Abstract
The interaction of phosphodiesterase 6 (PDE6) with its inhibitory Pγ-subunits is unparalleled among PDE families and is central for vertebrate phototransduction. The C-terminus of Pγ occludes the active site of PDE6 thereby preventing hydrolysis of cGMP. In this study we examined the determinants of this critical interaction using structure-based loss-of-function mutagenesis of a chimeric PDE5/PDE6 catalytic domain and a gain-of-function mutagenesis of the PDE5 catalytic domain. This analysis revealed the key role of PDE6-specific residues within the catalytic domain M-loop/α-helix15 region and suggested an important contribution of the H-M-loop interface to the PDE6 inhibition by the Pγ C-terminus. Identification of the determinants for the PDE6-Pγ interaction offers insights into the evolution of the visual effector enzyme.
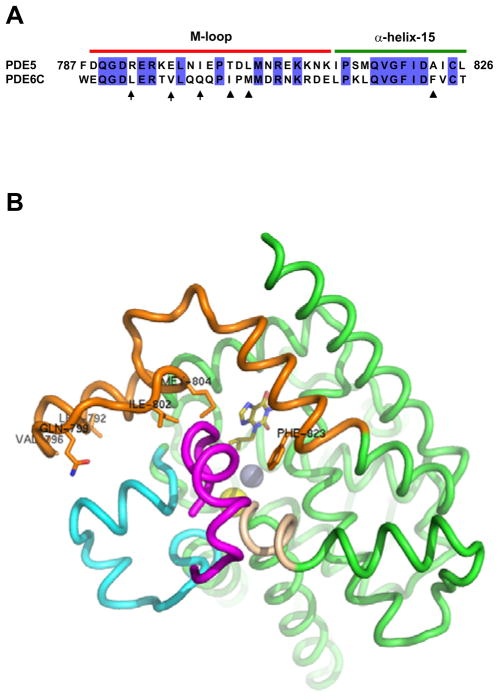
Photoreceptor cGMP phosphodiesterase (PDE6 family) is the effector enzyme in the vertebrate visual transduction cascade. The activity of rod and cone PDE6 catalytic subunits is blocked in the dark by the inhibitory Pγ-subunits. The inhibition is released upon light-stimulation of photoreceptor cells (1–3). The interaction of PDE6 with Pγ is critical for phototransduction and unique for the PDE6 family of phosphodiesterases (1–4). It involves at least two binding interfaces, one is between the regulatory GAF domain(s) of PDE6 and the central polycationic Pro-rich region of Pγ, and the second is between the catalytic domain of PDE6 and the Pγ C-terminus (5–7). The former site serves primarily to enhance the affinity of the interaction between PDE6 and Pγ, whereas the latter site constitutes the key inhibitory interaction. The C-terminus of Pγ occludes the active site of PDE6 thereby preventing hydrolysis of cGMP (8–10). Because of the failure of functional expression of PDE6 outside vertebrate photoreceptors, current data on the Pγ-contact sites on PDE6 have been developed with the use of chimeric enzymes between PDE6 and PDE5 (11–13). The catalytic domains of PDE5 (PDE5cd) and PDE6 (PDE6cd) display significant sequence homology, specificity for cGMP, and sensitivity to a number of common catalytic site inhibitors, but are different in that PDE5cd is not inhibited by Pγ (11, 14). However, a replacement of just one segment corresponding to the M-loop/α-helix15 region in PDE5cd with the corresponding sequence of cone PDE6C yields a chimeric catalytic domain, PDE5/6cd, capable of potent inhibition by Pγ or the Pγ C-terminal peptides (Figure 1A) (10, 15). The crystal structure of the complex between the IBMX-bound PDE5/6cd and the Pγ C-terminal peptide Pγ70-87 has been recently solved (10). This structure provided the first detailed picture of the binding site and suggested the determinants for the interaction with Pγ. The Pγ70-87 binding surface of PDE5/6cd is comprised of four structural elements, α-helices 12 and 15 and two variable H- and M-loops (Figure 1B) (10). The Pγ-binding segment in helix-12 is conserved between PDE5 and PDE6 and assumes similar conformations in the structures of PDE5cd and PDE5/6cd. Therefore, helix12 does not appear to contribute to the specificity of the Pγ-binding site. Although helix-15 is conformationally similar in the structures of PDE5cd and PDE5/6cd, it is important to the specificity of the Pγ/PDE6 interaction by providing essential PDE6-specific Pγ-contact residue Phe823. The H- and M-loop conformations are markedly different in the PDE5/6cd and PDE5cd structures (10, 16). In PDE5/6cd these loops are stabilized by extensive inter-loop interactions not seen in PDE5cd (10, 16). Since the H-loop sequence is identical in PDE5/6cd and PDE5cd, the ability to establish the inter-loop interface is dictated by the M-loop. Thus, the M-loop in PDE6 can contribute to the Pγ C-terminus binding directly by donating specific Pγ-contact residues or indirectly by inducing the H-M-loop interface to favorably position residues from both loops for the interaction with Pγ. Here, we performed mutational analysis of the Pγ contact residues and the H-M-loop interface. Using inhibition of PDE activity by the Pγ C-terminal peptide, Pγ63-87, as functional readout, loss-of-function and gain-of-function mutations were introduced into PDE5/6cd and PDE5cd, respectively. This analysis defined multiple structural determinants of PDE6 inhibition by the Pγ C-terminus.
