaction of FVIIa with relipidated TF [1], these methods are more complex and time-consuming compared to SPR analysis. Measurement of FVIIa binding interactions with TF in a lipid bilayer using SPR is more practical and also biologically appropriate than other methods that have been used currently for measuring this interaction.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (HL58869 to L.V.M. Rao).
Footnotes
Disclosure of Conflict of Interests
The authors state that they have no conflict of interests.
References
- 1.Waxman E, Ross JBA, Laue TM, Guha A, Thiruvikraman SV, Lin TC, Konigsberg WH, Nemerson Y. Tissue factor and its extracellular soluble domain: The relationship between intermolecular association with factor VIIa and enzymatic activity of the complex. Biochem. 1992;31:3998–4003. doi: 10.1021/bi00131a015. [DOI] [PubMed] [Google Scholar]
- 2.Kelley RF, Costas KE, O'Connell MP, Lazarus RA. Analysis of the factor VIIa binding site on human tissue factor: Effects of tissue factor mutations on the kinetics and thermodynamics of binding. Acta Chir Scand. 1995;434:10383–10392. doi: 10.1021/bi00033a009. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien DP, Kemball-Cook G, Hutchinson AM, Martin DMA, Johnson DJD, Byfield PGH, Takamiya O, Tuddenham EGD, McVey JH. Surface plasmon resonance studies of the interaction between factor VII and tissue factor. Demonstration of defective tissue factor binding in a variant FVII molecule (FVII-R79Q) Biochem. 1994;33:14162–14169. doi: 10.1021/bi00251a027. [DOI] [PubMed] [Google Scholar]
- 4.Petrovan RJ, Ruf W. Role of zymogenicity-determining residues of coagulation factor VII/VIIa in cofactor interaction and macromolecular substrate recognition. Biochem. 2002;41:9302–9309. doi: 10.1021/bi0202169. [DOI] [PubMed] [Google Scholar]
- 5.Sorensen BB, Persson E, Freskgard P-O, Kjalke M, Ezban M, Williams T, Rao LVM. Incorporation of an active site inhibitor in factor VIIa alters the affinity for tissue factor. J Biol Chem. 1997;272:11863–11868. doi: 10.1074/jbc.272.18.11863. [DOI] [PubMed] [Google Scholar]
- 6.Persson E, Bak H, Ostergaard A, Olsen OH. Augmented intrinsic activity of Factor VIIa by replacement of residues 305, 314, 337 and 374: evidence of two unique mutational mechanisms of activity enhancement. Biochem J. 2004;379:497–503. doi: 10.1042/BJ20031596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weimer T, Wormsbacher W, Kronthaler U, Lang W, Liebing U, Schulte S. Prolonged in-vivo half-life of factor VIIa by fusion to albumin. Thromb Haemost. 2008;99:659–667. doi: 10.1160/TH07-08-0525. [DOI] [PubMed] [Google Scholar]
- 8.Neuenschwander P, Morrissey JH. Roles of the membrane-interactive regions of factor VIIa and tissue factor. J Biol Chem. 1994;269:8007–8013. [PubMed] [Google Scholar]
- 9.Krishnaswamy S. The interaction of human factor VIIa with tissue factor. J Biol Chem. 1992;267:23696–23706. [PubMed] [Google Scholar]
- 10.Bach RR. Tissue factor encryption. Arterioscler Thromb Vasc Biol. 2006;26:456–461. doi: 10.1161/01.ATV.0000202656.53964.04. [DOI] [PubMed] [Google Scholar]
- 11.Rao LV, Pendurthi UR. Regulation of tissue factor-factor VIIa expression on cell surfaces: a role for tissue factor-factor VIIa endocytosis. Mol Cell Biochem. 2003;253:131–140. doi: 10.1023/a:1026004208822. [DOI] [PubMed] [Google Scholar]
- 12.Bach R, Gentry R, Nemerson Y. Factor VII binding to tissue factor in reconstituted phospholipid vesicles: induction of cooperativity by phosphatidylserine. Biochem. 1986;25:4007–4020. doi: 10.1021/bi00362a005. [DOI] [PubMed] [Google Scholar]
- 13.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 14.Mandal SK, Iakhiaev A, Pendurthi UR, Rao LV. Acute cholesterol depletion impairs functional expression of tissue factor in fibroblasts: modulation of tissue factor activity by membrane cholesterol. Blood. 2005;105:153–160. doi: 10.1182/blood-2004-03-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao LVM, Williams T, Rapaport SI. Studies of the activation of factor VII bound to tissue factor. Blood. 1996;87:3738–3748. [PubMed] [Google Scholar]
- 16.Petersen LC, Albrektsen T, Hjorto GM, Kjalke M, Bjorn SE, Sorensen BB. Factor VIIa/tissue factor-dependent gene regulation and pro-coagulant activity: effect of factor VIIa concentration. Thromb Haemost. 2007;98:909–911. [PubMed] [Google Scholar]
- 17.Ploplis VA, Edgington TS, Fair DS. Initiation of the extrinsic pathway of coagulation. Association of factor VIIa with a cell line expressing tissue factor. J Biol Chem. 1987;262:9503–9508. [PubMed] [Google Scholar]
- 18.Rodgers GM, Broze GJ, Jr, Shuman MA. The Number of Receptors for Factor VII Correlates With the Ability of Cultured Cells to Initiate Coagulation. Blood. 1984;63:434–438. [PubMed] [Google Scholar]
- 19.Sakai T, Lund-Hansen T, Paborsky L, Pedersen AH, Kisiel W. Binding of human factors VII and VIIa to a human bladder carcinoma cell line (J82) J Biol Chem. 1989;264:9980–9988. [PubMed] [Google Scholar]
- 20.Le DT, Rapaport SI, Rao LVM. Relations between factor VIIa binding and expression of factor VIIa/tissue factor catalytic activity on cell surfaces. J Biol Chem. 1992;267:15447–15454. [PubMed] [Google Scholar]
- 21.Ghosh S, Ezban M, Persson E, Pendurthi U, Hedner U, Rao LV. Activity and regulation of factor VIIa analogs with increased potency at the endothelial cell surface. J Thromb Haemost. 2006;5:336–346. doi: 10.1111/j.1538-7836.2007.02308.x. [DOI] [PubMed] [Google Scholar]
- 22.Rao LVM, Pendurthi UR. Tissue factor on cells. Blood Coag Fibrin. 1998;9:S27–S35. [PubMed] [Google Scholar]