Figure 1.
A. Sequence alignment of residues 787–826 of human PDE5 and corresponding residues 746–785 of human cone PDE6C. This region is replaced in PDE5cd (PDE5 residues 535–860) by PDE6C residues to yield the Pγ-sensitive catalytic domain PDE5/6cd. The numbering of residues for PDE5/6cd is the same as for PDE5. Arrows indicate PDE6-specific residues of PDE5/6cd involved in the H-M-loop interface (Leu792, Val796, and Gln799). Triangles indicate PDE6-specific residues of PDE5/6cd making direct contacts with the Pγ C-terminus (Ile802, Met804, and Phe823). B. Interactions of PDE5/6cd (green) with Pγ70-87 (magenta). Pγ70-87 interacts with the M-loop/helix-15 region (orange), the H-loop (cyan), and the N-terminal segment of α-helix-12 (wheat). Leu792, Val796, Gln799, Ile802, Met804, and Phe823 are shown as sticks. Olive – IBMX, yellow sphere – magnesium ion, grey sphere – zinc ion.
Experimental procedures
Materials
[3H]cGMP was purchased from Amersham Pharmacia Biotech. All restriction enzymes were purchased from NEB. Pfu turbo DNA polymerase was a product of Stratagene. Mini protease inhibitor cocktail tablets were from Roche Molecular Biochemicals. DNA miniprep kit was purchased from Qiagen. His-bind resin was obtained from Novagen. AG1-X2 cation exchange resin was a product of Bio-Rad. All other reagents were purchased from Sigma. Synthetic peptide corresponding to Pγ63–87 was custom-made by Sigma–Genosys and purified by reverse-phase HPLC.
Mutagenesis and isolation of mutant PDE5/6cd and PDE5cd
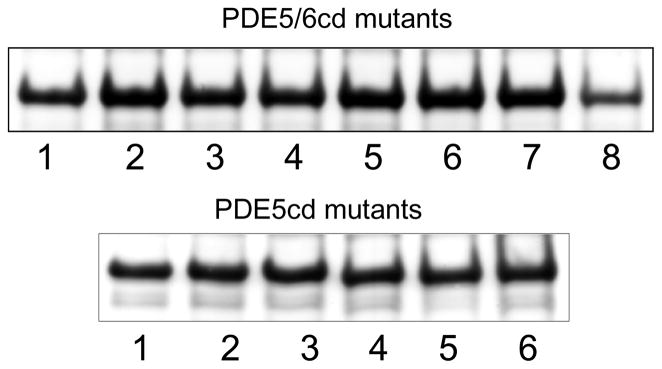
The DNA sequence corresponding to human PDE5 residues 535–860 was amplified by RT-PCR with primers containing NdeI and XhoI sites. Total RNA isolated from HEK293 cells was used as a template in the amplification. The PDE5cd construct was cloned into the pET15b vector (Novagen) using NdeI and XhoI sites. The PDE5/6cd chimera based on sequences of human PDE5 and PDE6C was generated as described previously (10). Mutations were introduced into PDE5/6cd and PDE5cd using the QuikChange mutagenesis protocol (Stratagene). Multiple mutations were produced by subsequent mutagenesis of single, double, or triple mutants. The sequences of all constructs were verified by automated DNA sequencing at the University of Iowa DNA Core Facility. The PDE5/6cd and PDE5cd mutant plasmids were transformed into BL21-codon plus competent cells (Stratagene). Expression and purification of mutant PDE5/6cd and PDE5cd proteins over His-bind resin (Novagen) were performed as previously described (15). Isolated recombinant proteins were analyzed by SDS-PAGE in 12% NuPage precast gels (Invitrogen). All PDE5/6cd and PDE5cd mutants, except PDE5/6cd I802A/P803D/M804L/F823A, were expressed as soluble proteins at comparable levels of 4–7 mg per 1 liter of culture (Figure 2). The quadrople mutant was expressed at ~2 mg/L.
Figure 2.
Coomassie blue-stained SDS-gel with isolated mutant PDE5/6cd and PDE5cd proteins. Equal fractions of the preparations from 1 liter cultures were loaded as samples. PDE5/6cd mutants: (1) PDE5/6cd; (2) I802A; (3) P803D; (4) M804A; (5) A823F; (6) I802A/ M804A; (7) L792R/V796E/Q799I; (8) I802A/P803D/M804L/F823A. PDE5cd mutants: (1) PDE5cd; (2) A823F; (3) R792L/E796V/I799Q/A823F; (4) T802I/D803P/L804M/A823F; (5) R792L/E796V/I799Q/T802I/D803P/L804M/A823F; (6) F787W/R792L/E796V/I799Q/T802I/D803P/L804M/A823F.
Characterization of mutant PDE5/6cd and PDE5cd
Mutant PDE5/6cd and PDE5cd proteins were analyzed as His6-tagged proteins. Control experiments showed no differences in stability, catalytic properties, or Pγ63-87-inhibition of these mutants prior to or after removing the His6-tag with thrombin. PDE activity was measured using 1 μM [3H]cGMP and 5 nM mutant PDE5/6cd or PDE5cd according to published protocols (15). The KM and kcat for cGMP hydrolysis and the Ki for inhibition by Pγ63-87 were determined as described (15). The KM , kcat and Ki values are expressed as Mean ± S.E. for three separate experiments.
Results
Mutational analysis of the PDE6-specific Pγ contact residues within the M-loop/a-helix15 region
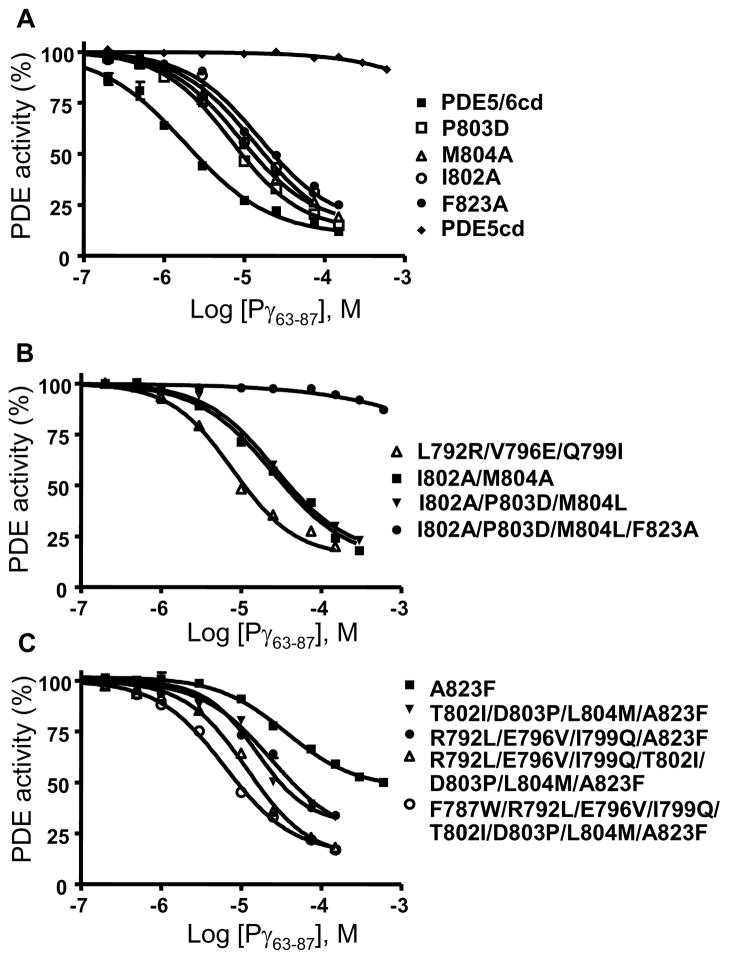
Replacement of the M-loop/helix-15 region of PDE5cd with the corresponding region of PDE6 allows for a potent inhibition of the chimeric catalytic domain PDE5/6cd by the Pγ C-terminal peptides (Figure 1) (10, 15). Accordingly, Pγ63-87 had no appreciable effect on the activity of PDE5cd when tested at concentrations up to 600 μM, and it inhibited PDE5/6cd with the Ki of 1.7±0.2 μM (Figure 3A, Table I). The KM and kcat values of PDE5cd (3.3±0.4 μM; 0.8±0.2 s−1) and PDE5/6cd (3.1±0.5 μM; 0.6±0.1 s−1) for cGMP hydrolysis were comparable. The crystal structure of PDE5/6cd in complex with IBMX and Pγ70-87 reveals three major PDE6-specific Pγ contact residues within this region, Ile802, Met804, and Phe823 (Figure 1) (10). These residues are absolutely conserved in the PDE6 family, and correspond to a nonconserved residue at position 802 (Ala, Thr, or Ile) and conserved Leu804 and Ala823 in the PDE5 family (Figure 1A, Figure S1 of the Supporting Information). The role for Met and Phe residues in the PDE6-Pγ interaction was previously recognized, whereas the significance of the Ile residue was unnoticed (12, 13). Indeed, the M804A and F823A mutations resulted in ~6- and ~9-fold reductions in the potency of PDE5/6cd inhibition by Pγ63-87, respectively (Figure 3A, Table I). The M804L is a conservative substitution. This mutation, unlike M804A, had no significant effect on the interaction of PDE5/6cd with the Pγ peptide (Table I). Two mutations, I802A and I802T, were introduced into PDE5/6cd to examine the contribution of Ile802. The I802A and I802T substitutions comparably increased the Ki value for the peptide inhibition of PDE5/6cd by ~7 fold and ~5 fold, respectively (Figure 3A, Table I). Replacement of both M-loop Pγ contacts Ile802 and Met804 by Ala residues cumulatively impaired the interaction of PDE5/6cd with Pγ63-87. The double PDE5/6cd mutant I802A//M804A was inhibited by the peptide with the Ki of 26.2±3.0 μM (Figure 3B, Table I).
Figure 3.
Inhibition of mutant PDE5/6cd and PDE5cd by Pγ63-87. The activities of PDE5/6cd and PDE5cd (A) or PDE5/6cd mutants (A,B) and PDE5cd mutants (C) were measured in the presence of 1 μM cGMP and increasing concentrations of Pγ63-87. PDE activity is expressed as a percentage of that in the absence of Pγ63-87. Results of representative experiments are shown. The Ki values are shown in Table I as the mean±SE from three similar experiments.
Table I.
Inhibition of mutant PDE5/6cd and PDE5cd by Pγ63-87.
| KM (μM) | Ki for Pγ63-87 (μM) | |
|---|---|---|
| PDE5/6cd and mutants | ||
| PDE5/6cd* | 3.1±0.5 | 1.7±0.2 |
| L792R | 3.4±0.6 | 3.2±0.3 |
| V796E | 4.1±0.2 | 1.8±0.1 |
| Q799I | 3.2±0.8 | 3.5±0.3 |
| L792R/V796E/Q799I | 3.5±0.9 | 6.0±0.6 |
| I802A | 2.9 ±0.2 | 12.1±2.1 |
| I802T | 3.9±0.6 | 8.6±0.3 |
| P803D | 3.0±0.2 | 6.7±0.5 |
| M804A | 2.8±0.7 | 10.3±1.6 |
| M804L | 2.9±0.6 | 2.0±0.1 |
| F823A | 3.0±0.7 | 15.1±1.7 |
| I802A//M804A | 3.2±0.7 | 26.2±3.0 |
| I802A/P803D/M804L | 3.8±0.1 | 25.5±2.6 |
| I802T/P803D/M804L | 2.5±0.2 | 21.8±1.8 |
| I802A/P803D/M804L/F823A | 4.0±0.3 | >3000 |
| PDE5cd and mutants | ||
| PDE5cd* | 3.3±0.4 | >10000 |
| A823F | 3.7±0.3 | 44.3±4.5 (52±4)** |
| A823F + Pγ63-87 (150 μM) | 4.0±0.4 | |
| R792L/E796V/I799Q/A823F | 4.0±0.4 | 23.5±3.4 (78±5)** |
| T802I/D803P/L804M/A823F | 2.8±0.6 | 19.4±0.1 (72±4)** |
| R792L/E796V/I799Q/T802I/ D803P/L804M/A823F |
3.2±0.7 | 10.2±0.7 |
| F787W/ R792L/E796V/I799Q/ T802I/D803P/L804M/A823F |
3.7±0.4 | 5.8±0.4 |
The kcat values for PDE5/6cd and PDE5cd were 0.6±0.1 s−1 and 0.8±0.2 s−1, respectively. The kcat values of the mutants were comparable to the parent protein kcat.
Values in parentheses indicate maximal inhibition (%). Maximal inhibition values for other mutants are greater than 85%.
Ile802 and Met804 in PDE5/6cd are separated by a conserved PDE6-specific Pro803 (Figure S1 of the Supporting Information). Substitution of Pro803 by a conserved PDE5 residue Asp803 was made to test its potential influence on the conformation of the Pγ contacts and the architecture of the M-loop. The P803D mutation led to ~4-fold decrease in the affinity of PDE5/6cd for Pγ63-87 (Figure 3A). The effects of combined replacement of the M-loop segment Ile802Pro803Met804 by the PDE5-specific residues Ala802Asp803Leu804 (bovine PDE5) or Thr802Asp803Leu804 (human PDE5) were ~15- and ~13-fold reductions in the efficiency of PDE5/6cd inhibition by Pγ63-87, respectively (Figure 3B, Table I). All three PDE6-specific Pγ-contact residues were swapped with PDE5 residues in a quadruple PDE5/6cd mutant I802A/P803D/M804L/F823A. This mutant was expressed at a lower level compared to the other PDE5/6cd mutants (Figure 2). However, the mutant folding was not significantly impaired, since its KM (4.0±0.3 μM) and kcat (0.5 s−1) values were similar to those of PDE5/6cd. The quadruple PDE5/6cd mutant was not appreciably inhibited by Pγ63-87 (Figure 3B, Table I).
Mutational analysis of the H-M-loop interface
Conformations of the H- and M-loops in the PDE5/6cd-IBMX-Pγ70-87 and PDE5cd-IBMX structures are distinctly different (10, 16). The H- and M-loops in PDE5/6cd are stabilized by an extensive inter-loop interface. Residues Val660 and Tyr664 of the H-loop make hydrophobic interactions with M-loop residues Leu792, Val796, and Leu797, whereas polar residues Asn661 and Arg667 in the H-loop form contacts with residues Gln789, Glu793, and Gln799 in the M-loop (10). Three of the M-loop residues involved in the interface with the H-loop, Leu792, Val796, and Gln799, are strongly conserved in PDE6 and differ from the corresponding residues in PDE5, Arg(Lys)792, Glu796, and Ile(Met)799 (Figure S1 of the Supporting Information). To probe the role of the H-M-loop interface in stabilizing the Pγ-binding surface, Leu792, Val796, and Gln799 of PDE5/6cd were mutated singly or in combination to PDE5-specific residues. The L792R and Q799I substitutions each resulted in a modest ~2-fold attenuation of the PDE5/6cd affinity for Pγ63-87, while the V796E mutation had no effect (Table I). Consistent with the effects of the single mutations, the triple mutant L792R/V796E/Q799I displayed a moderate 3.5 fold impairment of the affinity of the interaction with the Pγ peptide (Figure 3B).
Gain-of-function mutations of the M-loop/a-helix15 region in PDE5/6cd
To further examine the role of the M-loop and its interface with the H-loop for the PDE6-Pγ interaction in a gain-of-function approach, we first restored the critical α-helix-15 Pγ-contact residue by replacing PDE5cd Ala823 with Phe. The A823F mutant of PDE5cd acquired sensitivity to Pγ63-87 with the Ki of 44.3±4.5μM, although the maximal inhibition was only partial (~52%) (Figure 3C, Table I). The KM value of the mutant was unchanged in the presence of Pγ63-87 suggesting that the inhibition by the peptide was allosteric (Table I). The PDE5cdA823F mutant was then used as a template to introduce PDE6-specific M-loop residues essential for direct interactions with the H-loop (Leu792, Val796, and Gln799) or Pγ (Ile802, Pro803, and Met804). Both quadruple PDE5cd mutants, R792L/E796V/I799Q/A823F and T802I/D803P/L804M/A823F, showed improved maximal inhibition and a ~2-3 fold enhancement of the affinity of interaction with Pγ63-87 in comparison to the A823F mutant (Figure 3C, Table I). The effect of the former quadruple mutation matched the loss of affinity for Pγ63-87 in the reciprocal L792R/V796E/Q799I mutant of PDE5/6cd. However, the effect of the latter quadruple mutation was smaller than the ~13 fold increase in the Ki value for the reverse I802T/P803D/M804L mutant of PDE5/6cd, suggesting that residues Ile802Pro803Met804 are not as well conformationally positioned for the interaction with Pγ63-87 in the context of the PDE5cd mutant as they are in PDE5/6cd. Incorporation of the two clusters of mutations, R792L/E796V/I799Q and T802I/D803P/L804M, into the A823F mutant background had a cumulative effect (Figure 3C). The resulting PDE5cd mutant was inhibited by Pγ63-87 with the Ki of 10.2±0.7 μM (Figure 3C, Table I). Still, this PDE5cd mutant was less sensitive to Pγ than PDE5/6cd, indicating additional structural determinants for binding the Pγ C-terminus. In the PDE5/6cd structure, conserved PDE6-specific residue Trp787 buttresses the M-loop backbone at residues Asp805Arg806Asn807 thereby possibly affecting or stabilizing conformations of Ile802 and Met804 (10). Phe787, a PDE5-counterpart of Trp787 is less bulky and might provide less support for the M-loop. Indeed, the additional F787W mutation increased the potency of the inhibition by Pγ63-87 (Figure 3C, Table I).
Discussion
The unique ability of PDE6 enzymes to interact with the Pγ subunits is critical for vertebrate vision and may have evolved along with the evolution of the PDE6 family and the vertebrate phototransduction cascade (1–3, 17). The phylogenetic analysis of PDEs and the gene structure similarities indicate the common ancestry of PDE6 with PDE5 and PDE11 (17, 18). PDE5 is also similar to PDE6 in such biochemical characteristics as substrate specificity and sensitivity to common catalytic site inhibitors (14, 19). The common ancestry and biochemical similarities make PDE5cd perhaps the most useful template among all PDE families to study the determinants of the inhibitory interaction of PDE6cd with the Pγ C-terminus. Chimeric PDE5/6cd containing a relatively short 40 aa residue segment of PDE6 encompassing the M-loop and helix-15 was shown to be inhibited comparably to native PDE6 by the Pγ63-87 peptide (Figure 1A) (10, 15). Thus, the inhibition determinants are almost entirely confined to this segment of PDE6. The crystal structure of Pγ70-83 bound to PDE5/6cd reveals two Pγ-contact residues within the M-loop (Ile802and Met804) and one Pγ-contact within helix-15 (Phe823) that differ with the corresponding residues of PDE5cd (Figure 1B)(10). Consistent with the structure of the PDE5/6cd-Pγ70-87 complex, replacement of any of these residues with Ala significantly reduces the potency of PDE5/6cd inhibition by Pγ63-87. Nevertheless, PDE5-specific Leu804 is fully capable of substituting Met804 in the interaction with Pγ63-87 as evident from the analysis of the PDE5/6cd M804L mutant. Also, PDE5 in some lower vertebrate species contain Ile at position 802 (Figure S1 of the Supporting Information). An additional contributing factor to the M-loop/Pγ interaction is the PDE6-specific Pro803 that appears to be important in the positioning of Ile802 and Met804 (Figure 3A). It is unlikely that the effect of the P803D substitution in PDE5/6cd is due to repulsive interactions of the Asp residue with the negatively charged Pγ peptide. Based on the complex structure, the distance between Pro803 and the closest acidic residue Glu80 of Pγ exceeds 10 Å (10). The PDE5/6cd mutant I802A/P803D/M804L/F823A with all three Pγ-interacting residues and Pro803 substituted by PDE5 counterparts was insensitive to Pγ63-87 (Figure 3B). Thus, the lack of inhibition of PDE5cd by the Pγ C-terminal peptide can be largely attributed to the disruption of these contacts.
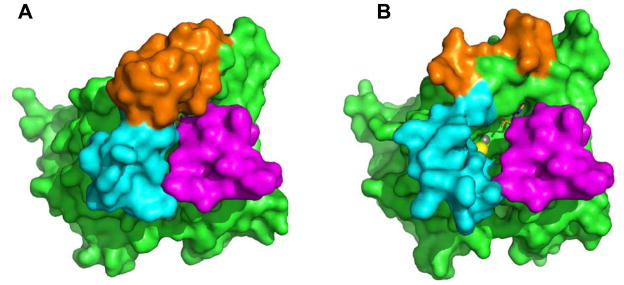
Comparison of the structures of PDE5/6cd and PDE5cd also suggests that conformational differences between the catalytic domains may constitute a major determinant for the interaction with the Pγ C-terminus. The structures of PDE5/6cd and PDE5cd are closely superimposable except for the H- and M-loops, both of which participate in the interaction with Pγ70-87 (10, 16). An extensive inter-loop interface in PDE5/6cd stabilizes the H- and M-loop in the conformations that favor Pγ binding. In structures of several PDE catalytic domains with bound IBMX, the H-loop and the M-loop interact with each other and adopt conformations reminiscent of the PDE5/6cd fold (20–22). In contrast, in the PDE5cd structure the M-loop points away from the H-loop, the catalytic cavity and the projected Pγ70-87 binding surface, and residues 793–807 are disordered (16). Surprisingly, the disruption of the H-M-loop interface in the PDE5/6cd L792R/V796E/Q799I mutant resulted in only a modest 3.5-fold decrease in the PDE5/6cd affinity for Pγ63-87. If the H- and M-loops of PDE5/6cd would assume the PDE5-like conformations, a much greater impairment of the interaction is predicted. We hypothesize that the lack of a stronger effect of the L792R/V796E/Q799I mutation is due to the ability of Pγ63-87 to induce conformations of the H- and M-loops that are similar to the inter-loop interface supported conformations. The inter-loop interface and induced fit binding of Pγ63-87 may cooperatively contribute to the interaction of the Pγ C-terminus with the catalytic domain of PDE6C. The results of gain-of-function experiments are in agreement with this hypothesis. The maximal inhibition of the PDE5cd A823F mutant by Pγ63-87 is only partial (Figure 3C), implying that cGMP can access the active site when the Pγ-peptide is bound to the catalytic domain. In the superimposed model of the PDE5cd-IBMX structure and the PDE5/6cd-IBMX-Pγ70-87 structure, there is a large opening between the H- and M-loops of PDE5cd and Pγ70-87 that allows diffusion of cGMP into the catalytic cavity (Figure 4). Bound Pγ63-87 does not affect the affinity (KM) of PDE5cdA823F for cGMP (Table 1). The noncompetitive mechanism of PDE5cdA823F inhibition by Pγ63-87 is in accord with the notion that Pγ is a noncompetitive inhibitor of PDE6. In the PDE5/6cd-IBMX-Pγ70-87 structure, the Pγ C-terminus does not interact or clash with IBMX (10). Similarly, in the superimposed model of the PDE5/6cd-IBMX-Pγ70-87 structure and the structure of PDE10A2 D674A mutant in complex with cGMP (23), there are no contacts or clashes between Pγ70-87 and cGMP (Figure S2 of the Supporting Information). Furthermore, a recent study directly demonstrated that the inhibiton of PDE6 by Pγ63-87 is noncompetitive when cGMP is used as a substrate (24). The allosteric inhibitory effect of Pγ63-87 on PDE5cdA823F may have resulted from conformational changes in the metal binding subpocket of the active site induced by the peptide interaction with α-helix-12 and/or the H-loop. Interestingly, an allosteric component to the Pγ-inhibition of PDE6 has been recently recognized (24). Using the PDE5cdA823F template, we have shown that mutations promoting formation of the inter-loop interface or introducing the PDE6-specific Pγ-binding motif Ile802Pro803Met804 into the M-loop increased the catalytic domain affinity for Pγ63-87 and augmented the maximal inhibition effect (Figure 3C). Thus, the interaction of Pγ with the M-loop apparently induces a conformational change in PDE5cd mutants similar to that of the inter-loop interface, i.e. a shift of the M-loop towards the catalytic cavity and the H-loop. Mutant PDE5cd catalytic domains containing the Pγ-contact residues from the M-loop/helix 15 and the inter-loop interface residues are inhibited by Pγ63-87 fully, but not as potently as PDE5/6cd. An additional Pγ interaction determinant was identified. A conserved PDE6-specific residue Trp787 is likely to support the M-loop conformation favorable for the binding of Pγ63-87. Among 22 residue positions that are different in PDE5/6cd and PDE5cd (Figure 1A), 8 positions were examined by mutagenesis in this study. The collective contribution of the remaining 14 PDE6-specific residues of PDE5/6cd to the interaction with Pγ is small, and the PDE5/6cd-Pγ70-87 structure does not indicate particular contributing residues. A number of the 14 uncharacterized residues may exert minor influences on the conformation of the M-loop/helix-15 region. In addition, the PDE5/6cd-Pγ70-87 structure apparently recaptures most, but not all, contacts between PDE6 and the Pγ C-terminus, because Pγ70-87 inhibits PDE5/6cd somewhat less potently than Pγ63-87 (10). Thus, additional unidentified PDE6 determinants for the interaction with the Pγ-C-terminus are possible.
Figure 4.
(A) Surface representation of PDE5/6cd (green) with IBMX and Pγ70-87 bound (magenta). Cyan – H-loop, orange – M-loop. Pγ70-87 entirely occludes the opening of the catalytic pocket. (B) The IBMX-bound PDE5cd (PDB ID: 1RKP) was structurally aligned with the PDE5/6cd (PDB ID: 3JWR, chain A) bound with Pγ70-87 (3JWR, chain C) using PyMOL 1.2r2 (26). The PDE5/6cd structure is omitted from the superimposed model, and the PDE5cd structure and Pγ70-87 are shown in surface representation. In the model, an opening exists between the H- and M-loops of PDE5cd and Pγ70-87 that allows an access to the catalytic pocket. Olive –IBMX, yellow sphere – magnesium ion, grey sphere – zinc ion.
When did the capacity of PDE6 enzymes for inhibition by Pγ evolve? PDE6 from lamprey Petromyzon marinus, representing the earliest surviving vertebrate class of jawless fish, is highly conserved with PDE6 from higher vertebrates (17). The catalytic domain of lamprey PDE6 is ~80% identical to the catalytic domains of human cone and rod PDE6 enzymes, and it contains all of the structural determinants for the interaction with the Pγ C-terminus identified in this study. The Pγ-subunits in P. marinus inhibit lamprey’s PDE6 equivalently to the PDE6 inhibition in higher vertebrates (17). A PDE6–related enzyme has been identified in tunicates Ciona savignyi and Ciona intestinalis. Ciona’s PDE6 is highly divergent from vertebrate PDE6s, but it groups together with the PDE6 family by phylogenetic analysis (17). Tunicates (urochordates) are the closest living relatives of vertebrates (25). The evolution of the PDE6 family appears to have begun prior to the last common ancestor of tunicates and vertebrates. Current genome databases provide no indication of the presence of Pγ genes in invertebrate species. Nonetheless, reconstruction of ancestral PDE6 and PDE5/6/11 enzymes suggests that they contained the M-loop Pγ-binding signature motif Ile-Pro-Met, and they may have been predisposed for the interaction with Pγ (Figure S3 of the Supporting Information). Intriguingly, prebilaterian cnidarians Nematostella vectensis and Hydra magnipapillata have PDE5/6-like enzymes that contain an essential Phe in α-helix-15 in addition to the Ile-Pro-Met motif (Figure S3 of the Supporting Information). Our analysis predicts that these enzymes are capable of effective interaction with the Pγ C-terminus. Thus, the PDE capacity for binding Pγ pre-existed the emergence of the inhibitory subunit, an ultimate step in the evolution of the visual effector.
Supplementary Material
Acknowledgments
We thank Dr. H. Muradov for cloning of the catalytic domain of human PDE5.
Abbreviations
- PDE
cGMP phosphodiesterase
- PDE6
photoreceptor PDE
- Pγ
γ-subunit of PDE6
- PDE5
cGMP-binding, cGMP-specific PDE (PDE5 family)
- PDE5/6cd
chimeric PDE5/PDE6 catalytic domain
- IBMX
3-isobutyl-1-methylxanthine
Footnotes
This work was supported by National Institutes of Health Grant EY-10843.
Supporting Information available
Figure S1 shows multiple sequence alignments of the M-loop/α-helix-15 regions of PDE6 and PDE5 from various species. Figure S2 shows the superimposed model of the PDE5/6cd-IBMX-Pγ70-87 structure and the structure of PDE10A2 D674A mutant in complex with cGMP. Figure S3 shows sequence alignment of the M-loop/α-helix-15 regions of the reconstructed ancestral PDE6 and PDE5/6/11 enzymes, as well as the PDE5/6-like enzymes from Nematostella vectensis and Hydra magnipapillata. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 2.Lamb TD, Pugh EN., Jr Phototransduction, dark adaptation, and rhodopsin regeneration the proctor lecture. Invest Ophthalmol Vis Sci. 2006;47:5137–5152. doi: 10.1167/iovs.06-0849. [DOI] [PubMed] [Google Scholar]
- 3.Fu Y, Yau KW. Phototransduction in mouse rods and cones. Pflugers Arch. 2007;454:805–819. doi: 10.1007/s00424-006-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 5.Artemyev NO, Hamm HE. Two-site high-affinity interaction between inhibitory and catalytic subunits of rod cyclic GMP phosphodiesterase. Biochem J. 1992;283:273–279. doi: 10.1042/bj2830273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mou H, Cote RH. The catalytic and GAF domains of the rod cGMP phosphodiesterase (PDE6) heterodimer are regulated by distinct regions of its inhibitory γ subunit. J Biol Chem. 2001;276:27527–27534. doi: 10.1074/jbc.M103316200. [DOI] [PubMed] [Google Scholar]
- 7.Guo LW, Muradov H, Hajipour AR, Sievert MK, Artemyev NO, Ruoho AE. The inhibitory γ subunit of the rod cGMP phosphodiesterase binds the catalytic subunits in an extended linear structure. J Biol Chem. 2006;281:15412–15422. doi: 10.1074/jbc.M600595200. [DOI] [PubMed] [Google Scholar]
- 8.Artemyev NO, Natochin M, Busman M, Schey KL, Hamm HE. Mechanism of photoreceptor PDE inhibition by its γ-subunits. Proc Natl Acad Sci USA. 1996;93:5407–5412. doi: 10.1073/pnas.93.11.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granovsky AE, Natochin M, Artemyev NO. The γ-subunit of rod cGMP-phosphodiesterase blocks the enzyme catalytic site. J Biol Chem. 1997;272:11686–11669. doi: 10.1074/jbc.272.18.11686. [DOI] [PubMed] [Google Scholar]
- 10.Barren B, Gakhar L, Muradov H, Boyd K, Ramaswamy S, Artemyev NO. Structural basis of phosphodiesterase 6 inhibition by the γ-subunit. EMBO J. 2009;28:3613–3622. doi: 10.1038/emboj.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granovsky AE, Natochin M, McEntaffer RL, Haik TL, Francis SH, Corbin JD, Artemyev NO. Probing domain functions of chimeric PDE6α’/PDE5 cGMP-phosphodiesterase. J Biol Chem. 1998;273:24485–24490. doi: 10.1074/jbc.273.38.24485. [DOI] [PubMed] [Google Scholar]
- 12.Granovsky AE, Artemyev NO. Identification of the γ-subunit interacting residues on photoreceptor cGMP phosphodiesterase, PDE6α’. J Biol Chem. 2000;275:41258–41262. doi: 10.1074/jbc.M008094200. [DOI] [PubMed] [Google Scholar]
- 13.Granovsky AE, Artemyev NO. Partial reconstitution of photoreceptor cGMP phosphodiesterase characteristics in cGMP phosphodiesterase-5. J Biol Chem. 2001;276:21698–21703. doi: 10.1074/jbc.M100626200. [DOI] [PubMed] [Google Scholar]
- 14.Cote RH. Characteristics of photoreceptor PDE (PDE6): similarities and differences to PDE5. Int J Impot Res. 2004;16(Suppl 1):S28–33. doi: 10.1038/sj.ijir.3901212. [DOI] [PubMed] [Google Scholar]
- 15.Muradov KG, Boyd KK, Artemyev NO. Analysis of PDE6 function using chimeric PDE5/6 catalytic domains. Vision Res. 2006;46:860–868. doi: 10.1016/j.visres.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Huai Q, Liu Y, Francis SH, Corbin JD, Ke H. Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor 3-isobutyl-1-methylxanthine suggest a conformation determinant of inhibitor selectivity. J Biol Chem. 2004;279:13095–13101. doi: 10.1074/jbc.M311556200. [DOI] [PubMed] [Google Scholar]
- 17.Muradov KG, Boyd KK, Kerov V, Artemyev NO. PDE6 in lamprey Petromyzon marinus: implications for the evolution of the visual effector in vertebrates. Biochemistry. 2007;46:9992–10000. doi: 10.1021/bi700535s. [DOI] [PubMed] [Google Scholar]
- 18.Yuasa K, Kanoh Y, Okumura K, Omori K. Genomic organization of the human phosphodiesterase PDE11A gene. Evolutionary relatedness with other PDEs containing GAF domains. Eur J Biochem. 2001;268:168–178. doi: 10.1046/j.1432-1327.2001.01866.x. [DOI] [PubMed] [Google Scholar]
- 19.Francis SH, Corbin JD, Bischoff E. Cyclic GMP-hydrolyzing phosphodiesterases. Handb Exp Pharmacol. 2009;191:367–408. doi: 10.1007/978-3-540-68964-5_16. [DOI] [PubMed] [Google Scholar]
- 20.Scapin G, Patel SB, Chung C, Varnerin JP, Edmondson SD, Mastracchio A, Parmee ER, Singh SB, Becker JW, Van der Ploeg LH, Tota MR. Crystal structure of human phosphodiesterase 3B: atomic basis for substrate and inhibitor specificity. Biochemistry. 2004;43:6091–60100. doi: 10.1021/bi049868i. [DOI] [PubMed] [Google Scholar]
- 21.Huai Q, Wang H, Zhang W, Colman RW, Robinson H, Ke H. Crystal structure of phosphodiesterase 9 shows orientation variation of inhibitor 3-isobutyl-1-methylxanthine binding. Proc Natl Acad Sci USA. 2004;101:9624–9629. doi: 10.1073/pnas.0401120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandit J, Forman MD, Fennell KF, Dillman KS, Menniti FS. Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc Natl Acad Sci USA. 2009;106:18225–182030. doi: 10.1073/pnas.0907635106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Liu Y, Hou J, Zheng M, Robinson H, Ke H. Structural insight into substrate specificity of phosphodiesterase 10. Proc Natl Acad Sci USA. 2007;104:5782–5787. doi: 10.1073/pnas.0700279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang XJ, Skiba NP, Cote RH. Structural requirements of the photoreceptor phosphodiesterase γ-subunit for inhibition of rod PDE6 holoenzyme and for its activation by transducin. J Biol Chem. 2010;285:4455–4463. doi: 10.1074/jbc.M109.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL molecular graphics system. 2004 http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.